Introduction
Esophageal cancer (EC) is a complex and common
cancer with a highly aggressive nature (1). Esophageal squamous cell carcinoma
(ESCC), a malignant epithelial tumor originating from EC (2), is one of the most common subtypes of
EC, which typically presents as progressive dysphagia. In 2020,
there were 604,000 new cases and 544,000 deaths from esophageal
cancer worldwide. By 2040, these numbers are expected to increase
to 957,000 cases and 880,000 deaths per year (3), constituting a serious public health
issue. The pathogenesis of EC is associated with dietary habits,
environmental factors, geographic location and gender (4). EC has a 5-year overall survival rate
of 15–25% in developing countries and is the sixth leading cause of
cancer-associated mortalities in men worldwide (5). Furthermore, the rates of EC mortality
and morbidity are higher in developing countries compared with in
developed countries (6).
Clinically, esophageal carcinoma is typically managed by drug
therapy, surgery, radiotherapy and chemotherapy. However, side
effects originating from these therapies and the poor prognosis of
patients remain a major concern, creating an urgent need to
evaluate new treatment modalities and improve the treatment and
prognosis of patients with EC (7).
Epithelial mesenchymal transformation (EMT) is a
process in which epithelial cells lose their cell polarity, lose
their connection to the basement membrane and gain a higher
interstitial phenotype such as migration and invasion,
anti-apoptosis, and degradation of the extracellular matrix
(8). According to the literature,
EMT progression will reduce the adhesion capacity of tumor cells,
make them more invasive and migratory, and accelerate tumor cell
invasion and metastasis (9). This
also suggests that the inhibition of the EMT process could hinder
the metastasis of cancer cells. In addition, cancer cells need a
large amount of energy to support the process of metastasis, and
the abnormal glucose metabolism of tumor cells can support this
requirement, assisting cancer cells in metastasis (10). Abnormal glucose metabolism is one of
the major changes seen in the tumor microenvironment (11). Thus, glucose metabolism-related gene
regulation has emerged as a new target for tumor therapy. The Janus
kinase (JAK)/signal transducer and activator (STAT) signaling
pathway is a ubiquitously expressed intracellular signal
transduction pathway that is involved in a number of key biological
processes, including cell proliferation, differentiation, apoptosis
and immune regulation (12).
Studies have reported that JAK/STAT signaling pathway can regulate
the glycolytic pathway in lung cancer (13), renal cell carcinoma (14) and breast cancer (15) cells, and so have the potential to
modulate cancer progression. Furthermore, high expression of
phosphorylated (p)-JAK1 and p-STAT3 indicates poor prognosis in
patients with EC (16). Zhao et
al (17) reported that STAT3
and hypoxia-inducible factor-1α (HIF-1α) were both expressed at
higher levels in ESCC tissues than in normal tissues, and the
addition of JAK2 inhibitors was effective in blocking the
proliferation of EC cells in vitro (18). It was also reported that the
inhibition of the JAK/STAT signaling pathway blocked the
angiogenesis of EC cells (19).
Accordingly, this evidence suggests that inhibition of the JAK/STAT
signaling pathway is a potential target for the treatment of EC.
Hexokinase (HK)1 and HK2 are the two major HKs and depletion of HK2
has been reported to reduce lactate and cellular HK activity. The
depletion of HK1 contributes little to the proliferative activity
of these cells, and the alteration of HK1 has a less significant
effect on lactate content compared with HK2. The role of HK2 in
tumor glucose metabolism is now widely reported (20,21).
Therefore, the present study focused on HK2.
Furthermore, studies have shown that dexmedetomidine
hydrochloride (DEX-HCl) alleviated neuropathic pain by modulating
the JAK/STAT signaling pathway (22) and prevented reperfusion injury
(23,24). During surgery in patients with
cancer, anesthetic drugs may affect the growth, proliferation and
metastasis of cancer cells (25,26).
However, these findings require further clinical trials and
validation, and currently it is not possible to determine whether
the effects of anesthetic drugs on cancer cells have clinical
application value. In order to further investigate the specific
effects of anesthetics on cancer cells, the present study
investigated the effects of DEX-HCl and sufentanil citrate (SFC) on
the JAK/STAT signaling pathway, and evaluated their effects on the
glucose metabolism, lactic acid production, ATP level, invasion and
migration of EC KYSE30 cells. This may provide a theoretical basis
for clinical cancer-associated anesthesia.
Materials and methods
Cell culture
Human ESCC cell lines KYSE30, KYSE520, KYSE140 and
KYSE410 and immortalized HEEC human normal esophageal epithelial
cells were purchased from the Qingqi (Shanghai) Biotechnology
Development Co., Ltd. The cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine
serum (Shanghai ExCell Biology, Inc.) and 1%
penicillin-streptomycin mixture (Beijing Solarbio Science &
Technology Co., Ltd.) in a 37°C, 5% CO2 incubator
(Zhejiang Jiemei Electronic & Technology Co., Ltd.). The medium
was changed every two days and cells were allowed to grow to the
exponential phase for use in subsequent experiments.
Cell counting kit-8 assay
KYSE30 cells at exponential culture stage were taken
out of the cell incubator. The medium was discarded and the cells
were washed with PBS three times. Digestion was performed in T25
culture flasks by adding 1 ml of trypsin solution (Beijing Solarbio
Science & Technology Co., Ltd.) for 2–3 min. Subsequently, 2 ml
of RPMI-1640 complete medium was added to terminate the digestion.
The cells were collected in a centrifuge tube and centrifuged at
1,200 × g for 3 min. RPMI-1640 complete medium was added to
resuspend the cells, and 10 µl of cell suspension was aspirated to
count the number of cells using a cell counter (Countess™ 3; cat.
no. AMQAX2000; Invitrogen; Thermo Fisher Scientific, Inc.). The
cells were transferred to 96-well plates at a concentration of
3,000 cells per well and incubated overnight in a cell incubator at
37°C and divided into four groups which were treated as follows: i)
Control (DMSO); ii) DEX-HCl (25, 50, 100 and 200 nmol/l); iii) SFC
(1.25, 2.5, 5 and 10 µmol/l); and iv) DEX-HCl (25 nmol/l) and SFC
(1.25 µmol/l) combined treatment. The 96-well plate was then
incubated for 48 h. A total of 10 µl of CCK-8 solution (Beijing
Solarbio Science & Technology Co., Ltd.) was added to each well
and incubated for 3 h. The absorbance at 450 nm was measured using
an enzyme immunoassay analyzer (ReadMax 1,200; Shanghai Shanpu
Biotechnology Co., Ltd.). IC50 values were calculated
using GraphPad Prism 9.5.0 (Dotmatics). DEX-HCI (cat. no. 22022431)
was purchased from Jiangsu Hengrui Pharmaceutical Co., Ltd., and
SFC (cat. no. 21A09311) was purchased from Yichang Renfu
Pharmaceutical Co., Ltd.
Clonogenic assay of cells in
vitro
KYSE30 cells at the exponential growth phase were
subjected to cell digestion and cell counting as described
previously for the CCK-8 assay. The cells were resuspended at 150
cells/ml. A total of 1 ml of cell suspension was mixed with 1 ml of
RPMI-1640 complete medium, transferred to a six-well plate and
incubated overnight in an incubator at 37°C with 5% CO2.
The medium was replenished every 2 days for 14 days. The medium was
then discarded, the cells were rinsed with PBS solution three times
and then stained using 0.1% crystal violet (Beijing Solarbio
Science & Technology Co., Ltd.) for 15 min, 25°C. The cells
were washed three times with PBS, air-dried naturally and imaged
with an SLR camera (Nikon D850; Nikon Corporation). The images were
analyzed using ImageJ version 1.52a (National Institutes of Health)
bundled with Java 8.
Wound healing assay
KYSE30 cells at the exponential growth phase were
subjected to cell digestion and cell counting as described
previously for the CCK-8 assay. The cell suspension was inoculated
into 6-well plates at 5×105 cells per well and incubated
overnight in a cell culture incubator (37°C and 5% CO2).
Three parallel vertical lines were drawn across the bottom of the
six-well plate with a 10 µl pipette tip. After discarding the
medium, the cells were washed three times with PBS, and serum-free
RPMI-1640 medium containing DEX-HCl (25 nmol/l) and SFC (1.25
µmol/l) drug solution was added to a 6-well plate and incubated in
an incubator. The six-well plate was imaged at 0, 24 and 48 h using
an inverted fluorescence microscope [Sunny Optical Technology
(Group) Co., Ltd.] at the intersection of the horizontal and
vertical lines to ensure that images were obtained from the same
location at different time points. The cell migration rate of each
group was analyzed by ImageJ version 1.52a (National Institutes of
Health) bundled with Java 8.
Transwell migration assay
Matrix-Gel™ Matrigel (cat. no. C0371; Beyotime
Institute of Biotechnology) was diluted 1:8 with serum-free
RPMI-1640 medium and spread on the upper part of the Transwell
chamber, and then left to set at 37°C for 2 h. Culture medium was
then added to hydrate the gel for 30 min before being discarded.
KYSE30 cells at the exponential phase were collected and counted as
described previously. The cell concentration was diluted to
1×106 cells/ml with serum-free RPMI-1640 medium and 50
µl of cell suspension was added to the upper chamber of the
Transwell. The upper chamber of the Transwell was supplemented with
50 µl of serum-free RPMI 1640 medium containing DEX-HCl and SFC to
give a final drug concentration of 25 nmol/l DEX-HCl and 1.25
µmol/l SFC. 500 ul of RPMI 1640 medium containing 20% FBS was added
to the lower chamber of the Transwell. The Transwell chambers were
then incubated in a cell culture incubator at 37°C for 12 h.
Subsequently, 500 µl of cell fixation solution (cat. no. P0099;
Beyotime Institute of Biotechnology) was added to each chamber for
25 min at room temperature, then washed with PBS. Finally, the
cells were stained with 0.1% crystal violet solution for 25 min at
room temperature before being imaged using a microscope.
Glucose, lactate and ATP content
assay
After adding DEX-HCI (25 nmol/l) or SFC (1.25
µmol/l) alone or in combination, KYSE30 cells were incubated in a
cell incubator at 37°C for 24 h. Cells were then collected and
transferred to a 1.5 ml centrifuge tube with a sterile cell
scraper. Cells were resuspended in 1 ml of distilled water and
sonicated (cat. no. E0380; Beyotime Institute of Biotechnology) at
room temperature for 1 min (40 Hz; ultrasound for 5 sec followed by
5 sec pauses). The cell lysate was incubated for 10 min in a water
bath at 4°C and centrifuged at 12,000 × g for 15 min. The
experiments were carried out according to the manufacturer's
protocols of each kit using a UV spectrophotometer (Cary 60 UV–Vis;
Agilent Technologies, Inc.) to determine the ATP, lactate and
glucose content. ATP (cat. no. S0026), glucose (cat. no. S0201S)
and lactate (cat. no. C0016) kits were purchased from Beyotime
Institute of Biotechnology.
Immunofluorescence
The RO8191 reverse validation experiment was divided
into four groups and were treated as follows: i) Control (DMSO);
ii) RO8191 (1 µmol/l); iii) DEX-HCl (25 µmol/l) combined with SFC
(1.25 µmol/l); and iv) combined treatment with DEX-HCl (25 µmol/l),
SFC (1.25 µmol/l) and RO8191 (1 µmol/l). Different drugs were added
to KYSE30 cells according to the requirements of the different
groups, and the cells were incubated in a cell incubator at 37°C
for 48 h, the cells were digested, centrifuged and resuspended as
previously described. The cell samples were incubated with 4%
paraformaldehyde at room temperature for 20 min, then incubated
with 0.5% Triton X-100 for 20 min. Subsequently, 5% bovine serum
albumin (cat. no. SW3015; Beijing Solarbio Science & Technology
Co., Ltd.) was added and the cells were incubated for 1 h at room
temperature. The samples were incubated with primary antibodies at
4°C overnight and the cells were then washed with 1% Tween TBST for
3 min. The primary antibodies used were: E-cadherin (cat. no.
ET1607-75; 1:100) and N-cadherin (cat. no. ET1607-37; 1:100), which
were purchased from Hangzhou HuaAn Biotechnology Co., Ltd.
Fluorescently labeled secondary antibodies (cat. no. ZF-0511;
1:500; Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.) were
added and incubated in the dark at room temperature for 1 h. The
slides were incubated with DAPI in the dark for 5 min, sealed with
an anti-fluorescence quencher (cat. no. P0131; Beyotime Institute
of Biotechnology), imaged using a confocal microscope and analyzed
using ImageJ version 1.52a (National Institutes of Health) bundled
with Java 8.
Reverse transcription-quantitative
(RT-q)PCR
After treating KYSE30 cells with DEX-HCI (25 nmol/l)
or SFC (1.25 µmol/l) alone or in combination for 24 h, the cells
were collected with a sterile cell scraper and transferred to a 1.5
ml centrifuge tube. The RNA was extracted using TRIzol (Beijing
Solarbio Science & Technology Co., Ltd.), according to the
manufacturer's instructions. Briefly, cells were resuspended in 1
ml of TRIzol and incubated at room temperature for 5 min. The RNA
sample was used immediately or stored at −80°C. RNA concentration
was measured using a Q5000 UV–Vis Spectrophotometer (Pono-550;
Prebo Instruments (Hangzhou) Co., Ltd.). RNA was reverse
transcribed (at 42°C for 15 min) using the FastKing gDNA Dispelling
RT SuperMix kit (Tiangen Biotech Co., Ltd.). The reverse
transcribed complementary DNA (cDNA) was used immediately or stored
in at −80°C. The cDNA concentration was measured using a Dalong
Gradient Thermal Cycler TC1000-G (Shaying Scientific Instruments
(Shanghai) Co., Ltd.) and amplified in a CFX Connect Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc.,) using FastKing One
Step RT-qPCR kit (SYBR Green) (Tiangen Biotech Co., Ltd.). The
primers used for RT-qPCR are listed in Table I. The thermal cycling conditions
used were: 50°C for 30 min; 95°C for 3 min; followed by 40 cycles
of 95°C for 15 s and 60°C for 30 s. By comparing the target gene Cq
with the internal reference Cq, ΔCq for each group, and the fold
difference was calculated by 2−ΔΔCq (27). GraphPad Prism version 9.5.0
(Dotmatics) was used for statistical analysis.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Primer (5′-3′) |
|---|
| HIF-1α | F:
GAACGTCGAAAAGAAAAGTCTCG |
|
| R:
CCTTATCAAGATGCGAACTCACA |
| HK2 | F:
GTGAATCGGAGAGGTCCCAC |
|
| R:
GCTAACTTCGGCCACAGGAT |
| LDHA | F:
CTGGCTGTGTCCTTGCTGTA |
|
| R:
TCACGTTACGCTGGACCAAA |
| GAPDH | F:
GATTCCACCCATGGCAAATTC |
|
| R:
CTGGAAGATGGTGATGGGATT |
Western blotting
DEX-HC (25 nmol/l), SFC (1.25 µmol/l) and RO819 (1
µmol/l) were added to KYSE30 cells according to the aforementioned
grouping requirements and incubated in a cell incubator at 37°C for
48 h. The cells were then digested and centrifuged according to the
aforementioned method. The RIPA solution (cat. no. R0010; Beijing
Solarbio Science & Technology Co., Ltd.) was added to the cell
precipitate and incubated on ice for 30 min. The lysate was cleared
by centrifugation at 12,000 × g for 10 min at 4°C. Protein
quantification was performed with bicinchoninic acid (BCA) protein
quantification kit. Protein samples (30 µg) were loaded on a 10%
polyacrylamide gel and separated by SDS-PAGE for 35 min at 80 V,
then 120 V for 60 min. Proteins were transferred to a PVDF membrane
using the Trans-Blot® Turbo™ Transfer System (Bio-Rad
Laboratories, Inc.) for 1 h, with a constant current of 260 mA. The
PVDF membrane was blocked using 5% skimmed milk powder for 2 h at
room temperature, and then washed with TBST (0.1% Tween) for 30
min. The membrane was then incubated with the corresponding primary
antibodies at 4°C overnight. The PVDF membrane was washed 3 times
with TBST (0.1% Tween) for 30 min, then blocked with 5% skim milk
powder and incubated with secondary antibodies for 1 h at room
temperature. Images were processed and analyzed using ImageJ
version 1.52a (National Institutes of Health) bundled with Java 8.
Antibodies used were as follows: β-actin (cat. no. EM21002; HUABIO;
1:2,000), anti-JAK2 (cat. no. M1501-8; HUABIO; 1:1,000), p-JAK2
(cat. no. ET1607-34; HUABIO; 1:1,000), STAT3 (cat. no. ET1607-38;
HUABIO; 1:1,000), p-STAT3 (cat. no. ET1603-40; HUABIO; 1:1,000),
HIF-1α (cat. no. R1510-5; HUABIO; 1:1,000), MMP2 (cat. no.
bs-4605R; BIOSS; 1:1,000), MMP9 (cat. no. bs-4593R; BIOSS;
1:1,000), E-cadherin (cat. no. ET1607-75; Hangzhou HuaAn
Biotechnology Co., Ltd.; 1:100), N-cadherin (cat. no. ET1607-37;
Hangzhou HuaAn Biotechnology Co., Ltd.; 1:100), HK2 (cat. no.
A0994; ABclonal Biotech Co., Ltd.; 1:1,000), lactate dehydrogenase
A (LDHA; cat. no. A1146; ABclonal Biotech Co., Ltd.; 1:1,000), HRP
Goat anti-rabbit (cat. no. AS014; ABclonal Biotech Co., Ltd.;
1:10,000) and HRP Goat anti-mouse (cat. no. AS003; ABclonal Biotech
Co., Ltd.; 1:10,000).
Statistical analysis
Statistical analysis was performed using SPSS 22.0
software (IBM Corp.). Measurement data are presented as mean ±
standard deviation. All data were analyzed using one-way ANOVA,
followed by Tukey's post hoc analysis. All parallel experiments
were repeated three or more times. P<0.05 was considered to
indicate a statistically significant difference.
Results
DEX-HCl and SFC inhibit KYSE30 cell
proliferation
The relative molecular masses and structural formula
of DEX-HCI and SFC are presented in Fig. 1A and B. Among the four esophageal
squamous cell carcinoma cell lines (KYSE30, KYSE520, KYSE140 and
KYSE410), p-STAT 3 and p-JAK were more significantly up-regulated
in KYSE30 cells compared with normal human esophageal epithelial
HEEC, and thus KYSE30 was chosen for use in further experiments
(Fig. S1). CCK-8 and clonogenic
assays of KYSE30 cells treated with DEX-HCl (25, 50, 100 and 200
nmol/l) and SFC (1.25, 2.5, 5 and 10 µmol/l) in vitro showed
a significant and concentration-dependent decrease in cell
viability compared with the control group after 48 h of treatment
(Fig. 1C and D). Compared with
DEX-HCL or SFC alone, the combination treatment demonstrated
enhanced inhibition of KYSE30 cell viability (Fig. 1E). The IC50 was
calculated to be 83.96 nmol/l for DEX-HCl and 2.173 µmol/l for SFC.
In subsequent experiments concentrations <IC50 values
were used, and thus, DEX-HCl was used at 25 nmol/l and SFC was used
at 1.25 µmol/l. To evaluate the effect of the co-administration of
DEX-HCl and SFC, the technique previously described by Chou
(28) was used. The combination
index (CI) theory provides quantitative definitions of additive
effect (CI, 1), synergistic effect (CI, <1) and antagonistic
effect (CI, >1) in drug combinations. The smaller the value of
CI, the stronger the synergistic effect of drugs. The CI of DEX-HCl
and SFC was 0.27, demonstrating a strong synergistic effect between
DEX-HCl and SFC (Fig. 1E). The
results of the clonogenic assay demonstrated that DEX-HCl and SFC
were able to inhibit the proliferation of KYSE30 cells and
demonstrated enhanced inhibition of KYSE30 cells in combination
compared with single dosing, which suggested that DEX-HCl and SFC
were more effective in combination (Fig. 1F and G).
Effects of DEX-HCl and SFC on the
invasion and migration ability of KYSE30 cells
Wound healing and Transwell assays were performed to
assess the effects of DEX-HCl and SFC on cell migration and
invasion. The wound healing assay results demonstrated that KYSE30
cells treated with DEX-HCl (25 nmol/l) or SFC (1.25 µmol/l) for 24
and 48 h demonstrated an inhibited migration, and demonstrated a
significantly greater inhibition after co-administration for both
24 and 48 h after treatment. Cells that were treated with DEX-HCl
and SFC in combination only healed 10% after 24 h of treatment, as
shown in Fig 2A. The wound healing
assay results demonstrated that DEX-HCI and SFC had an inhibitory
effect on cell migration, while the DEX-HCI combined with SFC group
had a stronger inhibitory effect on cell migration compared with
the DEX-HCI (25 nmol/l) or SFC (1.25 µmol/l) only groups. In the
DEX-HCI + SFC group there was also a time-dependent effect, and the
longer the migration time, the stronger the inhibition ability of
migration (Fig. 2A). The Transwell
migration assay demonstrated that the cell invasion ability of
KYSE30 cells treated with DEX-HCl (25 nmol/l) or SFC (1.25 µmol/l)
was significantly inhibited after 12 h compared with the control
group. The combination of DEX-HCl and SFC significantly increased
the inhibitory effect on the invasive ability of KYSE30 cells
compared with the single treatment groups (Fig. 2B).
DEX-HCl and SFC reduce glucose
metabolism in KYSE30 cells
Changes were observed in ATP levels, lactate
production and glucose uptake in KYSE30 cells after treatment with
DEX-HCl and SFC. The results showed that ATP levels, lactate
production and glucose uptake were significantly reduced in KYSE30
cells in the DEX-HCl group, SFC group and the combination group
compared with the control group (Fig.
2C). The combination group showed significantly decreased ATP
levels, lactate production and glucose uptake in KYSE30 cells
compared with the DEX-HCl and SFC alone groups (Fig. 2C).
DEX-HCl and SFC impact protein
expression in KYSE30 cells
To investigate the mechanism of action, the
expression of JAK/STAT3/HIF-1α pathway-related, invasion-related
marker proteins and glycolysis-related marker proteins were
assessed using qPCR and western blotting. The results of the qPCR
and western blotting experiments demonstrated that both DEX-HCI and
SFC were able to significantly reduce the expression and protein
levels of HIF-1α, HK2 and LDHA. Additionally, the combination group
of DEX-HCI and SCF inhibited the expression and protein levels of
HIF-1α, HK2 and LDHA more strongly compared with that of the drug
alone (Fig. 3A and B). It was also
demonstrated that DEX-HCl and SFC treatment significantly decreased
the protein expression levels of p-STAT3 and p-JAK compared with
the control group. The co-administration of DEX-HCl and SFC
demonstrated a further significant decrease in the protein
expression levels of p-STAT3 and p-JAK compared with both the
control and single treatment groups alone (Fig. 3C), without affecting the expression
levels of STAT3 and JAK proteins. The protein expression levels of
E-cadherin and N-cadherin were examined by immunofluorescence assay
and western blotting and the results showed that DEX-HCl and SFC
were able to significantly upregulate E-cadherin and significantly
downregulate N-cadherin protein expression compared with the
negative control in KYSE30 cells. SFC significantly reduced the
expression levels of MMP2 and MMP9 proteins in KYSE30 cells.
DEX-HCI significantly reduced the protein expression level of MMP9,
but the addition of DEX-HCI could not significantly reduce the
protein expression level of MMP2. The DEX-HCI + SFC group
significantly reduced the expression level of the MMP2 protein
compared with the DEX-HCI group and SFC group, which further
indicated that DEX-HCI and SFC combined have a drug synergistic
effect (Fig. 4A and B).
 | Figure 3.Effect of DEX-HCl and SFC on glucose
metabolism and JAK/STAT pathway-related protein expression in
KYSE30 cells. (A) RT-qPCR assay to detect changes in relative mRNA
expression of HIF-1α, HK2, and LDHA genes in KYSE30 cells. (B)
Semi-quantified protein expression levels assessed by western
blotting of HK2, LDHA and HIF-1α in KYSE30 cells. (C)
Semi-quantified protein expression levels assessed by western
blotting of STAT3, p-STAT3, JAK and p-JAK in KYSE30 cells.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; ns, not
significant (P>0.05). DEX-HCl, Dexmedetomidine hydrochloride;
SFC, sufentanil citrate; RT-qPCR, reverse transcription
quantitative PCR. |
 | Figure 4.Effects of DEX-HCl in combination
with SFC on invasion-related proteins. (A) The expression of the
invasion-associated proteins E-cadherin and N-cadherin was
determined by immunofluorescence (scale bar, 50 µm). (B)
Semi-quantified protein expression levels assessed by western
blotting of MMP 2, MMP 9, E-cadherin and N-cadherin. *P<0.05,
**P<0.01, ***P<0.001, ****P<0.0001. DEX-HCl,
Dexmedetomidine hydrochloride; SFC, sufentanil citrate; MMP2,
metalloproteinase 2; MMP9, metalloproteinase 9. |
DEX-HCI combined with SFC inhibited
the invasion of KYSE30 cells by inhibiting the JAK/STAT3/HIF-1α
pathway
To further assess whether DEX-HCl and SFC effect
KYSE30 cell glucose metabolism and invasion through the
JAK/STAT3/HIF-1α axis, reverse validation experiments were
performed by adding RO8191 to induce STAT3/JAK to undergo
phosphorylation.
RO8191 is an imidazolino pyridine compound that
activates phosphorylation of JAK and STAT proteins thereby
activating the JAK/STAT signaling pathway (29). Therefore, recovery of the invasion
and migration ability of KYSE30 with the addition of RO8191 would
suggest that DEX-HCl and SFC exhibit their function via the
JAK/STAT pathway. The addition of RO8191 in the Transwell migration
assay counteracted the inhibitory effect of DEX-HCl and SFC
treatment on KYSE30 cells, significantly increasing the number of
invaded cells compared with the DEX-HCl and SFC treatment group
(Fig. 5A). DEX-HCL combined with
SFC was demonstrated to reduce the expression levels of HIF-1α, HK2
and LDHA mRNA, whereas the addition of RO8191 was demonstrated to
upregulate the expression levels of HIF-1α, HK2 and LDHA mRNA,
counteracting the inhibitory effect of DEX-HCL combined with SFC
(Fig. 5B).
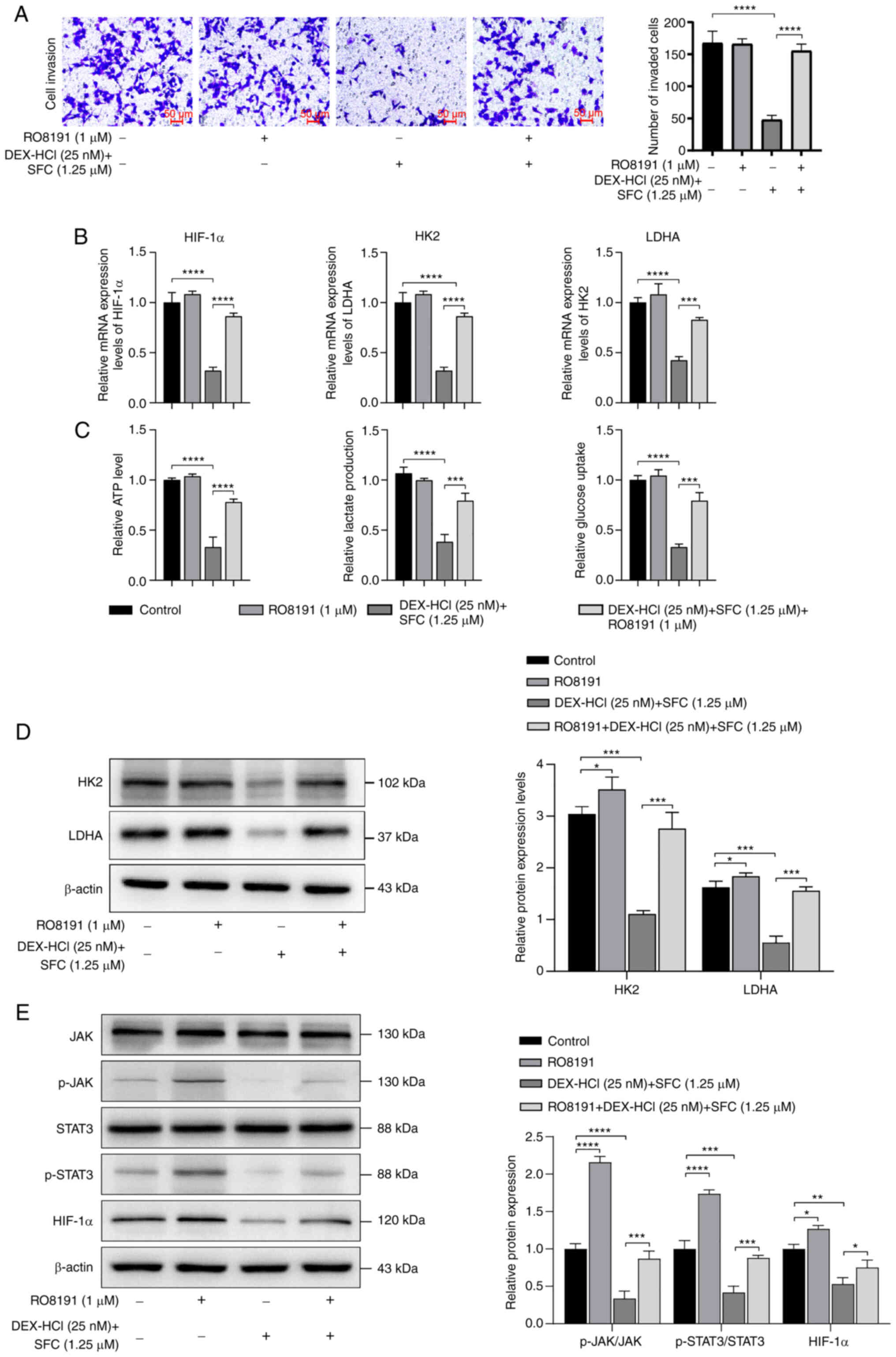 | Figure 5.RO8191 counteracts the effects of
DEX-HCl combined with SFC on KYSE30 cell invasion, migration and
glucose metabolism. (A) Transwell assay to test the effect of
RO8191 on the invasion ability of KYSE30 cells treated with DEX-HCl
and SFC. (B) RT-qPCR was used to assess changes in relative mRNA
expression of HIF-1α, HK2, and LDHA genes in KYSE30 cells. (C)
Detection of changes in lactate, glucose and ATP content in KYSE30
cells. (D) Semi-quantified protein expression levels assessed by
western blotting of glucose metabolism-related proteins HK2 and
LDHA. (E) Semi-quantified protein expression levels assessed by
western blotting of p-JAK, p-STAT 3 and HIF-1α proteins.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. DEX-HCl,
Dexmedetomidine hydrochloride; SFC, sufentanil citrate; HIF-1α,
Hypoxia-inducible factor-1α; p-JAK, phosphorylated Janus kinase;
p-STAT 3, phosphorylated signal transducer and activator 3;
RT-qPCR, reverse transcription-quantitative PCR. |
Glucose uptake, ATP levels and lactate production of
KEYSE30 cells significantly increased after treatment with RO8191
compared with the DEX-HCl and SFC co-administration group (Fig. 5C). DEX-HCI combined with SFC
significantly inhibited the expression levels of HK2, LDHA, p-JAK,
p-STAT3, and HIF-1α proteins, whereas RO8191 co-administration
increased the expression levels of HK2, LDHA, p-JAK, p-STAT3 and
HIF-1α proteins, counteracting the effect of DEX-HCI combined with
SFC (Fig. 5D and E). Likewise, the
addition of RO8191 in immunofluorescence experiments showed a
significant decrease in E-cadherin fluorescence intensity compared
with the DEX-HCl and SFC treatment group and a significant increase
in N-cadherin fluorescence compared with the DEX-HCl and SFC
treatment group (Fig. 6A). The
addition of RO8191 was also demonstrated to increase the protein
expression levels of MMP2, MMP9 and N-cadherin and decrease the
protein expression levels of E-cadherin compared with the DEX-HCl
and SFC treatment group (Fig. 6B).
In summary, DEX-HCI in combination with SFC was able to inhibit
KEYSE30 cell invasion and inhibit JAK/STAT3/HIF-1α axis activation,
and RO8191, as a JAK/STAT3 pathway activator, was able to reverse
this effect, which reinforces the conclusion that the inhibitory
effect of DEX-HCI in combination with SFC on KEYSE30 cell invasion
was mediated by the inhibition of JAK/STAT3/HIF-1α axis activation.
STAT3/HIF-1α axis to exert pharmacological effects.
 | Figure 6.Immunofluorescence and western
blotting of invasion-related proteins from KYSE30 cells treated
with RO8191 and DEX-HCl and SFC combined treatment. (A)
Immunofluorescence images and quantification of N-cadherin and
E-cadherin in KYSE30 cells after treatment with RO8191 and DEX-HCl
and SFC combined treatment (scale bar, 50 µm). (B) Semi-quantified
protein expression levels assessed by western blotting of MMP2,
MMP9, E-cadherin, and N-cadherin. *P<0.05, **P<0.01,
***P<0.001, ****P<0.0001. DEX-HCl, Dexmedetomidine
hydrochloride; SFC, sufentanil citrate; MMP2, metalloproteinase 2;
MMP9, metalloproteinase 9. |
Discussion
A major cause of the current low 5-year survival
rate after EC surgery is the epithelial-mesenchymal transition
(2). EMT serves a key role in skin
injury healing and tumor invasion and metastasis (30), and elevated expression of MMP2 and
MMP9 promotes EC proliferation, accelerates the EMT process and
facilitates tumor angiogenesis (31). MMP2 and MMP9 are risk factors for
metastasis and poor prognosis in EC (32). The results of the present study
demonstrated that DEX-HCl and SFC could inhibit the proliferation,
invasion and migration of KYSE30 cells and suppress the protein
expression levels of MMP2 and MMP9, and that the inhibitory effect
was increased following the co-administration of DEX-HCL and SFC.
Aberrant expression of N-cadherin and low expression of E-cadherin
are important indicators of poor prognosis and EMT in EC (33,34).
E-cadherin determines the adhesion ability between epithelial cells
during the development of EMT. Dysregulation of E-cadherin protein
expression leads to the loss of the adhesion ability of tumor cells
and contributes to the metastasis of tumor cells (35). Moreover, previous studies have
reported that N-cadherin can induce tumor cell invasion and
angiogenesis (36,37), and a number of N-cadherin
antagonists have been adopted for tumor therapy (38). The present study demonstrated that
DEX-HCl combined with SFC significantly upregulated E-cadherin and
significantly downregulated N-cadherin protein expression levels,
thereby inhibiting KYSE30 cell metastasis and EMT.
DEX-HCl has been reported to inhibit growth and
metastasis (39) and to induce
apoptosis in EC cells (40).
Previous studies have reported that SFC could also inhibit EC cell
metastasis (41), which was
consistent with the findings of this study. However, neither
elaborated on the effects of co-administration of DEX-HCl and SFC
on glucose metabolism in EC cells. Cancer cells require substantial
amounts of energy during metastasis, which is supplied directly
through the glycolytic pathway (42). The preferred metabolic pathway in
cancer cells under aerobic conditions is glycolysis for energy
supply rather than oxidative phosphorylation, a phenomenon known as
the Warburg effect (43).
Therefore, inhibition of tumor glycolysis is considered a new
strategy to inhibit tumor proliferation and metastasis (44). The results of the current study
indicated that DEX-HCl and SFC treatments reduced ATP production
and cellular glucose metabolism in KYSE30 cells. Furthermore,
co-administration of DEX-HCl and SFC enhanced their individual
inhibitory effects on proliferation, migration and invasion of
KYSE30 cells, as well as increasing the reduction in ATP production
and glucose metabolism in KYSE30 cells compared with single
treatment. Furthermore, overexpression of LDHA in tumor cells has
been frequently reported (45,46).
In addition to promoting glycolysis, elevated LDHA levels in tumors
also facilitates lactate production, thereby reconstituting the
tumor microenvironment and inhibiting the immune system to promote
immune escape (47). DEX-HCl and
SFC can decrease lactate production and downregulate mRNA and
protein expression levels of HK2 and LDHA, thereby suppressing
KYSE30 cell invasive migration.
The present study demonstrated that DEX-HCl combined
with SFC inhibited KYSE30 cell invasion and metastasis and blocked
KYSE30 cell glucose metabolism. Moreover, previous studies have
reported that JAK and STAT3 are involved in all stages of
development from cancer cell proliferation to cancer cell
metastasis (48,49). In rheumatoid arthritis, JAK/STAT3
exerts anti-inflammatory effects by regulating HIF-1α expression
(50), CXCL8 promotes melanoma
progression by activating the JAK/STAT1/HIF-1α axis (51), and ELTD1 promotes glioma
proliferation, migration and invasion by activating the
JAK/STAT3/HIF-1α signaling axis (52). Therefore, this pathway may be a
potential target for inhibiting the metastasis of cancer cells. The
present study demonstrated that DEX-HCl combined with SFC inhibited
the JAK/STAT1/HIF-1α axis and suppressed KYSE30 cell metastasis and
glucose metabolism, and that the inhibitory effect was counteracted
by the addition of RO8191, which activated the phosphorylation of
JAK and STAT proteins which increased the protein expression levels
of MMP2, MMP9, N-cadherin and E-cadherin, suggesting the results of
DEX-HCl and SFC treatment involve the modulation of the JAK/STAT
signaling pathway. Furthermore, in vitro experiments
demonstrated that DEX-HCl and SFC may regulate glucose
metabolism-related indicators through the JAK/STAT3/HIF-1α axis,
thereby inhibiting KYSE30 cell invasion and migration. However,
further studies are required to elucidate the in-depth mechanism by
which DEX-HCl combined with SFC affects glucose metabolism in
KYSE30 cells.
In the present study, DEX-HCl and SFC were
demonstrated to decrease the expression of EC and
metastasis-related proteins, significantly reducing glucose uptake,
ATP and lactate production in KYSE30 cells. The pharmacological
effects were significantly enhanced by the combined action of
DEX-HCI and SFC, and reverse validation experiments with RO8191
confirmed the possible involvement of the JAK/STAT3/HIF-1α axis in
this process.
Supplementary Material
Supporting Data
Acknowledgments
Not applicable.
Funding
This work was supported by the Natural Science Foundation of
Hebei Province (grant no. H2020206397).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WL, YW, XL, HW and LJ were involved in the design of
the project. WL wrote the first draft of the article. HW was
responsible for the final revision and layout of the article. XL
was responsible for the literature search and experimental methods.
YW was responsible for conducting the experiments. LJ was
responsible for the data processing and general notation series. WL
and YW confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu CQ, Ma YL, Qin Q, Wang PH, Luo Y, Xu
PF and Cui Y: Epidemiology of esophageal cancer in 2020 and
projections to 2030 and 2040. Thorac Cancer. 14:3–11. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Z, Zhao Y, Kong P, Liu Y, Huang J, Xu
E, Wei W, Li G, Cheng X, Xue L, et al: Integrated multi-omics
profiling yields a clinically relevant molecular classification for
esophageal squamous cell carcinoma. Cancer Cell. 41:181–195. 2023.
View Article : Google Scholar
|
|
3
|
Morgan E, Soerjomataram I, Rumgay H,
Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J and Arnold
M: The global landscape of esophageal squamous cell carcinoma and
esophageal adenocarcinoma incidence and mortality in 2020 and
projections to 2040: New estimates from GLOBOCAN 2020.
Gastroenterology. 163:649–658. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li P, Jing J, Liu W, Wang J, Qi X and
Zhang G: Spatiotemporal patterns of esophageal cancer burden
attributable to behavioral, metabolic, and dietary risk factors
from 1990 to 2019: Longitudinal observational study. JMIR Public
Health Surveill. 9:e460512023. View
Article : Google Scholar
|
|
5
|
Codipilly DC and Wang KK: Squamous cell
carcinoma of the esophagus. Gastroenterol Clin North Am.
51:457–484. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu D, Wu X, Wu W, Wu S, Li H, Zhang Y, Yan
X, Zhai J, Dong X, Feng S, et al: Plasma cell-free DNA
5-hydroxymethylcytosine and whole-genome sequencing signatures for
early detection of esophageal cancer. Cell Death Dis. 14:8432023.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang S, Zheng R, Arnold M, Abnet C, Zeng
H, Zhang S, Chen R, Sun K, Li L, An L, et al: Global and national
trends in the age-specific sex ratio of esophageal cancer and
gastric cancer by subtype. Int J Cancer. 151:1447–1461. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Manfioletti G and Fedele M:
Epithelial-mesenchymal transition (EMT). Int J Mol Sci.
24:113862023. View Article : Google Scholar
|
|
9
|
Lu W and Kang Y: Epithelial-mesenchymal
plasticity in cancer progression and metastasis. Dev Cell.
49:361–374. 2019. View Article : Google Scholar
|
|
10
|
Counihan JL, Grossman EA and Nomura DK:
Cancer metabolism: Current understanding and therapies. Chem Rev.
118:6893–6923. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang M, Wei T, Zhang X and Guo D:
Targeting lipid metabolism reprogramming of immunocytes in response
to the tumor microenvironment stressor: A potential approach for
tumor therapy. Front Immunol. 13:9374062022. View Article : Google Scholar
|
|
12
|
Xin P, Xu X, Deng C, Liu S, Wang Y, Zhou
X, Ma H, Wei D and Sun S: The role of JAK/STAT signaling pathway
and its inhibitors in diseases. Int Immunopharmacol. 80:1062102020.
View Article : Google Scholar
|
|
13
|
Zhang P, Li Z and Yang G: Silencing of
ISLR inhibits tumour progression and glycolysis by inactivating the
IL-6/JAK/STAT3 pathway in non-small cell lung cancer. Int J Mol
Med. 48:2222021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao C, Zhang W, Hua M, Chen H, Yang B,
Wang Y and Yang Q: RNF7 inhibits apoptosis and sunitinib
sensitivity and promotes glycolysis in renal cell carcinoma via the
SOCS1/JAK/STAT3 feedback loop. Cell Mol Biol Lett. 27:362022.
View Article : Google Scholar
|
|
15
|
Lei K, Du W, Lin S, Yang L, Xu Y, Gao Y,
Xu B, Tan S, Xu Y, Qian X, et al: 3B, a novel photosensitizer,
inhibits glycolysis and inflammation via miR-155-5p and breaks the
JAK/STAT3/SOCS1 feedback loop in human breast cancer cells. Biomed
Pharmacother. 82:141–150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
You Z, Xu D, Ji J, Guo W, Zhu W and He J:
JAK/STAT signal pathway activation promotes progression and
survival of human oesophageal squamous cell carcinoma. Clin Transl
Oncol. 14:143–149. 2012. View Article : Google Scholar
|
|
17
|
Zhao X, Tang YP, Wang CY, Wu JX and Ye F:
Prognostic values of STAT3 and HIF-1lα in esophageal squamous cell
carcinoma. Eur Rev Med Pharmacol Sci. 23:3351–3357. 2019.
|
|
18
|
Fang J, Chu L, Li C, Chen Y, Hu F, Zhang
X, Zhao H, Liu Z and Xu Q: JAK2 inhibitor blocks the inflammation
and growth of esophageal squamous cell carcinoma in vitro through
the JAK/STAT3 pathway. Oncol Rep. 33:494–502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Jin G, Liu H, Liu K, Zhao J, Chen
X, Wang D, Bai R, Li X, Jang Y, et al: Metformin inhibits
esophageal squamous cell carcinoma-induced angiogenesis by
suppressing JAK/STAT3 signaling pathway. Oncotarget. 8:74673–74687.
2017. View Article : Google Scholar
|
|
20
|
Garcia SN, Guedes RC and Marques MM:
Unlocking the potential of HK2 in cancer metabolism and
therapeutics. Curr Med Chem. 26:7285–7322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang L, Jiang C, Zhong Y, Sun K, Jing H,
Song J, Xie J, Zhou Y, Tian M, Zhang C, et al: STING is a
cell-intrinsic metabolic checkpoint restricting aerobic glycolysis
by targeting HK2. Nat Cell Biol. 25:1208–1222. 2023. View Article : Google Scholar
|
|
22
|
Xun S and Zheng R: Dexmedetomidine
alleviates neuropathic pain by regulating JAK/STAT pathway in rats.
J Cell Biochem. 121:2277–2283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Si Y, Bao H, Han L, Shi H, Zhang Y, Xu L,
Liu C, Wang J, Yang X, Vohra A and Ma D: Dexmedetomidine protects
against renal ischemia and reperfusion injury by inhibiting the
JAK/STAT signaling activation. J Transl Med. 11:1412013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Jiang W and Luo X: Remifentanil
combined with dexmedetomidine on the analgesic effect of breast
cancer patients undergoing modified radical mastectomy and the
influence of perioperative T lymphocyte subsets. Front Surg.
9:10166902022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cai Q, Liu G, Huang L, Guan Y, Wei H, Dou
Z, Liu D, Hu Y and Gao M: The role of dexmedetomidine in
tumor-progressive factors in the perioperative period and cancer
recurrence: A narrative review. Drug Des Devel Ther. 16:2161–2175.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Qu M, Guo K, Wang Y, Gu J, Wu H,
Zhu X, Sun Z, Cata JP, Chen W and Miao C: Intraoperative lidocaine
infusion in patients undergoing pancreatectomy for pancreatic
cancer: A mechanistic, multicentre randomised clinical trial. Br J
Anaesth. 129:244–253. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ruijter JM, Barnewall RJ, Marsh IB,
Szentirmay AN, Quinn JC, van Houdt R, Gunst QD and van den Hoff
MJB: Efficiency correction is required for accurate quantitative
PCR analysis and reporting. Clin Chem. 67:829–842. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chou TC: Drug combination studies and
their synergy quantification using the Chou-Talalay method. Cancer
Res. 70:440–446. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng J, Zhang C, Zhu N, Zhang C, Liu M,
Han Z and Li Y: EPN3 plays oncogenic role in non-small cell lung
cancer by activating the JAK1/2-STAT3 pathway. Environ Toxicol.
38:1968–1979. 2023. View Article : Google Scholar
|
|
30
|
Gundamaraju R, Lu W, Paul MK, Jha NK,
Gupta PK, Ojha S, Chattopadhyay I, Rao PV and Ghavami S: Autophagy
and EMT in cancer and metastasis: Who controls whom? Biochim
Biophys Acta Mol Basis Dis. 1868:1664312022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Shi G, Gao F, Liu P, Wang H and
Tan X: TSPAN1 upregulates MMP2 to promote pancreatic cancer cell
migration and invasion via PLCγ. Oncol Rep. 41:2117–2125.
2019.PubMed/NCBI
|
|
32
|
Zhang L, Xi RX and Zhang XZ: Matrix
metalloproteinase variants associated with risk and clinical
outcome of esophageal cancer. Genet Mol Res. 14:4616–4624. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu XL, Ling ZQ, Chen SZ, Li B, Ji WH and
Mao WM: The impact of E-cadherin expression on the prognosis of
esophageal cancer: A meta-analysis. Dis Esophagus. 27:79–86. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu S, Liu J, Min L, Sun X, Guo Q, Li H,
Zhang Z, Zhao Y, Gu J and Zhang S: Cadherin expression shift could
well distinguish esophageal squamous cell carcinoma from non-
cancerous esophageal tissues. Oncol Res Treat. 41:380–385. 2018.
View Article : Google Scholar
|
|
35
|
Mendonsa AM, Na TY and Gumbiner BM:
E-cadherin in contact inhibition and cancer. Oncogene.
37:4769–4780. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Parker J, Hockney S, Blaschuk OW and Pal
D: Targeting N-cadherin (CDH2) and the malignant bone marrow
microenvironment in acute leukaemia. Expert Rev Mol Med.
25:e162023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lou C, Wu K, Shi J, Dai Z and Xu Q:
N-cadherin protects oral cancer cells from NK cell killing in the
circulation by inducing NK cell functional exhaustion via the KLRG1
receptor. J Immunother Cancer. 10:e0050612022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mariotti A, Perotti A, Sessa C and Rüegg
C: N-cadherin as a therapeutic target in cancer. Expert Opin
Investig Drugs. 16:451–465. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshio T, Ishiyama A, Tsuchida T,
Yoshimizu S, Horiuchi Y, Omae M, Hirasawa T, Yamamoto Y, Sano H,
Yokota M and Fujisaki J: Efficacy of novel sedation using the
combination of dexmedetomidine and midazolam during endoscopic
submucosal dissection for esophageal squamous cell carcinoma.
Esophagus. 16:285–291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Che J, Liu M and Lv H: Dexmedetomidine
disrupts esophagus cancer tumorigenesis by modulating
circ_0003340/miR-198/HMGA2 axis. Anticancer Drugs. 33:448–458.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tang H, Li C, Wang Y and Deng L:
Sufentanil inhibits the proliferation and metastasis of esophageal
cancer by inhibiting the NF-κB and snail signaling pathways.
J Oncol. 2021:75861002021. View Article : Google Scholar
|
|
42
|
Kocianova E, Piatrikova V and Golias T:
Revisiting the warburg effect with focus on lactate. Cancers
(Basel). 14:60282022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao
S, Wei P and Li D: Warburg effect in colorectal cancer: The
emerging roles in tumor microenvironment and therapeutic
implications. J Hematol Oncol. 15:1602022. View Article : Google Scholar
|
|
44
|
Chelakkot C, Chelakkot VS, Shin Y and Song
K: Modulating glycolysis to improve cancer therapy. Int J Mol Sci.
24:26062023. View Article : Google Scholar
|
|
45
|
Zhang K, Zhang T, Yang Y, Tu W, Huang H,
Wang Y, Chen Y, Pan K and Chen Z: N(6)-methyladenosine-mediated
LDHA induction potentiates chemoresistance of colorectal cancer
cells through metabolic reprogramming. Theranostics. 12:4802–4817.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jacquet P and Stephanou A: Searching for
the metabolic signature of cancer: A review from warburg's time to
now. Biomolecules. 12:14122022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Brand A, Singer K, Koehl GE, Kolitzus M,
Schoenhammer G, Thiel A, Matos C, Bruss C, Klobuch S, Peter K, et
al: LDHA-associated lactic acid production blunts tumor
immunosurveillance by T and NK cells. Cell Metab. 24:657–671. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dinakar YH, Kumar H, Mudavath SL, Jain R,
Ajmeer R and Jain V: Role of STAT3 in the initiation, progression,
proliferation and metastasis of breast cancer and strategies to
deliver JAK and STAT3 inhibitors. Life Sci. 309:1209962022.
View Article : Google Scholar
|
|
49
|
Malekan M, Ebrahimzadeh MA and Sheida F:
The role of Hypoxia-Inducible Factor-1alpha and its signaling in
melanoma. Biomed Pharmacother. 141:1118732021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu L, Liu R and Zhang L: Advance in bone
destruction participated by JAK/STAT in rheumatoid arthritis and
therapeutic effect of JAK/STAT inhibitors. Int Immunopharmacol.
111:1090952022. View Article : Google Scholar
|
|
51
|
Hu X, Yuan L and Ma T: Mechanisms of
JAK-STAT signaling pathway mediated by CXCL8 gene silencing on
epithelial-mesenchymal transition of human cutaneous melanoma
cells. Oncol Lett. 20:1973–1981. 2020. View Article : Google Scholar
|
|
52
|
Li J, Shen J, Wang Z, Xu H, Wang Q, Chai
S, Fu P, Huang T, Anas O, Zhao H, et al: ELTD1 facilitates glioma
proliferation, migration and invasion by activating
JAK/STAT3/HIF-1alpha signaling axis. Sci Rep. 9:139042019.
View Article : Google Scholar : PubMed/NCBI
|




















