MicroRNAs (miRNAs/miRs) are small, single-stranded,
non-coding RNAs comprising ~22 nucleotides. miRNAs typically bind
to the 3′-untranslated region (UTR) of genes by recruiting
Argonaute (AGO) protein complexes of messenger RNA (mRNA),
resulting in the downregulation of gene expression (1-3).
Growing evidence indicates that miRNAs have critical roles in the
progression of various disorders, such as viral infections, cancers
and neurodegenerative diseases (4-10).
Mature miRNAs are thought to localize in multiple
subcellular sites in the cytoplasm, such as the mitochondria, rough
endoplasmic reticulum, lysosomes and endosomes, and are secreted
outside cells via vesicles, such as exosomes (11). Furthermore, miRNAs have been found
in the nucleus, where they exert biological functions in regulating
gene transcription (12).
Molecular mechanisms underlying miRNA-mediated transcriptional gene
silencing or activation are complex. Gene activation requires an
intact transcript for recognition and a small RNA with
complementary pairing to its own sequence, which is capable of
recruiting AGO2 to the target (13). In addition, miRNAs and their
premature hairpins promote gene transcription by targeting specific
sites that are highly complementary to miRNAs in their promoters
(14). Nuclear activating miRNAs
facilitate gene transcription by binding and activating targeted
enhancers (15).
In the present review, the roles of nuclear miRNAs
in the progression of different types of tumor were discussed. It
focused on the function of these nuclear miRNAs as transcriptional
regulators through targeting promoters or enhancers, which results
in the alteration of tumor cell proliferation, invasion,
metastasis, migration, apoptosis and angiogenesis. This review
provides insight into the potential clinical utility of nuclear
miRNAs in cancer treatment.
In this section, the roles of nuclear miRNAs that
influence gene transcription in cancers of the urinary system,
including prostate cancer (PCa), bladder cancer (BCa), renal cell
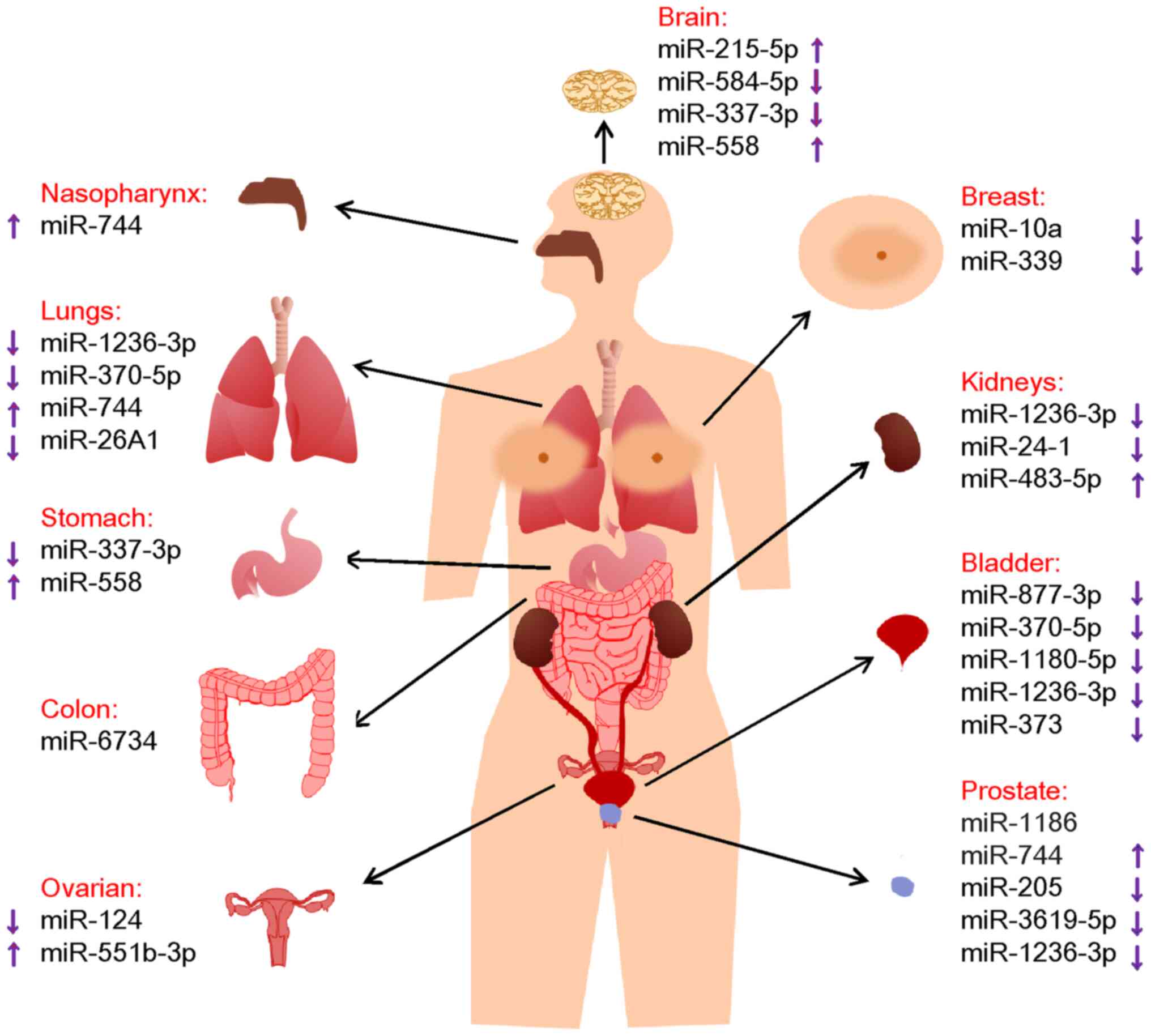
carcinoma (RCC) and Wilms' tumor (WT), are discussed (Table I).
PCa is the most common malignancy in males and one
of the leading causes of cancer-related mortality worldwide
(16). In a study investigating
the underlying mechanisms of miRNAs in the development of PCa,
miRNAs were found to affect the viability, migration and
invasiveness of PCa cells by downregulating and potentially
upregulating target genes. Cyclin B1 (CCNB1) overexpression is
associated with an aggressive phenotype and functions as an
independent prognostic factor in various cancers (17,18). miRNA target prediction analysis
and chromatin immunoprecipitation (ChIP) assay revealed that
miRNAs, including miR-744 and miR-1186, regulate the transcription
of CCNB1 by interacting with distinct binding sites in the CCNB1
promoter and increasing the enrichment of RNA polymerase II (PolII)
and trimethylation of histone 3 at lysine 4, thus enhancing the
proliferation of PCa cells (19).
However, prolonged overexpression of miR-744 and miR-1186
negatively regulates tumor growth in PCa cells as the chromosome
composition is altered, consequently leading to chromosomal
instability (19). Majid et
al (20) reported that
miR-205 inhibited PCa cell proliferation and impaired cell
viability, at least in part by binding to the promoters of
interleukin (IL)24 and IL32 and inducing their gene expression, as
determined through sequence scanning analysis and luciferase
reporter assay. Furthermore, a ChIP assay with biotinylated miRNA
confirmed that miR-3619-5p enhanced cyclin-dependent kinase (CDK)
inhibitor 1A transcription by interacting with its promoter,
suppressing the growth of PCa cells (21). In addition, Zhang et al
(22) reported that miR-1236-3p
activates the expression of P21, a tumor suppressor or oncogene, by
interacting with the promoter region, as determined with a sequence
scanning analysis, consequently inhibiting the proliferation and
metastasis of PCa cells.
BCa is the most common urinary tract malignancy and
has a high incidence worldwide (23,24). Li et al (25) performed a luciferase reporter
assay and revealed that miR-877-3p increased the expression of P16
by binding to its promoter and significantly suppressing the
proliferation and tumorigenicity of BCa cells via inducing G1-phase
arrest. This process was mediated by the inhibition of CDKs, such
as CDK4 and CDK6 (26). Wang
et al (27) performed a
ChIP assay via biotinylated miRNAs and found that P21 was inhibited
by three miRNAs (miR-370-5p, miR-1180-5p and miR-1236-3p) at
diverse sites on its promoter to inhibit BCa cell migration and
invasion and induce apoptosis. As a subclass of the cadherin
family, epithelial cadherin (E-cadherin) has an essential role in
maintaining epithelial cell-cell adhesion and intercellular
junctions (28). In a ChIP assay
with biotinylated miR-373, miR-373 was revealed to facilitate
E-cadherin expression by interacting with its gene promoter, with
the effect of inhibiting BCa cell proliferation (29). Taken together, these findings
support the development of novel therapies based on potentially
targeted genes for BCa.
WT is the most common pediatric renal tumor and is
associated with nephrogenesis, which may lead to malformations or
overgrowth (34). Liu et
al (35) showed that nuclear
miR-483, which is overexpressed in WT, enhanced insulin-like growth
factor 2 (IGF2) transcription through interaction with the 5′-UTR
of IGF2, as indicated using a biotinylated miRNA-RNA affinity
pulldown assay.
In this section, the roles of miRNAs via their
nuclear functions to influence gene transcription in diseases of
the reproductive system were summarized, including breast cancer
(BC) and ovarian cancer (OC) (Table
II).
BC is one of the most lethal and widespread cancers
among females worldwide. It occurs in the epithelial tissue of the
mammary gland and comprises diverse morphological characteristics
(36,37). In three studies on the role of
miRNAs in human BC, the authors performed a luciferase assay to
investigate the roles of nuclear miRNAs in gene transcription. Tan
et al (38) revealed that
miRNA-10a mediates the repression of target promoters on homeobox
D4 by interfering with promoter-associated transcripts to impact
the development of BC. Liang et al (39) demonstrated that miR-339 represses
the proliferation of BC cells, leading to the upregulation of tumor
suppressor gene G protein-coupled estrogen receptor 1 expression by
enhancer switching, indicating that miR-339 is a potential target
and novel approach for the treatment of BC, particularly for
triple-negative BC. In addition, Seviour et al (40) verified that, by binding to the P27
promoter, miR-124 induced the transcription of the P27 gene,
leading to a subsequent G1-phase arrest. As a tumor suppressor,
miR-124 was able to impair the growth of BC and OC cells and
sensitize cells to etoposide.
OC is associated with the highest mortality rate
among gynecological malignancies (41). Forkhead box (Fox)o3 regulates
ovarian follicular development and atresia (42). A luciferase reporter assay and a
ChIP assay indicated that the interaction between miR-195-5p and
the Foxo3 promoter may be associated with AGO2 recruitment and
histone modification, which affect follicular development (43). In addition, Chaluvally-Raghavan
et al (44) found that
miR-551b-3p interacted directly with a complementary sequence
within the signal transducer and activator 3 (STAT3) promoter by
recruiting PolII, thus facilitating STAT3 transcription. By
contrast, silencing miR-551b-3p inhibited the growth of OC cells,
as shown using luciferase reporter and DNA pull-down assays.
In this section, the roles of miRNAs in nervous
system diseases, including glioma and neuroblastoma (NB), were
summarized (Table III).
Gliomas account for the majority of primary
malignant and central nervous system tumors and are associated with
high morbidity and mortality (45,46). Wang et al (47) reported that miR-215-5p increases
the development of aggressive phenotypes, facilitates cell
proliferation and migration, and represses apoptosis in gliomas at
least in part by negatively regulating the expression of
protocadherin 9 via targeting both its promoter and 3′-UTR, as
determined by sequence scanning analysis and a luciferase reporter
assay.
NB is a common extracranial solid tumor in children,
mostly under the age of 10 years (48), and is an embryonal neoplasm of the
sympathetic nervous system (49,50). NB exhibits clinical heterogeneity
in response to current monotherapies (51,52). Researchers have studied the
mechanisms underlying the role of miRNAs in NB. Luciferase reporter
and ChIP assays demonstrated that miR-584-5p (53) and miR-337-3p (54) are involved in repressing the
transcription of matrix metalloproteinase (MMP-14) by binding to
different binding sites of its promoter, recruiting AGO2 and
attenuating the proliferation, invasion, metastasis and
angiogenesis of NB (55). By
contrast, using a sequence scanning analysis and luciferase
reporter assay, Qu et al (56) found that miR-558 facilitates the
transcription of heparinase (HPSE) by binding to the MMP-14
promoter, recruiting AGO1 to promote the proliferation, invasion,
metastasis and angiogenesis of NB.
In this section, the roles of miRNAs via their
nuclear functions that influence gene transcription in digestive
system diseases, including colon cancer and gastric cancer (GC),
were summarized (Table IV).
Colon cancer is a malignant tumor and a common cause
of cancer-related death worldwide, which may spread to organs such
as the lymph nodes, liver, lungs and ovaries (57). P21 is a CDK inhibitor that is
frequently deregulated in cancers, depending on the cellular
context (58). Kang et al
(59) performed a ChIP assay and
found that miR-6734 induced the transcription of P21 and attenuated
the proliferation and survival of HCT-116 cells by facilitating
cell cycle arrest and apoptosis.
In this section, the roles of miRNAs in respiratory
system diseases were summarized, including non-small-cell lung
cancer (NSCLC) and nasopharyngeal carcinoma (NPC) (Table V).
Lung cancer is the leading cause of cancer-related
morbidity and mortality in China, and NSCLC accounts for the
overwhelming majority of lung cancer cases (67). miR-1236-3p and miR-370-5p
negatively regulate the proliferation, migration and invasiveness
of NSCLC cells at least in part by targeting the promoter of the
P21 gene to upregulate its expression, as indicated using sequence
scanning analysis (68). As a
major component of the activator protein-1 complex, c-Fos has been
implicated in cell differentiation, proliferation, motility, cancer
growth, angiogenesis, invasion and metastasis (69). The results of a luciferase
reporter assay indicated that miR-744 acts as an oncogene in NSCLC
cell growth and metastasis at least in part by directly binding to
the promoter of c-Fos (70). Li
et al (71) found that
miR-26A1 is decreased in NSCLC and used luciferase reporter and
ChIP assays to demonstrate that miR-26A1 acts as a key regulator in
reactivating villin 1-like protein by targeting its enhancer,
leading to the inhibition of the proliferation and metastasis of
NSCLC cells.
NPC is a malignant tumor associated with a specific
geographical distribution in Southeast Asia and North Africa
(72,73). Rho GTPase activating protein 5
(ARHGAP5), a proto-oncogene, is a direct target of miR-744. Using a
luciferase reporter assay, Fang et al (74) found that miR-744 facilitates
expression at the transcriptional level by directly targeting the
ARHGAP5 promoter, consequently enhancing the progression and
metastasis of NPC.
Cancers are devastating to all physiological
systems. Growth, invasion, metastasis and angiogenesis of cancer
cells affect all organ systems and subsequently, morbidity and
mortality (75). The effects of
miRNAs in cancer through their action on the 3′-UTRs of relevant
genes at the post-transcriptional level have been well-established.
However, nuclear miRNAs have been reported to be aberrantly
expressed in various cancers and other diseases. The present review
outlines the current understanding of the contribution of nuclear
miRNAs to cancer progression of different organs at the
transcriptional level, as they mostly interact with gene promoters
or enhancers (Fig. 1), suggesting
that nuclear miRNAs may serve as promising candidate biomarkers for
predicting prognosis and enhancing therapeutic strategies for
patients with cancer.
Over the past decade, non-coding RNAs, including
long non-coding RNAs and miRNAs, have emerged as powerful
regulatory molecules in human diseases (76-84). Among them, nuclear miRNAs have
been reported to be upregulated or downregulated in cancers and
promote or inhibit the transcription of cancer progression-related
genes. However, the roles and underlying molecular mechanisms of a
specific nuclear miRNA vary greatly from cancer to cancer. For
instance, miR-744 expression has been shown to be upregulated in
PCa, NSCLC and NPC. In PCa, miR-744 targets the CCNB1 gene
promoter to initiate gene transcription by increasing the
enrichment of PolII and trimethylation of histone 3 at lysine 4 at
the CCNB1 gene transcription start site (19). In NSCLC, miR-744 promoted the
growth, invasion and metastasis of NSCLC cells at least in part by
binding to the c-FOS gene promoter and induce gene
transcription (70). In NPC,
miR-744 was observed to be associated with TNM stage, cancer
progression and metastasis at least in part by targeting the
ARHGAP5 gene and activating gene transcription (74).
At present, several miRNA-targeted therapeutics are
in clinical development for the treatment of diseases, including
keloids, hepatitis C virus infection, nonalcoholic fatty liver
disease, type II diabetes, Huntington's disease and cancer;
however, none has been approved by the Food and Drug Administration
or the European Medicines Agency to treat cancers (85). Key challenges faced by miRNA-based
therapeutics are the hurdles of specific delivery, immune responses
and low specificity. Therefore, technical advancements in
pharmacology, molecular biology, immunology and nanotechnology are
needed to improve specificity, tolerance and delivery. In addition,
more studies are required to reveal the underlying mechanisms by
which nuclear miRNAs regulate gene transcription, such as the
mechanisms by which nuclear miRNAs can be recruited to and retained
in the nucleus, whether AGO proteins are required to the effects of
nuclear miRNAs on gene transcription, and whether the interaction
between nuclear miRNAs and promoters or enhancers influences the
transcription of surrounding genes.
In summary, the present review summarizes and
discusses nuclear miRNAs' centric regulation of gene transcription
and their roles in cancer of various bodily systems, indicating
that nuclear miRNAs may be potent targets for the treatment of
cancers and other pathological disorders.
Not applicable.
ZW: Conceptualization, writing-original draft,
supervision, writing-review & editing. YZ: Writing-original
draft. KL: Conceptualization, supervision, writing-review &
editing. All authors have read and approved the final version of
the manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the Shandong Provincial Natural
Science Foundation (grant nos. ZR2020LZL008 and ZR2021LSW017).
|
1
|
Omer AD, Janas MM and Novina CD: The
chicken or the egg: microRNA-mediated regulation of mRNA
translation or mRNA stability. Mol Cell. 35:739–740. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Z, Lu Y, Zhang X, Ren X, Wang Y, Li
Z, Xu C and Han J: Serum microRNA is a promising biomarker for
osteogenesis imperfecta. Intractable Rare Dis Res. 1:81–85.
2012.PubMed/NCBI
|
|
5
|
Wang ZQ, Lu YQ and Han JX: MicroRNAs:
Important mediators of ossification. Chin Med J (Engl).
125:4111–4116. 2012.PubMed/NCBI
|
|
6
|
Wang Z, Li K, Wang X and Huang W:
MiR-155-5p modulates HSV-1 replication via the epigenetic
regulation of SRSF2 gene expression. Epigenetics. 14:494–503. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li K, Yao T, Zhang Y, Li W and Wang Z:
NEAT1 as a competing endogenous RNA in tumorigenesis of various
cancers: Role, mechanism and therapeutic potential. Int J Biol Sci.
17:3428–3440. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li K and Wang Z: Non-coding RNAs: Key
players in T cell exhaustion. Front Immunol. 13:9597292022.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li K, Yao T and Wang Z: lncRNA-mediated
ceRNA network in bladder cancer. Noncoding RNA Res. 8:135–145.
2022. View Article : Google Scholar
|
|
10
|
Wang Z, Lu Y and Han J: Peripheral blood
microRNAs: A novel tool for diagnosing disease? Intractable Rare
Dis Res. 1:98–102. 2012.PubMed/NCBI
|
|
11
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Taft RJ, Simons C, Nahkuri S, Oey H,
Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJL, Rasko JEJ, et
al: Nuclear-localized tiny RNAs are associated with transcription
initiation and splice sites in metazoans. Nat Struct Mol Biol.
17:1030–1034. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matsui M, Chu Y, Zhang H, Gagnon KT,
Shaikh S, Kuchimanchi S, Manoharan M, Corey DR and Janowski BA:
Promoter RNA links transcriptional regulation of inflammatory
pathway genes. Nucleic Acids Res. 41:10086–10109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Place RF, Li LC, Pookot D, Noonan EJ and
Dahiya R: MicroRNA-373 induces expression of genes with
complementary promoter sequences. Proc Natl Acad Sci USA.
105:1608–1613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zou Q, Liang Y, Luo H and Yu W:
miRNA-mediated RNAa by targeting enhancers. Adv Exp Med Biol.
983:113–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aaltonen K, Amini RM, Heikkilä P,
Aittomäki K, Tamminen A, Nevanlinna H and Blomqvist C: High cyclin
B1 expression is associated with poor survival in breast cancer. Br
J Cancer. 100:1055–1060. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan J, Yan R, Krämer A, Eckerdt F, Roller
M, Kaufmann M and Strebhardt K: Cyclin B1 depletion inhibits
proliferation and induces apoptosis in human tumor cells. Oncogene.
23:5843–5852. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang V, Place RF, Portnoy V, Wang J, Qi
Z, Jia Z, Yu A, Shuman M, Yu J and Li LC: Upregulation of cyclin B1
by miRNA and its implications in cancer. Nucleic Acids Res.
40:1695–1707. 2012. View Article : Google Scholar :
|
|
20
|
Majid S, Dar AA, Saini S, Yamamura S,
Hirata H, Tanaka Y, Deng G and Dahiya R: MicroRNA-205-directed
transcriptional activation of tumor suppressor genes in prostate
cancer. Cancer. 116:5637–5649. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S, Wang C, Yu X, Wu H, Hu J, Wang S and
Ye Z: miR-3619-5p inhibits prostate cancer cell growth by
activating CDKN1A expression. Oncol Rep. 37:241–248. 2017.
View Article : Google Scholar
|
|
22
|
Zhang Q, Yang X, Luo L, Ma X, Jiao W, Li
B, Zhang M, Zhao K and Niu H: Targeted p21 activation by a new
double stranded RNA suppresses human prostate cancer cells growth
and metastasis. Am J Transl Res. 12:4175–4188. 2020.PubMed/NCBI
|
|
23
|
Dobruch J and Oszczudłowski M: Bladder
cancer: Current challenges and future directions. Medicina
(Kaunas). 57:7492021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN Estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li S, Zhu Y, Liang Z, Wang X, Meng S, Xu
X, Xu X, Wu J, Ji A, Hu Z, et al: Up-regulation of p16 by
miR-877-3p inhibits proliferation of bladder cancer. Oncotarget.
7:51773–51783. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liggett WH Jr and Sidransky D: Role of the
p16 tumor suppressor gene in cancer. J Clin Oncol. 16:1197–1206.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang C, Chen Z, Ge Q, Hu J, Li F, Hu J, Xu
H, Ye Z and Li LC: Up-regulation of p21(WAF1/CIP1) by miRNAs and
its implications in bladder cancer cells. FEBS Lett. 588:4654–4664.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Oka H, Shiozaki H, Kobayashi K, Inoue M,
Tahara H, Kobayashi T, Takatsuka Y, Matsuyoshi N, Hirano S,
Takeichi M, et al: Expression of E-cadherin cell adhesion molecules
in human breast cancer tissues and its relationship to metastasis.
Cancer Res. 53:1696–1701. 1993.PubMed/NCBI
|
|
29
|
Zhang Q, Wang C, Miao S, Li C, Chen Z and
Li F: Enhancing E-cadherin expression via promoter-targeted miR-373
suppresses bladder cancer cells growth and metastasis. Oncotarget.
8:93969–93983. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siddiqi A, Rani M, Bansal P and Rizvi MMA:
Renal cell carcinoma management: A step to nano-chemoprevention.
Life Sci. 308:1209222022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
Update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang C, Tang K, Li Z, Chen Z, Xu H and Ye
Z: Targeted p21(WAF1/CIP1) activation by miR-1236 inhibits cell
proliferation and correlates with favorable survival in renal cell
carcinoma. Urol Oncol. 34:59.e23–e34. 2016. View Article : Google Scholar
|
|
33
|
Ju D, Liang Y, Hou G, Zheng W, Zhang G,
Dun X, Wei D, Yan F, Zhang L, Lai D, et al: FBP1/miR-24-1/enhancer
axis activation blocks renal cell carcinoma progression via Warburg
effect. Front Oncol. 12:9283732022. View Article : Google Scholar
|
|
34
|
Treger TD, Chowdhury T, Pritchard-Jones K
and Behjati S: The genetic changes of Wilms tumour. Nat Rev
Nephrol. 15:240–251. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu M, Roth A, Yu M, Morris R, Bersani F,
Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, et al: The
IGF2 intronic miR-483 selectively enhances transcription from IGF2
fetal promoters and enhances tumorigenesis. Genes Dev.
27:2543–2548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Giaquinto AN, Sung H, Miller KD, Kramer
JL, Newman LA, Minihan A, Jemal A and Siegel RL: Breast cancer
statistics, 2022. CA Cancer J Clin. 72:524–541. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X,
Guo X, He S and Chen R: Transcriptional inhibiton of Hoxd4
expression by miRNA-10a in human breast cancer cells. BMC Mol Biol.
10:122009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang Y, Lu Q, Li W, Zhang D, Zhang F, Zou
Q, Chen L, Tong Y, Liu M, Wang S, et al: Reactivation of tumour
suppressor in breast cancer by enhancer switching through NamiRNA
network. Nucleic Acids Res. 49:8556–8572. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T,
Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, et al: Functional
proteomics identifies miRNAs to target a p27/Myc/phospho-Rb
signature in breast and ovarian cancer. Oncogene. 35:691–701. 2016.
View Article : Google Scholar
|
|
41
|
Coburn SB, Bray F, Sherman ME and Trabert
B: International patterns and trends in ovarian cancer incidence,
overall and by histologic subtype. Int J Cancer. 140:2451–2460.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cui C, Han S, Yin H, Luo B, Shen X, Yang
F, Liu Z, Zhu Q, Li D and Wang Y: FOXO3 is expressed in ovarian
tissues and acts as an apoptosis initiator in granulosa cells of
chickens. Biomed Res Int. 2019:69029062019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bai Y, Pan B, Zhan X, Silver H and Li J:
MicroRNA 195-5p targets Foxo3 promoter region to regulate its
expression in granulosa cells. Int J Mol Sci. 22:67212021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chaluvally-Raghavan P, Jeong KJ, Pradeep
S, Silva AM, Yu S, Liu W, Moss T, Rodriguez-Aguayo C, Zhang D, Ram
P, et al: Direct upregulation of STAT3 by MicroRNA-551b-3p
deregulates growth and metastasis of ovarian cancer. Cell Rep.
15:1493–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miller KD, Ostrom QT, Kruchko C, Patil N,
Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL and
Barnholtz-Sloan JS: Brain and other central nervous system tumor
statistics, 2021. CA Cancer J Clin. 71:381–406. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fang Y, Zhang G, Bai Z, Yan Y, Song X,
Zhao X, Yang P and Zhang Z: Low-intensity ultrasound: A novel
technique for adjuvant treatment of gliomas. Biomed Pharmacother.
153:1133942022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang C, Chen Q, Li S, Li S, Zhao Z, Gao H,
Wang X, Li B, Zhang W, Yuan Y, et al: Dual inhibition of PCDH9
expression by miR-215-5p up-regulation in gliomas. Oncotarget.
8:10287–10297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cheung NK and Dyer MA: Neuroblastoma:
Developmental biology, cancer genomics and immunotherapy. Nat Rev
Cancer. 13:397–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qiu B and Matthay KK: Advancing therapy
for neuroblastoma. Nat Rev Clin Oncol. 19:515–533. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gomez RL, Ibragimova S, Ramachandran R,
Philpott A and Ali FR: Tumoral heterogeneity in neuroblastoma.
Biochim Biophys Acta Rev Cancer. 1877:1888052022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Johnsen JI, Dyberg C and Wickström M:
Neuroblastoma-a neural crest derived embryonal malignancy. Front
Mol Neurosci. 12:92019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lin L, Miao L, Lin H, Cheng J, Li M, Zhuo
Z and He J: Targeting RAS in neuroblastoma: Is it possible?
Pharmacol Ther. 236:1080542022. View Article : Google Scholar
|
|
53
|
Xiang X, Mei H, Qu H, Zhao X, Li D, Song
H, Jiao W, Pu J, Huang K, Zheng L and Tong Q: miRNA-584-5p exerts
tumor suppressive functions in human neuroblastoma through
repressing transcription of matrix metalloproteinase 14. Biochim
Biophys Acta. 1852:1743–1754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H,
Jiao W, Zhao J, Huang K, Zheng L and Tong Q: miRNA-337-3p
suppresses neuroblastoma progression by repressing the
transcription of matrix metalloproteinase 14. Oncotarget.
6:22452–22466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mei H, Lin ZY and Tong QS: The roles of
microRNAs in neuroblastoma. World J Pediatr. 10:10–16. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao
X, Li D, Li S, Mao L, Huang K and Tong Q: miRNA-558 promotes
tumorigenesis and aggressiveness of neuroblastoma cells through
activating the transcription of heparanase. Hum Mol Genet.
24:2539–2551. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
PDQ Adult Treatment Editorial Board: Colon
Cancer Treatment (PDQ®): Patient version. 2022 Apr 6. PDQ Cancer
Information Summaries (Internet) Bethesda (MD): National Cancer
Institute (US); 2002
|
|
58
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kang MR, Park KH, Yang JO, Lee CW, Oh SJ,
Yun J, Lee MY, Han SB and Kang JS: miR-6734 Up-regulates p21 gene
expression and induces cell cycle arrest and apoptosis in colon
cancer cells. PLoS One. 11:e01609612016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Watari J, Chen N, Amenta PS, Fukui H,
Oshima T, Tomita T, Miwa H, Lim KJ and Das KM: Helicobacter pylori
associated chronic gastritis, clinical syndromes, precancerous
lesions, and pathogenesis of gastric cancer development. World J
Gastroenterol. 20:5461–5473. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
He L, Chu D, Li X, Zheng J, Liu S, Li J,
Zhao Q and Ji G: Matrix metalloproteinase-14 is a negative
prognostic marker for patients with gastric cancer. Dig Dis Sci.
58:1264–1270. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Imanishi Y, Fujii M, Tokumaru Y, Tomita T,
Kanke M, Kanzaki J, Kameyama K, Otani Y and Sato H: Clinical
significance of expression of membrane type 1 matrix
metalloproteinase and matrix metalloproteinase-2 in human head and
neck squamous cell carcinoma. Hum Pathol. 31:895–904. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zheng L, Jiao W, Mei H, Song H, Li D,
Xiang X, Chen Y, Yang F, Li H, Huang K and Tong Q: miRNA-337-3p
inhibits gastric cancer progression through repressing myeloid zinc
finger 1-facilitated expression of matrix metalloproteinase 14.
Oncotarget. 7:40314–40328. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Takaoka M, Naomoto Y, Ohkawa T, Uetsuka H,
Shirakawa Y, Uno F, Fujiwara T, Gunduz M, Nagatsuka H, Nakajima M,
et al: Heparanase expression correlates with invasion and poor
prognosis in gastric cancers. Lab Invest. 83:613–622. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hulett MD, Freeman C, Hamdorf BJ, Baker
RT, Harris MJ and Parish CR: Cloning of mammalian heparanase, an
important enzyme in tumor invasion and metastasis. Nat Med.
5:803–809. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zheng L, Jiao W, Song H, Qu H, Li D, Mei
H, Chen Y, Yang F, Li H, Huang K and Tong Q: miRNA-558 promotes
gastric cancer progression through attenuating Smad4-mediated
repression of heparanase expression. Cell Death Dis. 7:e23822016.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gao S, Li N, Wang S, Zhang F, Wei W, Li N,
Bi N, Wang Z and He J: Lung cancer in People's Republic of China. J
Thorac Oncol. 15:1567–1576. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li C, Ge Q, Liu J, Zhang Q, Wang C, Cui K
and Chen Z: Effects of miR-1236-3p and miR-370-5p on activation of
p21 in various tumors and its inhibition on the growth of lung
cancer cells. Tumour Biol. 39:10104283177108242017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu ZG, Jiang G, Tang J, Wang H, Feng G,
Chen F, Tu Z, Liu G, Zhao Y, Peng MJ, et al: c-Fos over-expression
promotes radioresistance and predicts poor prognosis in malignant
glioma. Oncotarget. 7:65946–65956. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li S, Qiao S, Li N and Zhu X: MiR-744
functions as an oncogene through direct binding to c-Fos promoter
and facilitates non-small cell lung cancer progression. Ann Surg
Oncol. 29:1465–1475. 2022. View Article : Google Scholar
|
|
71
|
Li H, Da D, Yu W, Chen L, Yang S, Zhang B,
Wang Y, Li L and Dang C: Tumor suppressor genes are reactivated by
miR-26A1 via enhancer reprogramming in NSCLC. Hum Mol Genet.
32:79–92. 2023. View Article : Google Scholar :
|
|
72
|
Chen YP, Chan ATC, Le QT, Blanchard P, Sun
Y and Ma J: Nasopharyngeal carcinoma. Lancet. 394:64–80. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
McDermott AL, Dutt SN and Watkinson JC:
The aetiology of nasopharyngeal carcinoma. Clin Otolaryngol Allied
Sci. 26:82–92. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Fang Y, Zhu X, Wang J, Li N, Li D, Sakib
N, Sha Z and Song W: MiR-744 functions as a proto-oncogene in
nasopharyngeal carcinoma progression and metastasis via
transcriptional control of ARHGAP5. Oncotarget. 6:13164–13175.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fares J, Fares MY, Khachfe HH, Salhab HA
and Fares Y: Molecular principles of metastasis: A hallmark of
cancer revisited. Signal Transduct Target Ther. 5:282020.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li K and Wang Z: lncRNA NEAT1: Key player
in neurodegenerative diseases. Ageing Res Rev. 86:1018782023.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang T, Wang Y, Liao W, Zhang S, Wang S,
Xu N, Xie W, Luo C, Wang Y, Wang Z and Zhang Y: Down-regulation of
EPB41L4A-AS1 mediated the brain aging and neurodegenerative
diseases via damaging synthesis of NAD+ and ATP. Cell
Biosci. 11:1922021. View Article : Google Scholar
|
|
78
|
Li K and Wang Z: Speckles and paraspeckles
coordinate to regulate HSV-1 genes transcription. Commun Biol.
4:12072021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wang Z, Zhang S and Li K: LncRNA NEAT1
induces autophagy through epigenetic regulation of
autophagy-related gene expression in neuroglial cells. J Cell
Physiol. 237:824–832. 2022. View Article : Google Scholar
|
|
80
|
Zhao Y, Wang Z, Mao Y, Li B, Zhu Y, Zhang
S, Wang S, Jiang Y, Xu N, Xie Y, et al: NEAT1 regulates microtubule
stabilization via FZD3/GSK3β/P-tau pathway in SH-SY5Y cells and
APP/PS1 mice. Aging (Albany NY). 12:23233–23250. 2020.PubMed/NCBI
|
|
81
|
Wang Z, Zhao Y, Xu N, Zhang S, Wang S, Mao
Y, Zhu Y, Li B, Jiang Y, Tan Y, et al: NEAT1 regulates neuroglial
cell mediating Aβ clearance via the epigenetic regulation of
endocytosis-related genes expression. Cell Mol Life Sci.
76:3005–3018. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang Z, Zhao Y and Zhang Y: Viral lncRNA:
A regulatory molecule for controlling virus life cycle. Noncoding
RNA Res. 2:38–44. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wang Z, Fan P, Zhao Y, Zhang S, Lu J, Xie
W, Jiang Y, Lei F, Xu N and Zhang Y: NEAT1 modulates herpes simplex
virus-1 replication by regulating viral gene transcription. Cell
Mol Life Sci. 74:1117–1131. 2017. View Article : Google Scholar :
|
|
84
|
Wang Z, Li K and Huang W: Long non-coding
RNA NEAT1-centric gene regulation. Cell Mol Life Sci. 77:3769–3779.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Winkle M, El-Daly SM, Fabbri M and Calin
GA: Noncoding RNA therapeutics-challenges and potential solutions.
Nat Rev Drug Discov. 20:629–651. 2021. View Article : Google Scholar : PubMed/NCBI
|