Introduction
Breast cancer is a common malignant disease in the
female population with high numbers of mortality (1–3).
Although significant progress has been made in breast cancer
research and anti-hormone treatment in recent years, the survival
rate of patients with advanced-stage disease remains poor (4,5). The
identification of new biomarkers of prognosis and therapeutic
target molecules and the development of effective therapeutic drugs
for advanced breast cancer are necessary for improving the survival
rate of patients with this disease, particularly with advanced
breast cancer.
Nuclear receptor co-activator 7 (NCOA7, also named
ERAP140) is a member of the NCOA family and binds to the estrogen
receptor (ER) (6). NCOA7 also
belongs to the oxidation-resistant 1 (OXR1) protein family that
contains the Tre2/Bub2/Cdc16, lysin motif, domain catalytic (TLDc)
domain and may serve as an anti-oxidation protein against oxidative
stress (7,8). Previous studies have shown that NCOA7
interacts with [H+]-V-ATPase through its TLDc domain and
may regulate endosomal vesicle trafficking and viral entry
(9–12). In addition, NCOA7 inhibited the
MAPK/ERK pathway to regulate epithelial-mesenchymal transition and
apoptosis, thereby inhibiting the progression of clear-cell renal
cell carcinoma (13); it is a
potential biomarker in oral squamous cell carcinoma and a member of
prognostic indicators for neuroblastoma and colon adenocarcinoma
(14–16). Genetic variations of the
NCOA7 gene are associated with a reduced risk of breast
cancer (17). However, the role of
NCOA7, as an ER-binding protein, in regulating breast cancer
progression has remained elusive. To the best of our knowledge, the
expression of NCOA7 in breast tumor tissues and its association
with clinicopathological characteristics and the survival rate of
patients with breast cancer have remained to be investigated.
To fully understand the role of NCOA7 in breast
cancer progression, the current study set out to determine the
expression levels of NCOA7 in tumors from patients with breast
cancer and the association of NCOA7 expression with the survival of
these patients. The results indicated that NCOA7 was overexpressed
in breast tumors and that its expression was reversely associated
with the survival of patients with breast cancer, suggesting that
it may be a biomarker for poor prognosis of breast cancer.
Patients and methods
Materials and patient samples
The following materials were used: Anti-NCOA7
antibody (cat. no. 393427; 1:300 dilution for immunohistochemistry
and 1:500 dilution for western blot analysis; Santa Cruz
Biotechnology, Inc.), anti-β-actin antibody (cat. no. 100166-MM10;
1:500 dilution; Zoonbio Biotechnology), immunohistochemical (IHC)
reagents including formalin, hydrogen peroxide, xylene, ethanol,
trisodium citrate and citric acid (Shanghai Sinopharm Chemical
Co.), Cell Counting Kit-8 (CCK-8; Vazyme Biotech Co. Ltd.), crystal
violet (Beyotime Institute of Biotechnology), diaminobenzidine
(DAB) color development (ZSGB-BIO; OriGene Technologies, Inc.), the
human breast cancer cell lines T47D (cat. no. HTB-133) and MCF7
(cat. no. HTB-22) and the human embryo kidney cell line 293T (cat.
no. CRL-3216; American Type Culture Collection). The breast cancer
tissues used in the present study were selected from a breast
cancer sample deposit library from patients with newly diagnosed
breast cancer who visited the Breast Surgery Department of Jiangsu
University Affiliated People's Hospital (Zhenjiang, China) between
December 2010 and October 2016. The paraffin-embedded breast cancer
tissue microarray (TMA) chips containing 241 tumor tissue samples
and 163 adjacent normal tissue samples and related
clinicopathological information of the patients used in the present
study were obtained from the Department of Pathology of Jiangsu
University Affiliated People's Hospital (Zhenjiang, China). All
tissue specimens collected were from patients who provided written
informed consent. The study protocol was approved by the
Institutional Review Board of Jiangsu University Affiliated
People's Hospital (Zhenjiang, China; approval no. LLYW20210004).
All breast cancer tissue donors were female and did not receive any
treatment prior to the surgery. The follow-up for patient survival
information was performed by telephone. Information regarding
patient age, tumor size, lymph node metastasis (N-stage), and
tumor, node and metastasis (TNM) stages is presented in Tables I and SI. The median age of all patients was
53.0 years. Furthermore, the median age was 53.0 years in patients
with triple negative breast cancer (TNBC) and 53.5 years in
patients with non-TNBC (N-TNBC). There was no significant age
difference (P=0.938) between the TNBC and the N-TNBC groups.
Overall survival was defined as the time from surgery to patient
death from any cause.
 | Table I.Association of NCOA7 expression with
clinicopathological parameters in total breast cancer tumor
samples. |
Table I.
Association of NCOA7 expression with
clinicopathological parameters in total breast cancer tumor
samples.
|
|
| NCOA7 status, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Pathological
parameter | Total | Positive | Negative | P-value |
|---|
| Age, years |
|
|
| 0.914 |
| ≤50 | 91 | 40 (44) | 51 (56) |
|
|
>50 | 150 | 67 (45) | 83 (55) |
|
| Tumor size, cm |
|
|
| 0.020 |
| ≤3 | 194 | 79 (41) | 115 (59) |
|
|
>3 | 47 | 28 (60) | 19 (40) |
|
| T stage |
|
|
| 0.005 |
| T1 | 126 | 45 (36) | 81 (64) |
|
|
T2/T3 | 115 | 62 (54) | 53 (46) |
|
| N stage |
|
|
| 0.008 |
| N0 | 155 | 59 (38) | 96 (62) |
|
|
N1-3 | 86 | 48 (56) | 38 (44) |
|
| TNM stage |
|
|
| 0.299 |
|
I/II | 213 | 92 (43) | 121 (57) |
|
|
IIIA/B | 28 | 15 (54) | 13 (46) |
|
IHC staining of the TMA
The paraffin-embedded breast cancer or normal tissue
microarray chips were baked at 65°C for 1 to 2 h. The specimens
were subsequently dewaxed by immersion in xylene for 30 min,
followed by hydration using gradients of alcohol (concentrations of
100, 95, 80, 70, 50%) for 5 min each at room temperature. The
specimens were completely immersed in 800 ml sodium citrate buffer
(pH 6.0) for antigen repair, boiled in a microwave oven for 20 min
and then cooled naturally to room temperature. Incubation with 3%
hydrogen peroxide at room temperature for 10 min was performed to
block endogenous peroxidase activity and non-specific protein
interactions. The samples were subsequently incubated with NCOA7
antibody (rabbit; cat. no. 393427; 1:300 dilution; Santa Cruz
Biotechnology, Inc.) overnight at 4°C and then with biotinylated
goat anti-rabbit secondary antibody working solution (cat. no.
SA1020; Boster Biological Technology Co. Ltd.) at 37°C for 30 min,
followed by color development with DAB. The slides were visualized
under light microscopy and the reaction was terminated immediately
when a yellow pellet was present. The staining was repeated with
hematoxylin for 30 sec at room temperature. The specimens were
subsequently dehydrated in a gradient of alcohol (50, 70, 80, 95
and 100%) for 5 min each, followed by immersion in xylene for 30
min for transparency at room temperature. Finally, the films were
sealed with resin glue. Phosphate buffer (5% BSA; Sigma-Aldrich;
Merck KGaA) was used instead of primary antibodies to prepare the
negative controls and positive tissue sections with specific
staining expression were used as positive controls. The IHC
staining assay was performed as previously described (18).
IHC staining scoring
Semi-quantitative scoring was performed by two
independent observers. The immunostaining score was determined
according to the positivity rate and staining intensity and was
scored according to the immuno-reactive score (19), based on the percentage of positive
cells: 0 (≤5%), 1 (5–25%), 2 (25–50%), 3 (50–75%) and 4 (≥75%). The
staining intensity was scored as follows: 0 (no staining), 1
(yellowish), 2 (yellow) and 3 (brownish). The NCOA7 immunostaining
score was then calculated as follows: Percentage positive score ×
staining intensity score. A score ≥4 was considered to indicate
positive expression.
Cell culture
T47D and MCF7 cells were cultured in RPMI-1640 (cat.
no. 350-006-CL) and 293T cells were cultured in DMEM [cat. no.
319-005-CL; both from Wisent Biotechnology (Nanjing) Co. Ltd.]
supplemented with 10% FBS (cat. no. 40130ES76; Yeasen Biotechnology
Co., Ltd.), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C
in the presence of 5% CO2 and 90–95% relative
humidity.
Knockdown of NCOA7 expression in
breast cancer cells by lentiviral vector-loaded short hairpin RNAs
(shRNAs)
shRNA of luciferase (shLuc; target sequence:
5′-CGCTGAGTACTTCGAAATGTC-3′) was used as a control. The nucleotide
oligos of two NCOA7 shRNAs (Sangon Biotech Co., Ltd.) were
synthesized and cloned into the lentiviral shRNA expression vector
TETO-pLKO.1-TRC (Addgene, Inc.), in which expression of shRNA was
induced by doxycycline. The shRNA oligos were inserted into the
AgeI/EcoRI sites of the vector (20).
Two NCOA7 shRNA sequences were used as follows:
shNCOA7-1 forward,
5′-CCGGTTGCGCTCTACAATGACATTTCTCGAGAAATGTCATTGTAGAGCGCAATTTTTG-3′
and reverse,
5′-AATTCAAAAATTGCGCTCTACAATGACATTCTCGAGAAATGTCATTGTAGAGCGCAA-3′;
shNCOA7-2 forward,
5′-CCGGCCTGTGAGAAGCAAGATATAACTCGAGTTATATCTTGCTTCTCACAGGTTTTTG-3′
and reverse,
5′-AATTCAAAAACCTGTGAGAAGCAAGATATAACTCGAGTTATATCTTGCTTCTCACAGG-3′.
The lentiviral shRNA plasmid was co-transfected with
psPAX2 (cat. no. 12260; Addgene, Inc.) and pMD2.G (cat. no. 12259;
Addgene, Inc.) in 293T cells for 8–10 h. The transfection medium
was then replaced with 2 ml culture medium. Following 24 h, the
culture medium containing viral particles was collected every day
for three days. The collected medium was cleared by centrifugation
at 1,250 × g for 5 min at 4°C and stored at 4°C for use. To
transfect T47D and MCF7 cells, 6 µg/ml polybrene was added along
with the viral particle-containing medium (multiplicity of
infection, 12) and selected with puromycin. The NCOA7 knockdown
effect was determined by western blot analysis with an anti-NCOA7
antibody (1:500 dilution). Lentiviral construction and infection
were performed as previously described (21).
Preparation of breast cancer cell
lysates, gel electrophoresis and western blot analysis
The cells were washed with precooled PBS, lysed by
adding an appropriate amount of mammalian cell lysis buffer [40 mM
Hepes (pH 7.4), 100 mM NaCl, 1% Triton X-100, 25 mM glycerol
phosphate, 1 mM EDTA, 1 mM sodium orthovanadate, 10 µg/ml leupeptin
and 10 µg/ml aprotinin] and shaken slowly at 4°C for 30 min. The
cell lysates were transferred to an Eppendorf (EP) tube and
centrifuged for 15 min at 12,000 g at 4°C. The cleared lysates were
transferred to a clean EP tube, mixed with 5X SDS buffer and boiled
at 100°C for 8 min. The proteins of the lysates were separated by
10% SDS-PAGE and subsequently transferred to a polyvinylidene
fluoride membrane (cat. no. IPFL00010; EMD Millipore). The
transferred membrane was blocked with 1% BSA for 1 h at room
temperature and incubated with primary antibodies (NCOA7 or
β-actin; 1:500 dilution) at 4°C overnight. Following washing with
1X Tris-buffered saline containing Tween-20, the membrane was
incubated with horseradish peroxidase-conjugated secondary antibody
(anti-mouse; cat. no. 31430; 1:10,000 dilution; Thermo Fisher
Scientific, Inc.) at room temperature for 1–2 h. The protein bands
on the membrane were detected by an electrochemiluminescence
detection kit (cat. no. P0018M; Beyotime Institute of
Biotechnology). Western blot analysis was performed as previously
described (22).
Cell proliferation assay
Cell proliferation was determined by the CCK-8
assay. The control or NCOA7-knockdown T47D cells
(1×104/well) or MCF7 cells (8×103/well) were
seeded in a 96-well plate. Following cell culture for the indicated
durations, the CCK-8 reagent was added. The cells were cultured for
2 h and the cell density was detected by measuring the absorbance
at 450 nm using a microplate reader.
Colony-formation assay
The control or NCOA7-knockdown T47D
(1×103/well) or MCF7 cells (0.5×103/well)
were seeded in a 6-well culture plate. Following culture for 14
days, the cell colonies were fixed with 4% paraformaldehyde and
stained with 0.1% crystal violet solution for 8 min at room
temperature. Following washing three times with PBS, the stained
cell colonies were counted.
Wound-healing assay
Cell migration was determined by the wound-healing
assay. T47D cells (6×105/well) or MCF7 cells
(5×105/well) were seeded in a 12-well plate. Following
culture for 12 h, a straight scratch line was performed at the
center area of the cell monolayer using a pipette tip. The cell
layers were then rinsed to remove any detached cells and debris.
The cells were then cultured with fresh DMEM. Images of the scratch
line were captured following cell culture for 24 or 48 h after the
scratch line was created under a phase-contrast microscope. The
migrated area of the cells was calculated from the images using the
imaging software Image J (version 1.53e; National Institutes of
Health).
Transwell assay
Transwell chambers (cat. no. 3422; Corning, Inc.)
were used for migration assays. The cells (2×105
cells/ml) in 200 µl serum-free DMEM were seeded in each top well of
the chamber. DMEM with a migration attractant (10% FBS) was added
to the bottom well of the chamber. Following incubation under
normal culture conditions for 24 h, the cells on the top side of
the membrane between the top and the bottom chambers were carefully
removed and the cells that had migrated to the bottom side of the
membrane were fixed with 4% paraformaldehyde for 30 min and stained
with 0.1% crystal violet solution for 8 min at room temperature.
The stained cells were washed with PBS three times, visualized
under a phase-contrast microscope and counted. Cell proliferation
and migration assays were performed as previously described
(20).
Statistical analysis
SPSS 22.0 statistical software (IBM Corp.) was used
for statistical analysis of the data. The association of NCOA7
expression with the survival of patients with breast cancer was
statistically analyzed by Kaplan-Meier (K-M) survival analysis with
the log-rank test. An independent-samples t-test was used to assess
differences between two groups. Differences between multiple groups
were analyzed using one-way ANOVA followed by Tukey's honestly
significant difference post-hoc test. Data are expressed as the
mean ± standard error of the mean. The χ2 test was used
to analyze the clinicopathological data of the patients in Table I, Table
II, Table III and SII. P≤0.05 was considered to indicate a
statistically significant difference.
 | Table II.Association of NCOA7 expression with
clinicopathological parameters in triple-negative breast cancer
tumor samples. |
Table II.
Association of NCOA7 expression with
clinicopathological parameters in triple-negative breast cancer
tumor samples.
|
|
| NCOA7 status, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Pathological
parameter | Total | Positive | Negative | P-value |
|---|
| Age, years |
|
|
| 0.683 |
|
≤50 | 35 | 17 (49) | 18 (51) |
|
|
>50 | 72 | 38 (53) | 34 (47) |
|
| Tumor size, cm |
|
|
| 0.060 |
| ≤3 | 73 | 33 (45) | 40 (55) |
|
|
>3 | 34 | 22 (65) | 12 (35) |
|
| T stage |
|
|
| 0.104 |
| T1 | 39 | 16 (41) | 23 (59) |
|
|
T2/T3 | 68 | 39 (57) | 29 (43) |
|
| N stage |
|
|
| 0.032 |
| N0 | 65 | 28 (43) | 37 (57) |
|
|
N1-3 | 42 | 27 (64) | 15 (36) |
|
| TNM stage |
|
|
| 0.674 |
|
I/II | 91 | 46 (51) | 45 (49) |
|
|
IIIA/B | 16 | 9 (56) | 7 (44) |
|
 | Table III.Association of NCOA7 expression with
clinicopathological parameters in non-triple negative breast cancer
tumor samples. |
Table III.
Association of NCOA7 expression with
clinicopathological parameters in non-triple negative breast cancer
tumor samples.
|
|
| NCOA7 status, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Pathological
parameter | Total | Positive | Negative | P-value |
|---|
| Age, years |
|
|
| 0.648 |
|
≤50 | 56 | 23 (41) | 33 (59) |
|
|
>50 | 78 | 29 (37) | 49 (63) |
|
| Tumor size, cm |
|
|
| 0.567 |
| ≤3 | 121 | 46 (38) | 75 (62) |
|
|
>3 | 13 | 6 (46) | 7 (54) |
|
| T stage |
|
|
| 0.077 |
| T1 | 87 | 29 (33) | 58 (67) |
|
|
T2/T3 | 47 | 23 (49) | 24 (51) |
|
| N stage |
|
|
| 0.138 |
| N0 | 90 | 31 (34) | 59 (66) |
|
|
N1-3 | 44 | 21 (48) | 23 (52) |
|
| TNM stage |
|
|
| 0.601 |
|
I/II | 122 | 46 (38) | 76 (62) |
|
|
IIIA/B | 12 | 6 (50) | 6 (50) |
|
Results
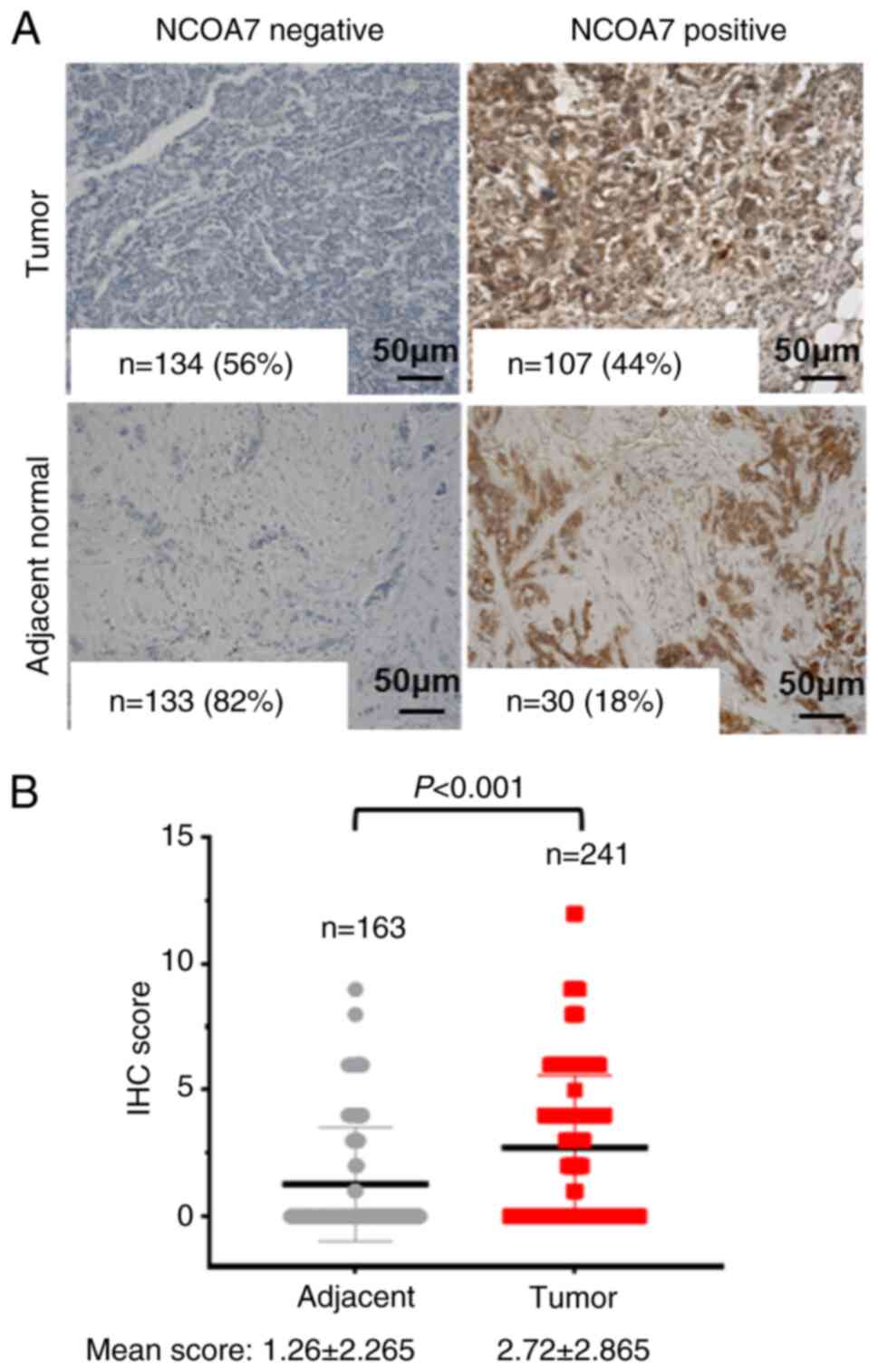
NCOA7 is overexpressed in breast tumor
tissues
To address the role of NCOA7 in breast cancer
progression, its expression was examined in a TMA containing 241
breast tumor tissues and 163 normal tissue samples (adjacent to the
tumor tissue) by IHC staining with an anti-NCOA7 antibody. The
results indicated that NCOA7 was expressed in 107 out of 241 breast
cancer tumor samples (44%) and in only 30 out of 163 adjacent
normal tissue samples (18%; Fig.
1A). The mean score of the IHC staining in the breast tumor
samples was 2.72±2.865, while in the normal tissue samples, it was
1.26±2.265 (Fig. 1B). Statistical
analysis of the difference in expression of NCOA7 between the
breast tumor and the adjacent normal tissue samples indicated that
the expression of NCOA7 in breast tumor tissues was significantly
higher than that in the adjacent normal tissues (P<0.001;
Fig. 1B).
Expression of NCOA7 is positively
associated with tumor size, N-stage and T-stage of breast
cancer
Subsequently, the association of the expression of
NCOA7 with the clinicopathological parameters was determined by
statistical analysis of the TMA data. As presented in Table I, the expression of NCOA7 was
positively associated with the tumor size (P=0.020), T-stage
(P=0.005) and N-stage (P=0.008). However, no significant
association was observed for the age of the patients (P=0.914) and
the TNM stage (P=0.299). Among the samples of TNBC, the expression
levels of NCOA7 were significantly associated with the N stage
(P=0.032), while they were not associated with any other parameters
(Table II). However, the
expression levels of NCOA7 in samples of N-TNBC did not exhibit any
significant association with any of the clinicopathological
parameters examined (Table
III).
Furthermore, the expression levels of NCOA7 were
compared in each pathological parameter of TNBC and N-TNBC. As
shown in Table SII, the expression
of NCOA7 was significantly higher in patients with TNBC than in
patients with N-TNBC who were aged >50 years (P=0.023), with a
tumor size of >3 cm (P=0.007) and those who had lymph node
metastasis stages N1-3 (P=0.023). These data suggested that the
expression levels of NCOA7 were preferentially associated with the
pathological parameters related to advanced breast cancer in TNBC
compared with N-TNBC.
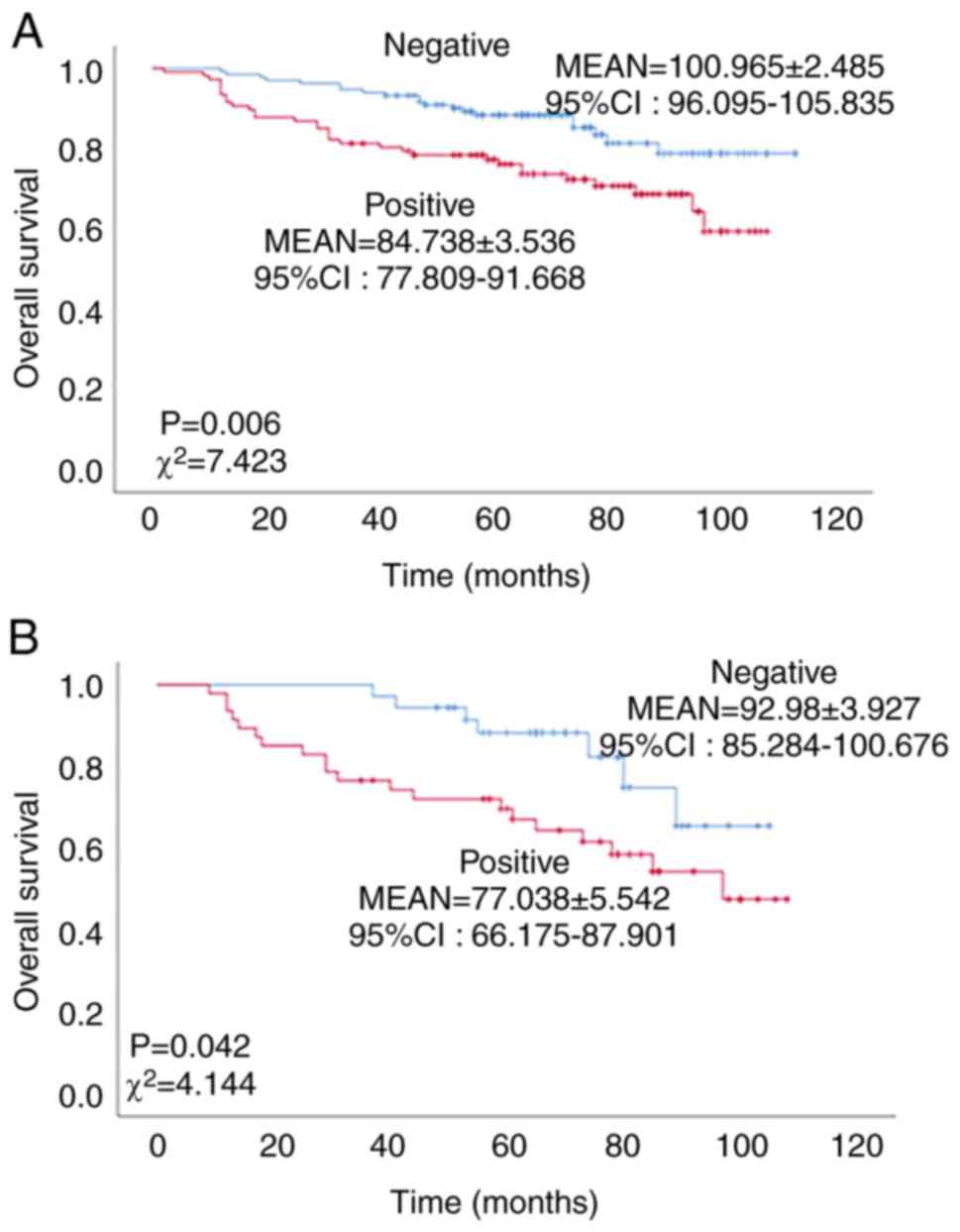
Expression of NCOA7 is inversely
associated with overall survival of patients with breast
cancer
The association between the expression of NCOA7 and
the overall survival of patients with breast cancer was determined
by K-M analysis. As shown in Fig.
2A, the overall survival of the NCOA7-positive patients with
breast cancer was significantly lower than that of the
NCOA7-negative patients (χ2=7.423, P=0.006). The mean
survival time of NCOA7-positive patients with breast cancer was
84.738±3.536 months, while that of the NCOA7-negative patients was
100.965±2.485 months. These data indicate that the expression of
NCOA7 is inversely associated with overall survival of patients
with breast cancer. To determine the role of NCOA7 in breast cancer
metastasis, the association of NCOA7 expression with the overall
survival of patients with lymph node metastasis (N1-3; total cases,
n=86) was statistically analyzed. As shown in Fig. 2B, the expression of NCOA7 in
patients with stage N1-3 was significantly associated with lower
overall survival (P=0.042, χ2=4.144). The mean survival
time of NCOA7-positive N1-3 patients was 77.038±5.542 [95% CI:
66.175–87.901] months, while in NCOA7-negative N1-3 patients, it
was 92.980±3.927 [95% CI: 85.284–100.676] months.
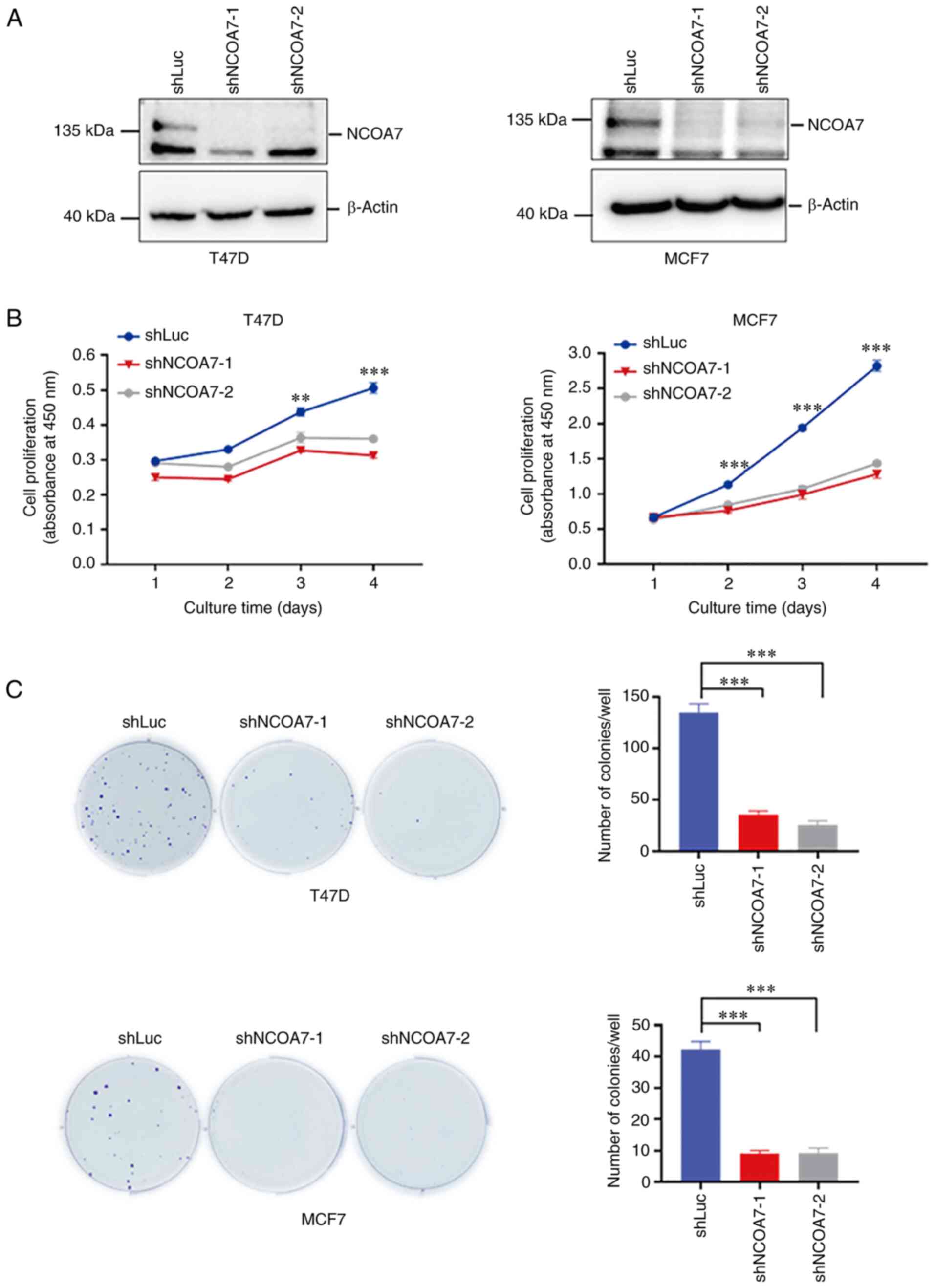
Knockdown of NCOA7 expression inhibits
breast cancer-cell proliferation
To confirm the role of NCOA7 in promoting breast
cancer progression (noted in the breast tumor IHC staining
analysis), the effect of knockdown of NCOA7 expression on the
proliferation of breast cancer T47D and MCF7 cells was examined.
The expression of NCOA7 was knocked down in the breast cancer cell
lines T47D and MCF7 by the lentiviral vector loaded with NCOA7
shRNAs along with the control shRNA-transfected cell line that
expressed a luciferase shRNA (shLuc) (Fig. 3A). The shNCOA7s were able to deplete
~90% of NCOA7 in both T47D and MCF7 cells (Fig. 3A). CCK-8 and colony-formation assays
were performed to determine the effect of knockdown of NCOA7
expression on the proliferation in these cell lines. As shown in
Fig. 3B, knockdown of NCOA7
expression by both shNCOA7s consistently inhibited ~80% of cell
proliferation following 4 days of culture in both T47D and MCF7
cells, as determined by the CCK-8 assay. However, in the
colony-formation assay, knockdown of NCOA7 expression eliminated
~80% of the colony formation in both T47D and MCF7 cells (Fig. 3C), which was consistent with the
results obtained by the CCK-8 assay.
Knockdown of NCOA7 expression severely
impairs breast cancer cell migration
To examine the role of NCOA7 in promoting breast
cancer metastasis, the effect of NCOA7 on breast cancer cell
migration was determined using the wound-healing and Transwell
assays. As shown in Fig. 4A,
knockdown of NCOA7 expression by both shNCOA7s reduced the
migration rate of T47D and MCF7 cells by ~50%, as determined by the
wound-healing assay. Consistent with this, the Transwell assay
indicated that knockdown of NCOA7 expression by both shNCOA7s
inhibited ~40-50% of cell migration in MCF7 cells, while this
percentage was somewhat higher in T47D cells (~60-80%; Fig. 4B).
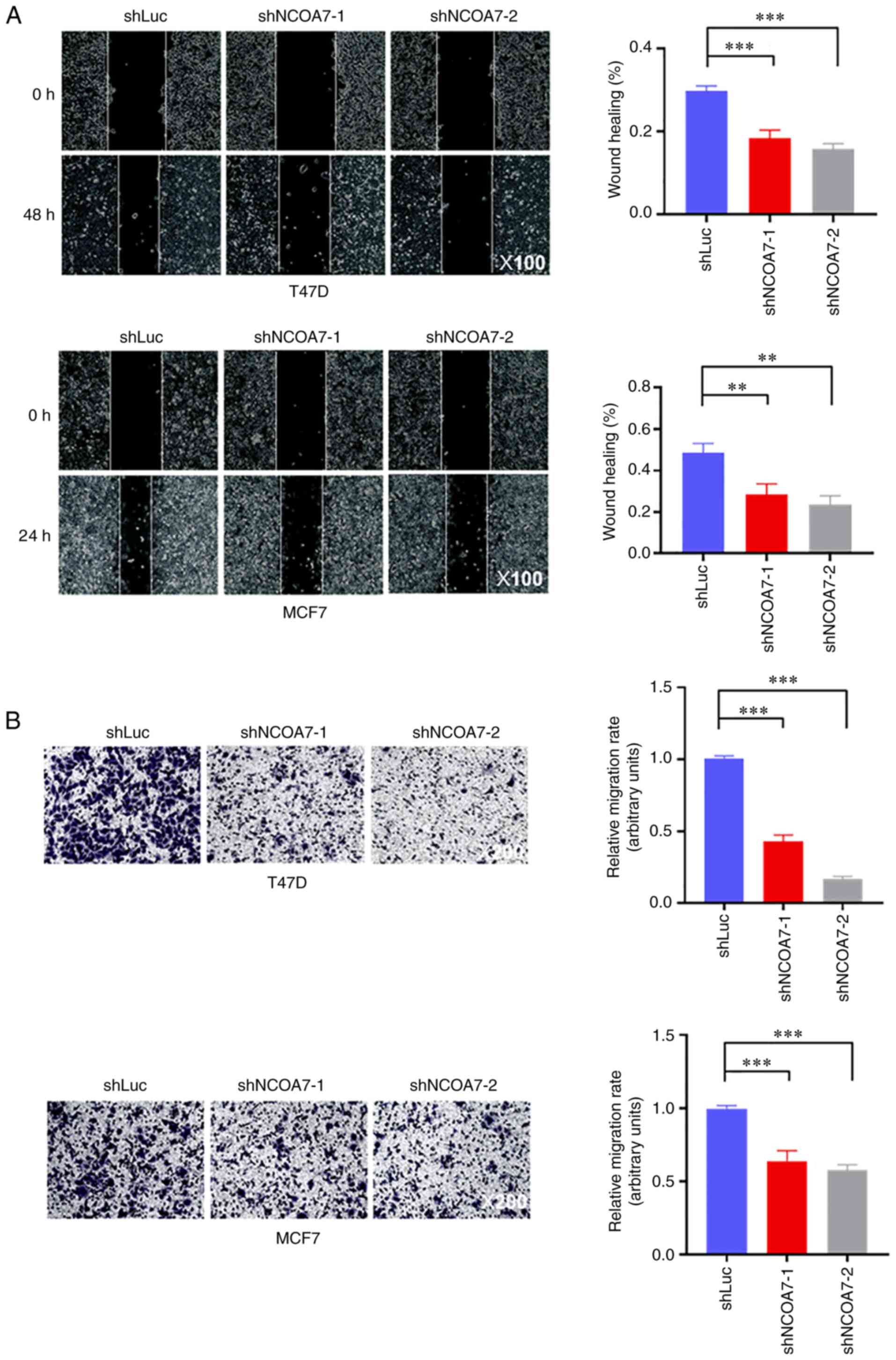 | Figure 4.Knockdown of NCOA7 expression in
breast cancer T47D and MCF7 cells inhibits their migration. (A)
Cell migration was detected by the wound-healing assay. Left
panels, representative images of wound healing of confluent control
and knockdown cell line layers (magnification, ×100). Right panels,
quantification of cell migration by the wound-healing assay with
T47D and MCF7 cells. The ratio of the wound-healing area at 24 or
48 h to the scratch area at 0 h was presented. (B) Cell migration
was detected by the Transwell assay. Left panels, representative
crystal violet staining images of migrated cells on the bottom side
of the membrane in the Transwell assays (magnification, ×200).
Right panels, quantification of cell migration by the Transwell
assay with T47D and MCF7 cells. **P<0.01, ***P<0.001. NCOA7,
nuclear receptor coactivator 7; sh, short hairpin RNA; Luc,
luciferase (control). |
Discussion
Establishing prognostic biomarker molecules for
patients with postoperative breast cancer is important for the
treatment of patients with breast cancer, notably for those with
advanced stages of the disease (23,24).
In the present study, it was shown that the nuclear receptor
(particularly ER) co-activator NCOA7 was overexpressed in breast
tumors and its expression was reversely associated with the overall
survival of patients with breast cancer. Studies using IHC staining
of NCOA7 in breast tumor tissue samples indicated that its
expression was associated with tumor size and lymph node metastasis
of breast cancer. The data from the breast cancer cell studies
(knockdown of NCOA7 expression) indicated that diminishing NCOA7
expression in breast cancer cells inhibited cell proliferation and
migration. This confirmed the results obtained from the breast
tumor IHC staining studies indicating that the expression levels of
NCOA7 were associated with breast tumor growth and metastasis.
Therefore, it is proposed that NCOA7 is a prognostic biomarker
associated with poor survival of patients with breast cancer.
Furthermore, NCOA7 may be considered to be a driver protein for
breast tumor growth and metastasis and a potential therapeutic
target for breast cancer, notably for advanced breast cancer.
Currently, the molecular mechanism by which NCOA7
promotes breast cancer progression remains to be elucidated.
Previous studies have shown that NCOA7 interacts with ERα and
potentially functions as an ERα co-activator (17). It was initially expected that NCOA7
may promote breast cancer progression via ER signaling. However,
the results of the present study indicate that the expression
levels of NCOA7 are preferentially associated with the
clinicopathological parameters of TNBC, suggesting that the
oncogenic effect of NCOA7 on breast cancer, at least in TNBC, may
not be mediated via ER signaling. Notably, in addition to a
potential co-activator of ER, NCOA7 may function as an
anti-oxidation and vesicle trafficking protein. NCOA7 contains the
TLDc domain that is the signature domain of members of the
anti-oxidation OXR1 family of proteins (7). Therefore, NCOA7 may exert an
anti-oxidative function and protect breast cancer cells from the
damage of oxidative stress. However, limited experimental data have
shown the role of NCOA7 in oxidation resistance or have examined
the TLDc domain required for NCOA7 to promote breast cancer cell
proliferation or migration. Recent studies discovered that the TLDc
domain of NCOA7 interacted with [H+]-V-ATPase and
regulated vesicle acidification (9,10).
NCOA7 may utilize its function in vesicle trafficking to regulate
intracellular oncogenic signaling, particularly endocytotic
receptor signaling or secretory signaling. Given these known
molecular interactions of NCOA7, it is speculated that it promotes
breast cancer progression through a complex regulatory network, not
just its ER co-activator activity. To fully understand the role of
NCOA7 in breast cancer progression, notably in TNBC progression,
additional investigation is required on the functions of NCOA7
other than those required for activation of ER transcription. More
importantly, its functions for anti-oxidation and vesicle
trafficking in breast cancer cells must be examined.
Therefore, future studies will focus on the role of
NCOA7 in regulating ER activation, anti-oxidation and vesicle
trafficking processes. This will explore the association of
NCOA7-mediated ER regulation, anti-oxidation and/or vesicle
trafficking signaling pathways with breast tumor growth and
metastasis. By performing these studies, it is expected that the
molecular mechanism underlying the promoting effect of NCOA7 on
breast cancer progression will be revealed. In addition, NCOA7 will
be established as a poor prognostic biomarker and a target molecule
for breast cancer therapy.
The nuclear receptor co-activator protein NCOA7 is a
potential prognostic biomarker of breast cancer, a possible driver
protein for breast cancer progression and a potential target for
anti-metastatic therapy for advanced breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Aiqin Sun
(Jiangsu University School of Medicine, Zhenjiang, China) for their
helpful comments on this research.
Funding
This work is supported by National Natural Science Foundation of
China (grant nos. 82172942 and 81871888 and 81472558), the Clinical
Research Project of the Jiangsu University Affiliated People's
Hospital (grant no. Y2022019) and Medical Education Collaborative
Innovation Fund of Jiangsu University (grant no JDYY2023018).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
QL designed the study; ZP and YL performed the
research; SX, QD, LY and MC were involved in the acquisition of
data; ZP, YL and WY analyzed the data; ZP and YL were involved in
writing-original draft preparation; QL, WY and GS were involved in
writing-review and editing; QL supervised the study; QL and WY
acquired funding. ZP and YL confirm the authenticity of all the raw
data. All authors have read and agreed to the published version of
the manuscript.
Ethics approval and consent to
participate
All tissue specimens were collected after written
informed consent was obtained from each subject for this study and
approved for use by the Institutional Review Board of Jiangsu
University (Zhenjiang, China; approval no. LLYW20210004),
affiliated with the People's Hospital. All experiments with the
human tissue samples conform to the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Houghton SC and Hankinson SE: Cancer
progress and priorities: Breast cancer. Cancer Epidemiol Biomarkers
Prev. 30:822–844. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fahad Ullah M: Breast cancer: Current
perspectives on the disease status. Adv Exp Med Biol. 1152:51–64.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taskindoust M, Thomas SM, Sammons SL,
Fayanju OM, DiLalla G, Hwang ES and Plichta JK: Survival outcomes
among patients with metastatic breast cancer: Review of 47,000
patients. Ann Surg Oncol. 28:7441–7449. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, Shao X, Huang J, Lei L, Huang Y,
Zheng Y, Cao W and Chen Z: Exploring the concepts and practices of
advanced breast cancer treatment: A narrative review. Ann Transl
Med. 9:7212021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shao W, Halachmi S and Brown M: ERAP140, a
conserved tissue-specific nuclear receptor coactivator. Mol Cell
Biol. 22:3358–3372. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Durand M, Kolpak A, Farrell T, Elliott NA,
Shao W, Brown M and Volkert MR: The OXR domain defines a conserved
family of eukaryotic oxidation resistance proteins. BMC Cell Biol.
8:132007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Finelli MJ, Sanchez-Pulido L, Liu KX,
Davies KE and Oliver PL: The evolutionarily conserved
Tre2/Bub2/Cdc16 (TBC), lysin motif (LysM), domain catalytic (TLDc)
domain is neuroprotective against oxidative stress. J Biol Chem.
291:2751–2763. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eaton AF, Brown D and Merkulova M: The
evolutionary conserved TLDc domain defines a new class of
(H+)V-ATPase interacting proteins. Sci Rep. 11:226542021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Castroflorio E, den Hoed J, Svistunova D,
Finelli MJ, Cebrian-Serrano A, Corrochano S, Bassett AR, Davies B
and Oliver PL: The Ncoa7 locus regulates V-ATPase formation and
function, neurodevelopment and behaviour. Cell Mol Life Sci.
78:3503–3524. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khan H, Winstone H, Jimenez-Guardeño JM,
Graham C, Doores KJ, Goujon C, Matthews DA, Davidson AD, Rihn SJ,
Palmarini M, et al: TMPRSS2 promotes SARS-CoV-2 evasion from
NCOA7-mediated restriction. PLoS Pathog. 17:e10098202021.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Doyle T, Moncorgé O, Bonaventure B,
Pollpeter D, Lussignol M, Tauziet M, Apolonia L, Catanese MT,
Goujon C and Malim MH: The interferon-inducible isoform of NCOA7
inhibits endosome-mediated viral entry. Nat Microbiol. 3:1369–1376.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guo J, Ke S, Chen Q, Zhou J, Guo J and Qiu
T: NCOA7 regulates growth and metastasis of clear cell renal cell
carcinoma via MAPK/ERK signaling pathway. Int J Mol Sci.
24:115842023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arai H, Ozaki T, Niizuma H, Nakamura Y,
Ohira M, Takano K, Matsumoto M and Nakagawara A: ERAP140/Nbla10993
is a novel favorable prognostic indicator for neuroblastoma induced
in response to retinoic acid. Oncol Rep. 19:1381–1388.
2008.PubMed/NCBI
|
|
15
|
Lu J, Annunziata F, Sirvinskas D, Omrani
O, Li H, Rasa SMM, Krepelova A, Adam L and Neri F: Establishment
and evaluation of module-based immune-associated gene signature to
predict overall survival in patients of colon adenocarcinoma. J
Biomed Sci. 29:812022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie X, Jiang Y, Yuan Y, Wang P, Li X, Chen
F, Sun C, Zhao H, Zeng X, Jiang L, et al: MALDI imaging reveals
NCOA7 as a potential biomarker in oral squamous cell carcinoma
arising from oral submucous fibrosis. Oncotarget. 7:59987–60004.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Higginbotham KSP, Breyer JP, Bradley KM,
Schuyler PA, Plummer WD Jr, Freudenthal ME, Trentham-Dietz A,
Newcomb PA, Sanders ME, Page DL, et al: A multistage association
study identifies a breast cancer genetic locus at NCOA7. Cancer
Res. 71:3881–3888. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Wu T, Peng Z, Tian X, Dai Q, Chen M,
Zhu J, Xia S, Sun A, Yang W and Lin Q: ETS1 is a prognostic
biomarker of triple-negative breast cancer and promotes the
triple-negative breast cancer progression through the YAP
signaling. Am J Cancer Res. 12:5074–5084. 2022.PubMed/NCBI
|
|
19
|
Umemura S, Kurosumi M, Moriya T, Oyama T,
Arihiro K, Yamashita H, Umekita Y, Komoike Y, Shimizu C, Fukushima
H, et al: Immunohistochemical evaluation for hormone receptors in
breast cancer: A practically useful evaluation system and handling
protocol. Breast Cancer. 13:2322006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun A, Tian X, Yang W and Lin Q:
Overexpression of SCYL1 is associated with progression of breast
cancer. Curr Oncol. 29:6922–6932. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu J, Peng Z, Tian X, Wu T, Sun A, Yang W
and Lin Q: Activation of E3 ubiquitin ligase WWP2 by non-receptor
tyrosine kinase ACK1. IUBMB Life. 75:595–608. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng K, Sun A, Zhu J, Gao J, Li Y, Shao G,
Yang W and Lin Q: Restoration of the ATG5-dependent autophagy
sensitizes DU145 prostate cancer cells to chemotherapeutic drugs.
Oncol Lett. 22:6382021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saez RA, McGuire WL and Clark GM:
Prognostic factors in breast cancer. Semin Surg Oncol. 5:102–110.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Esteva FJ, Sahin AA, Cristofanilli M, Arun
B and Hortobagyi GN: Molecular prognostic factors for breast cancer
metastasis and survival. Semin Radiat Oncol. 12:319–328. 2002.
View Article : Google Scholar : PubMed/NCBI
|