Introduction
As the second most frequent cancer in the world,
hepatocellular carcinoma (HCC) causes 788,000 deaths each year
(1). In addition, HCC is also the
main cause of tumor death in China. At the time of diagnosis, most
HCC patients are at more advanced stages, which are accompanied by
intrahepatic and extrahepatic metastases, and the prognosis is
poor. At present, there is a lack of effective prevention and
treatment measures clinically. The 5-year metastasis and recurrence
rate after radical resection is as high as 61.5% (2). Recurrence and metastasis of tumors are
still the main causes of treatment failure in HCC, causing a heavy
burden on families and society (3).
Therefore, it is urgent to elucidate the mechanism of recurrence
and metastasis, and provide new treatment strategies for HCC.
MicroRNAs (miRNAs) are single-stranded noncoding
small RNA with a length of 21–23 nt (4). When miRNAs are loaded into the
RNA-induced silencing complex (RISC), they can play a role in
regulating gene expression. Through their seed sequence (5-terminal
2–8 nucleotides), miRNAs can identify binding sites on target gene
mRNA 3′-UTRs and carry recruit RISC to play its role, which results
in transcriptional inhibition, cleavage and degradation of Mrna
(5). It has been reported that
miRNA dysfunction is closely related to the invasion and metastasis
of tumors (6,7). In HCC, miR-424-5p was revealed to
inhibit TRIM29 expression, and then regulate HCC cell proliferation
and invasion by inhibiting gene expression of proliferation- and
apoptosis-related indicators (8). A
recent study revealed that miR-505 inhibited the malignant
development of non-small cell lung cancer via the MAP3K3-mediated
AKT-NFκB signaling pathway (9). In
gastric cancer, miR-125a inhibited the invasion and migration of
tumor cells by regulating the expression of HAS1 by targeting STAT3
(10). Recent studies have revealed
that miR-29c-3p is abnormally expressed in many malignant tumors
including gastric cancer (11),
colon cancer (12) and pancreatic
cancer (13). A previous study
revealed that miR-29c-3p was expressed at low levels in laryngeal
squamous cell carcinoma (14). It
was reported that miR-29c-3p overexpression led to decreased
migration of GC cells in vitro and in vivo by
suppressing the expression of KIAA1199 and several key proteins in
the Wnt/β-catenin and EGFR signaling pathways (15). miR-29c-3p regulates CRC cell
proliferation and migration by regulating SPARC expression
(16). A study also revealed that
miR-29c-3p promoted the malignant development of HCC by regulating
the methylation of LATS1 caused by DNMT3B and inhibiting the
anticancer function of the Hippo signaling pathway (17). However, a single miRNA can regulate
the expression of hundreds of target gene mRNAs after
transcription. Therefore, the specific roles and molecular
mechanisms of miR-29c-3p in HCC have not been fully elucidated
(18,19).
In the present study, the expression of miR-29c-3p
in HCC was revealed to be significantly decreased, and its low
expression was closely related to the poor prognosis of HCC
patients. Overexpression of miR-29c-3p could significantly inhibit
the proliferation and migration of HCC cells. It was also confirmed
that miR-29c-3p could inhibit the malignant progression of HCC by
directly acting on tripartite motif containing 31 (TRIM31) to
regulate tumor proliferation and migration-related factors.
Materials and methods
HCC patients and specimens
A total of 60 HCC tissue samples were collected in
this study, including tumor tissues and paired normal adjacent
tissues, which were collected from January 2006 to July 2011 at the
West China Hospital of Sichuan University and sample collection
used liquid nitrogen for preservation. The histological diagnosis
of all HCC samples was performed independently by two pathologists.
All patients signed an informed consent form. The present study was
approved by the Ethics Committee of West China Hospital, Sichuan
University.
Cell culture and transfection
The liver cancer cells (HepG2 and MHCC-97H), which
were assessed by STR profiling, used in the present study were
obtained from the Institute of Biochemistry and Cell Biology
(Chinese Academy of Sciences). All cell lines were cultured with
high-glucose DMEM containing 10% FBS (Hyclone; GE Healthcare Life
Sciences) and 1% penicillin/streptomycin.
miR-29c-3p mimic (miR-29c-3p), miR-29c-3p inhibitor
and miR-29c-3p control were obtained from Guangzhou RiboBio Co.,
Ltd. The TRIM31 overexpression vector pcDNA-TRIM31 and empty
control vector pcDNA were constructed by Shanghai GenePharma Co.,
Ltd. Lipofectamine 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for liver cancer cell (HepG2 and
MHCC-97H) transfection according to the manufacturer's
instructions.
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total RNA was extracted from HCC tissue samples and
cell lines using TRIzol reagent (Takara Bio, Inc.) according to the
manufacturer's protocol. miR-29c-3p expression was determined by a
TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Total RNA was reverse-transcribed into cDNA
using PrimeScript RT Reagent (Takara Bio, Inc.). qRT-PCR was
performed using SYBR Premix Ex Taq II (Takara Bio, Inc.). The
temperature protocol for qRT-PCR was as follows: 35°C for 5 min,
followed by 45°C for 40 min and 70°C for 5 min. U6 and GAPDH were
used as internal references. The sequences of the primers used for
each gene are presented in Table
SI. The mRNA expression of miR-29c-3p and TRIM31 was determined
using the 2−ΔΔCq method (20).
Western blotting
Total protein was extracted using RIPA lysis buffer
(Beyotime Institute of Biotechnology). Protein was quantified using
the Bradford protein assay (Bio-Rad Laboratories, Inc.) with a
NanoDrop spectrophotometer. A total of 25 µg/well of protein was
electrophoresed on 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE). After the transfer was completed, the
PVDF membranes were blocked with 5% non-fat powdered milk at 37°C
for 1 h. Next, the membranes were incubated with anti-TRIM31
(1:2,000; ab98207; Abcam) and anti-β-actin antibody (1:5,000;
ab179467; Abcam) at 4°C overnight. Subsequently, the PVDF membranes
were incubated with horseradish peroxidase-conjugated secondary
antibody (1:5,000; ab6721; Abcam) at room temperature for 1 h.
β-Actin was used as an internal reference. Finally, enhanced
chemiluminescence (ECL) (Thermo Fisher Scientific, Inc.) was used
to detect the expression of the target proteins. Quantity One
software v4.6.5 (Bio-Rad Laboratories, Inc.) was used for
densitometry, and the values are expressed relative to β-actin.
Cell Counting Kit-8 (CCK-8) assay
The transfected cells were inoculated into 96-well
plates. After adding 10 µl/well of CCK-8 solution (Dojindo
Molecular Technologies, Inc.), the absorption was determined at 450
nm by microplate spectrophotometer. The OD values at 450 nm were
detected at 0, 6, 12, 24, 48 and 72 h according to the
manufacturer's instructions.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
In brief, 5×103 cells/wells were plated
in 96-well plates and cultured for 24 h. Next, 4% ice formaldehyde
was added for cell fixation for 30 min at room temperature, and the
cells were permeabilized with 0.5% Triton X-100 solution for 20
min. Cells were incubated with EdU (50 µM) for 2 h at 37°C. After
washing the cells three times with PBS, 1X ApolloR reaction
cocktail (100 µl) was added, and the reaction proceeded for 30 min
at 37°C in the dark. Subsequently, 1X Hoechst 33342 (100 µl) was
used to stain the cell nuclei for 30 min at room temperature. Cell
proliferation was analyzed using the mean number of the cells in
each sample using a fluorescence microscope (Lionheart; BioTek
Instruments, Inc.; magnification, ×100).
Wound healing assay
The cells (1×105 cells/wells) were seeded
into 6 empty plates, and after the cells were confluent (80–90%), 3
lines were scratched with a 1 ml pipette tip on the plate wells.
Non-adherent cells were washed off with PBS. After 0, 24 and 48 h,
the distance between the wound edges was measured. Cells were
examined under a Leica light microscope (Olympus Corp.;
magnification, ×40).
Cell migration assay
After 48 h of cell transfection, the cells were
collected in a cell suspension and added to the upper chamber
(Corning, Inc.) with 200 µl of serum-free medium. Next, 500 µl of
medium containing 10% FBS was added to the lower chamber. After 24
h of culture, the non-migrating cells were wiped off with a cotton
swab. The cells were fixed with 4% formaldehyde and then stained
with 0.1% crystal violet, both at room temperature. The cells were
washed three times with PBS and counted under a microscope.
Luciferase reporter assay
miRanda (http://www.microrna.org/microrna/home.do) and
TargetScan (http://www.targetscan.org) were used
to identify downstream target genes of miR-29c-3p. The wild-type
TRIM31-3′UTR (WT) and mutant TRIM31-3′UTR (MUT) containing the
putative binding site of miR-29c-3p were amplified by Shanghai
GenePharma Co., Ltd., and cloned into the firefly
luciferase-expressing pMIR-REPORT vector (OBiO Technology Corp.,
Ltd.). The luciferase reporter vector and miR-29c-3p mimic or
miR-NC were transiently co-transfected using Lipofectamine 3000.
After transfection for 24 h, luciferase assays were performed using
the Luciferase Reporter Assay System (GloMax) according to the
manufacturer's protocol.
Mouse xenograft tumor model
BALB/c-nu mice (age, 5 weeks; sex, male; weight,
20–22 g) were purchased from Shanghai Experimental Animal Center
and housed in a sterile room at the Experimental Animal Center of
West China Hospital of Sichuan University at 25°C and 40–70%
humidity, with a 12-h light/dark cycle and free access to food and
water. All animal experiments were performed in accordance with the
institutional guidelines, and the method of euthanasia was cervical
dislocation (when the heart stopped completely, the mouse was
determined as dead). Body weight loss >20% was assumed to be a
humane endpoint for euthanasia. After transfecting the miR-29c-3p
mimic into liver cancer cells (HepG2 and MHCC-97H) and culturing
for 48 h, the transfected liver cancer cells (HepG2 and MHCC-97H)
(5×106) were subcutaneously injected into the left hip
flanks of the mice. The tumor volume was calculated according to
the following formula: Volume = (length × width2)/2.
Tumor sizes (the length and width of tumor nodules) were measured
every 5 days. All animal experiments were approved by the Animal
Care Ethics Review Committee of West China Hospital of Sichuan
University. Then, 50 days after injection, the mice were
sacrificed, and tumors were collected for analysis. The tumor
experiments ended when tumor diameters were <20 mm (the maximum
tumor volume was 397 mm3).
Statistical analysis
All data are presented as the means ± standard
deviation (SD). Statistical analysis of data was performed using
GraphPad Prism version 6.0 (GraphPad Software, Inc.) or SPSS 20.0
software (IBM Corp.). Statistical differences were analyzed by
Student's t-test, while the significance of differences between
multiple groups was determined by one-way analysis of variance
(ANOVA), followed by the Newman-Keuls test, and repeated measures
ANOVA. The Kaplan-Meier method was used to assess disease-specific
survival (DSS) and recurrence-free survival (RFS), the log-rank
test and chi-squared analysis were used to analyze the differences
between the curves. Univariate and multivariate Cox regression
analyses were carried out to determine the prognostic significance
of miR-29c-3p and TRIM31 expression. The correlation between
miR-29c-3p and TRIM31 expression was evaluated by Spearman's
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-29c-3p is reduced in
HCC tissues and the low expression of miR-29c-3p is closely related
to poor prognosis in HCC
To study the expression of miR-29c-3p in HCC,
qRT-PCR analysis of 60 HCC and paired normal samples was performed.
The results revealed that the expression of miR-29c-3p was
downregulated in the HCC tissues compared with the paired normal
samples (Fig. 1A).
Through the analysis of clinical prognosis in HCC,
it was revealed that HCC patients with low miR-29c-3p expression
had significantly shorter DSS than those with high miR-29c-3p
expression (Fig. 1B). Moreover, HCC
patients with low miR-29c-3p expression had significantly shorter
RFS than those with high miR-29c-3p expression (Fig. 1C).
In addition, miR-29c-3p expression was positively
correlated with tumor size (P=0.013), TNM stage (P=0.003) and
multiplicity (P=0.047) (Table I).
By univariate analysis, TNM stage (P=0.014), tumor size (P=0.009),
multiplicity (P=0.046) and miR-29c-3p (P=0.012) were significantly
associated with DSS, and TNM stage (P=0.010), tumor size (P=0.005),
multiplicity (P=0.034) and miR-29c-3p (P=0.019) were significantly
associated with RFS (Tables II and
III). The multivariate model
revealed that DSS was significantly dependent on TNM stage
(P=0.020), tumor size (P=0.016) multiplicity (P=0.035) and
miR-29c-3p (P=0.017), while TNM stage (P=0.015), tumor size
(P=0.013), multiplicity (P=0.027) and miR-29c-3p (P=0.014) were
significantly associated with RFS (Tables II and III), which indicated that miR-29c-3p was
an independent prognostic factor for DSS and RFS in patients with
HCC.
 | Table I.Correlations between miR-29c-3p and
clinicopathological features of HCC patients (n=60). |
Table I.
Correlations between miR-29c-3p and
clinicopathological features of HCC patients (n=60).
|
|
| miR-29c-3p
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. of cases | Low (n=39) | High (n=21) | P-value |
|---|
| Age (years) |
|
|
| 0.515 |
|
<50 | 28 | 17 | 11 |
|
|
≥50 | 32 | 22 | 10 |
|
| Sex |
|
|
| 0.548 |
|
Male | 34 | 21 | 13 |
|
|
Female | 26 | 18 | 8 |
|
| Tumor size
(cm) |
|
|
| 0.013 |
| ≤5 | 27 | 13 | 14 |
|
|
>5 | 33 | 26 | 7 |
|
| AFP (ng/ml) |
|
|
| 0.399 |
|
≤20 | 27 | 16 | 11 |
|
|
>20 | 33 | 23 | 10 |
|
| TNM stage |
|
|
| 0.003 |
|
I/II | 27 | 12 | 15 |
|
|
III/IV | 33 | 27 | 6 |
|
| Liver
cirrhosis |
|
|
| 0.664 |
|
Presence | 32 | 20 | 12 |
|
|
Absence | 28 | 19 | 9 |
|
| HBsAg |
|
|
| 0.254 |
|
Positive | 37 | 22 | 15 |
|
|
Negative | 23 | 17 | 6 |
|
| Vascular
invasion |
|
|
| 0.183 |
|
Presence | 27 | 20 | 7 |
|
|
Absence | 33 | 19 | 14 |
|
| Multiplicity |
|
|
| 0.047 |
|
Single | 24 | 12 | 12 |
|
|
Multiple (≥2) | 36 | 27 | 9 |
|
| Intrahepatic
metastasis |
|
|
| 0.217 |
|
Presence | 25 | 14 | 11 |
|
|
Absence | 35 | 25 | 10 |
|
 | Table II.Univariate and multivariate analysis
of various prognostic variables influencing DSS in HCC
patients. |
Table II.
Univariate and multivariate analysis
of various prognostic variables influencing DSS in HCC
patients.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex |
| 0.958
(1.306–3.485) | 0.754 |
|
|
|
Male | 34 |
|
|
|
|
|
Female | 26 |
|
|
|
|
| Age (years) |
| 0.648
(0.594–1.751) | 0.392 |
|
|
|
<50 | 28 |
|
|
|
|
|
≥50 | 32 |
|
|
|
|
| Tumor size
(cm) |
| 0.598
(0.678–1.985) | 0.009 | 0.757
(1.032–2.485) | 0.016 |
| ≤5 | 27 |
|
|
|
|
|
>5 | 33 |
|
|
|
|
| AFP (ng/ml) |
| 1.864
(1.358–2.603) | 0.561 |
|
|
|
≤20 | 27 |
|
|
|
|
|
>20 | 33 |
|
|
|
|
| HBsAg |
| 0.894
(0.754–2.625) | 0.435 |
|
|
|
Positive | 37 |
|
|
|
|
|
Negative | 23 |
|
|
|
|
| TNM stage |
| 1.385
(0.677–1.807) | 0.014 | 1.048
(1.986–3.842) | 0.020 |
|
I/II | 27 |
|
|
|
|
|
III/IV | 33 |
|
|
|
|
| Multiplicity |
| 0.481
(0.539–1.750) | 0.046 | 0.780
(0.954–1.874) | 0.035 |
|
Single | 24 |
|
|
|
|
|
Multiple (≥2) | 36 |
|
|
|
|
| Vascular
invasion |
| 1.235
(0.953–4.473) | 0.842 |
|
|
|
Presence | 27 |
|
|
|
|
|
Absence | 33 |
|
|
|
|
| Liver
cirrhosis |
| 0.684
(0.465–1.383) | 0.796 |
|
|
|
Presence | 32 |
|
|
|
|
|
Absence | 28 |
|
|
|
|
| Intrahepatic
metastasis |
| 0.597
(1.346–3.846) | 0.480 |
|
|
|
Presence | 25 |
|
|
|
|
|
Absence | 35 |
|
|
|
|
| miR-29c-3p
expression |
| 1.459
(0.734–1.975) | 0.012 | 1.103
(1.657–3.189) | 0.017 |
|
Low | 39 |
|
|
|
|
|
High | 21 |
|
|
|
|
 | Table III.Univariate and multivariate analysis
of various prognostic variables influencing RFS in HCC
patients. |
Table III.
Univariate and multivariate analysis
of various prognostic variables influencing RFS in HCC
patients.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Variables | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Sex |
| 0.672
(0.486–1.485) | 0.509 |
|
|
|
Male | 34 |
|
|
|
|
|
Female | 26 |
|
|
|
|
| Age (years) |
| 0.537
(0.389–1.059) | 0.543 |
|
|
|
<50 | 28 |
|
|
|
|
|
≥50 | 32 |
|
|
|
|
| Tumor size
(cm) |
| 0.473
(0.571–1.839) | 0.005 | 0.634
(1.007–2.310) | 0.013 |
| ≤5 | 27 |
|
|
|
|
|
>5 | 33 |
|
|
|
|
| AFP (ng/ml) |
| 1.539
(1.409–2.847) | 0.733 |
|
|
|
≤20 | 27 |
|
|
|
|
|
>20 | 33 |
|
|
|
|
| HBsAg |
| 1.003
(0.984–1.530) | 0.527 |
|
|
|
Positive | 37 |
|
|
|
|
|
Negative | 23 |
|
|
|
|
| TNM stage |
| 1.219
(0.347–1.595) | 0.010 | 1.180
(1.038–2.641) | 0.015 |
|
I/II | 27 |
|
|
|
|
|
III/IV | 33 |
|
|
|
|
| Multiplicity |
| 0.495
(0.671–1.734) | 0.034 | 0.834
(0.734–1.995) | 0.027 |
|
Single | 24 |
|
|
|
|
|
Multiple (≥2) | 36 |
|
|
|
|
| Vascular
invasion |
| 1.045
(1.048–3.618) | 0.649 |
|
|
|
Presence | 27 |
|
|
|
|
|
Absence | 33 |
|
|
|
|
| Liver
cirrhosis |
| 1.217
(1.257–2.804) | 0.806 |
|
|
|
Presence | 32 |
|
|
|
|
|
Absence | 28 |
|
|
|
|
| Intrahepatic
metastasis |
| 0.673
(0.758–2.645) | 0.587 |
|
|
|
Presence | 25 |
|
|
|
|
|
Absence | 35 |
|
|
|
|
| miR-29c-3p
expression |
| 1.148
(0.687–1.694) | 0.019 | 1.327
(1.224–2.647) | 0.014 |
|
Low | 39 |
|
|
|
|
|
High | 21 |
|
|
|
|
Upregulated expression of miR-29c-3p
suppresses cell proliferation and migration in HCC cells
Gain-of-function experiments were used for the
analysis of miR-29c-3p expression in liver cancer cells (MHCC-97H
and HepG2). qRT-PCR results confirmed that the expression of
miR-29c-3p was significantly increased after transfection of the
miR-29c-3p mimic in liver cancer cells (MHCC-97H and HepG2)
(Fig. 2A). The CCK-8 results
revealed that overexpression of miR-29c-3p inhibited the
proliferation of liver cancer cells (MHCC-97H and HepG2) (Fig. 2B). As revealed in Fig. 2C, EdU assays indicated that
overexpression of miR-29c-3p significantly inhibited the uptake of
EdU in liver cancer cells (MHCC-97H and HepG2). Wound healing
assays revealed that overexpression of miR-29c-3p suppressed the
migration of liver cancer cells (MHCC-97H and HepG2) (Fig. 2D). Moreover, upregulated expression
of miR-29c-3p inhibited liver cancer cell (MHCC-97H and HepG2)
migration (Fig. 2E).
miR-29c-3p suppresses tumorigenicity
in HCC
The role of miR-29c-3p on proliferation in
vivo was further studied. In the subcutaneous tumor formation
model of nude mice, overexpression of miR-29c-3p inhibited the
growth of subcutaneous tumors, which were thinner and lighter in
liver cancer cells (MHCC-97H and HepG2) (Fig. 3A-D).
miR-29c-3p directly interacts and
inhibits TRIM31
To further understand the specific mechanism of
miR-29c-3p in HCC, TargetScan and miRanda were used to predict the
target gene of miR-29c-3p, and the results revealed that miR-29c-3p
could partially bind to the 3′-UTR of TRIM31 (Fig. 4A). The luciferase reporter assay
demonstrated that overexpression of miR-29c-3p significantly
inhibited luciferase activity in cells transfected with the
wt-3′-UTR of TRIM31, while no significant inhibition was revealed
in cells transfected with the mt-3′-UTR of TRIM31 (Fig. 4B). In addition, qRT-PCR and western
blot assay results revealed that overexpression of miR-29c-3p
inhibited the mRNA and protein expression of TRIM31 in MHCC-97H
cells (Fig. 4C and D). The
expression of miR-29c-3p was significantly decreased after
transfection of the miR-29c-3p inhibitor in MHCC-97H cells
(Fig. S1).
miR-29c-3p is highly expressed and
negatively correlated with TRIM31 in HCC
The role of TRIM31 in HCC was further explored.
qRT-PCR results revealed that TRIM31 was upregulated in HCC tumors
compared with paired normal samples (Fig. 5A). Notably, Spearman's correlation
analysis revealed that TRIM31 had a negative correlation with
miR-29c-3p expression (Fig. 5B).
For further experiments, pcDNA-TRIM31 was used to overexpress the
expression of TRIM31. qRT-PCR results confirmed that the expression
of TRIM31 was significantly increased after transfection of the
pcDNA-TRIM31 in liver cancer cells (MHCC-97H and HepG2) (Fig. 5C).
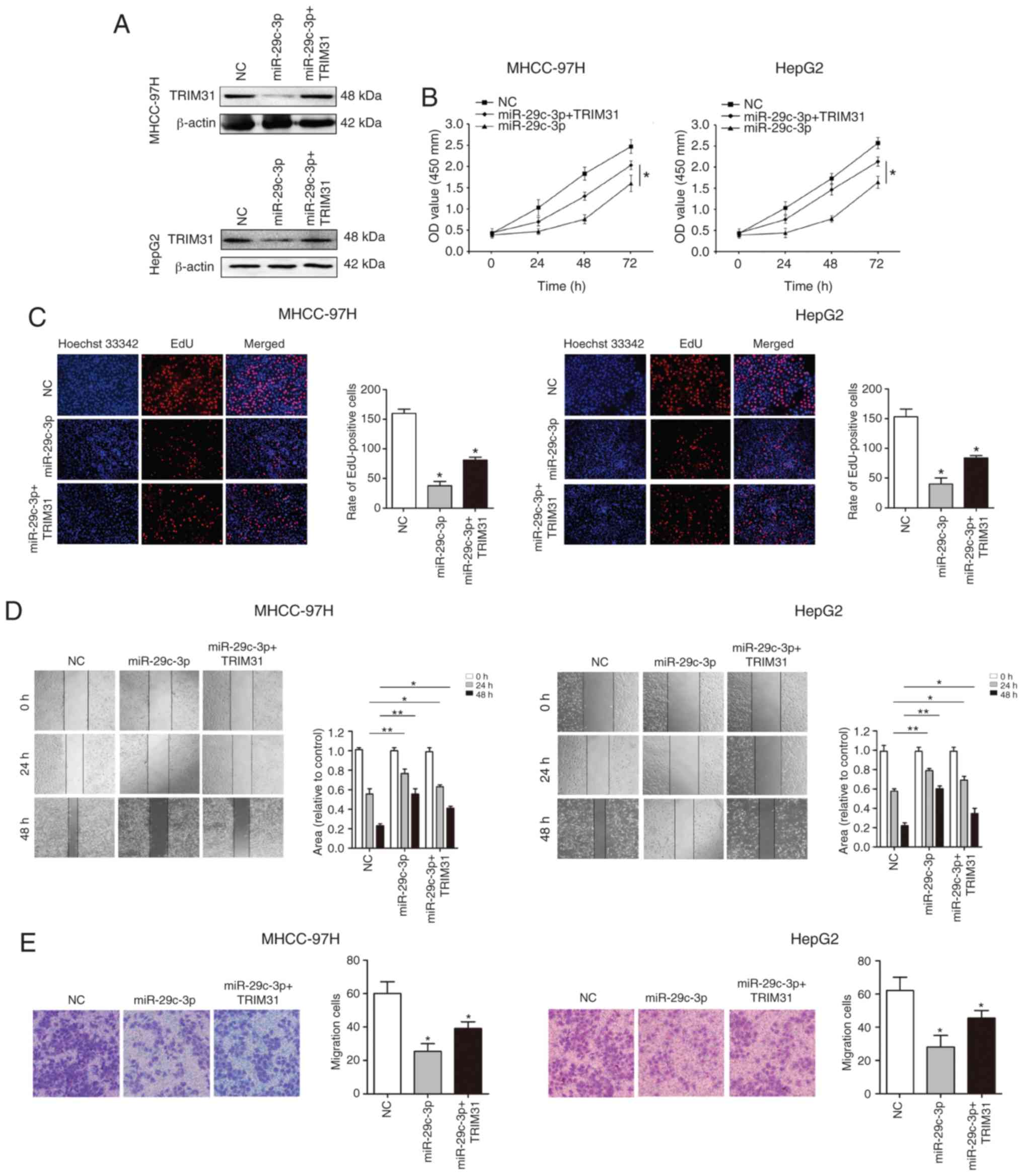
Overexpression of TRIM31 partially
abrogates the inhibitory effect of miR-29c-3p in HCC
To verify whether miR-29c-3p plays a biological
function through its target gene TRIM31, TRIM31 was overexpressed
using pcDNA-TRIM31 in liver cancer cells (MHCC-97H and HepG2)
overexpressing miR-29c-3p. Western blot analysis revealed that
TRIM31 expression was restored (Fig.
6A). Cellular functional studies reveealed that overexpression
of TRIM31 partially abolished the inhibitory effect of miR-29c-3p
on the malignant biological behavior of HCC (Fig. 6B-E).
Discussion
As one of the most significant malignant tumors that
threaten human health, HCC causes a large number of deaths every
year (2). Therefore, it is urgent
to find effective treatments to accurately treat the malignant
progression of HCC and improve the quality of life of HCC patients.
Numerous studies have confirmed that miRNAs are involved in the
malignant progression of HCC and have the ability to predict poor
prognosis in HCC patients (21,22).
The present study sought to elucidate the potential mechanism of
action of miR-29c-3p in HCC. The results revealed that miR-29c-3p
was expressed at low levels in HCC and was closely related to the
poor prognosis of patients. Moreover, miR-29c-3p inhibited the
proliferation and migration of HCC by directly acting on TRIM31.
Collectively, the present findings indicated that miR-29c-3p acts
as a tumor suppressor gene by targeting TRIM31 in HCC.
There is evidence that dysfunction of miR-29c-3p
promotes malignant development of tumors (4,23). Wu
et al revealed that miR-29c-3p regulates the DNA methylation
of LATS1 via DNMT3B to influence the Hippo signaling pathway and
inhibit the malignant development of HCC (17). Another study also revealed that
miR-29c-3p has a binding site in the 3′UTR of KDM5B and that
miR-29c-3p reduces the resistance of endometrial cancer to
paclitaxel through KDM5B (24). In
gastric cancer, miR-29c-3p regulated malignant development of
cancer cells by regulating KIAA1199 expression and activating the
FGFR4/Wnt/β-catenin and EGFR signaling pathways (15). In the present research results, it
was revealed that miR-29c-3p was expressed at low levels in HCC and
that patients with decreased expression of miR-29c-3p had a poor
prognosis. In addition, overexpression of miR-29c-3p could
significantly inhibit the proliferation and migration of HCC.
Previous studies have reported that miR-29c-3p can
regulate its expression levels by binding to multiple target genes
(25,26). In the present study, the molecular
mechanism of miR-29c-3p in the development of HCC was further
explored. TRIM31 is a member of the tripartite motif (TRIM) family,
and the motif includes three zinc-binding domains, a RING, a B-box
type 1 and a B-box type 2, and a coiled-coil region (27). TRIM31 has been revealed to be widely
involved in cell proliferation, cell cycle regulation and cell
response to viruses and other life processes (28). Through bioinformatics software
analysis, it was revealed that TRIM31 is a direct target gene of
miR-29c-3p. A recent study reported that TRIM31 improved the
resistance of pancreatic cancer to gemcitabine by increasing the
K63-linked polyubiquitination of TRAF2 and maintaining the
activation of NF-κB to upregulate the level of p65 (29). In addition, high expression of
TRIM31 was revealed to activate the PI3K/Akt signaling pathway to
promote tumor cell proliferation and invasion in gallbladder cancer
(30). In intestinal epithelial
cells, TRIM31 interacted directly with phosphatidylethanolamine in
a palmitoylation-dependent manner, resulting in the formation of
autolysosomes and providing a preventable pathway for the study of
intestinal pathogen infections (31). Notably, it was revealed that TRIM31
was highly expressed in HCC, and overexpression of TRIM31 could
partially abrogate the inhibitory effect of miR-29c-3p on HCC. In
addition, studies have revealed that TRIMs can participate in tumor
development in many ways. Most members of the TRIM family have E3
ligase activity. They are directly involved in the specific
recognition of target molecules by acting on the skeleton protein
between E3 and the enzyme (32).
TRIMs play an important role in numerous tumor-related signaling
pathways, such as the NF-kB signaling pathway and MAPK signaling
pathway (33,34). Based on the aforementioned results
and previous studies, we will further explore the specific
molecular mechanism of miR-29c-3p and TRIM31in HCC.
In conclusion, the present results demonstrated that
miR-29c-3p could inhibit the proliferation and migration of HCC by
targeting TRIM31, and may be used as an important prognostic
indicator for HCC patients. Further study of the specific molecular
mechanism of the miR-29c-3p/TRIM31 axis will provide new hope for
the diagnosis and treatment of HCC.
Supplementary Material
Supporting Data
Acknowledgements
We thank Dr Zhenru Wu and Dr Yongjie Zhou for
pathology technique assistance and animal model establishing.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (NM. 81470037 and
817700653), the ‘1.3.5 Project for Disciplines of Excellence, West
China Hospital, Sichuan University (ZY2017308), and the Projects of
the Ministry of Science and Technology
(2017ZX10203205-005-002).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
TL and JY acquired the data and created a draft of
the manuscript. TL and LJ prepared the experimental materials and
performed the in vitro assays. TL, LJ and LK interpreted the
data, performed the statistical analysis and analyzed the results.
TL and JY revised and approved the final version of the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Institutional Research Ethics Committee of West China Hospital
of Sichuan University and informed consent was obtained from every
patient enrolled in this study. The protocols regarding the in
vivo manipulations were approved by the Animal Care Ethics
Committee of West China Hospital of Sichuan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greten TF, Wang XW and Korangy F: Current
concepts of immune based treatments for patients with HCC: From
basic science to novel treatment approaches. Gut. 64:842–848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Z and Rana TM: Therapeutic targeting of
microRNAs: Current status and future challenges. Nat Rev Drug
Discov. 13:622–638. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dragomir MP, Knutsen E and Calin GA:
SnapShot: Unconventional miRNA functions. Cell. 174:1038–1038.e1.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hayes CN and Chayama K: MicroRNAs as
biomarkers for liver disease and hepatocellular carcinoma. Int J
Mol Sci. 17:2802016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C,
Ren G and Wu H: MicroRNA-424-5p acts as a potential biomarker and
inhibits proliferation and invasion in hepatocellular carcinoma by
targeting TRIM29. Life Sci. 224:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang H, Lv W, Sun W, Bi Q and Hao Y:
miR-505 inhibits cell growth and EMT by targeting MAP3K3 through
the AKT-NFκB pathway in NSCLC cells. Int J Mol Med. 43:1203–1216.
2019.PubMed/NCBI
|
|
10
|
Yang L, Zhang S, Guo K, Huang H, Qi S, Yao
J and Zhang Z: miR-125a restrains cell migration and invasion by
targeting STAT3 in gastric cancer cells. Onco Targets Ther.
12:205–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong XZ, Song Y, Lu YP, Hu Y, Liu P and
Zhang L: Sanguinarine inhibits the proliferation of BGC-823 gastric
cancer cells via regulating miR-96-5p/miR-29c-3p and the MAPK/JNK
signaling pathway. J Nat Med. 73:777–788. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen G, Zhou T, Li Y, Yu Z and Sun L: p53
target miR-29c-3p suppresses colon cancer cell invasion and
migration through inhibition of PHLDB2. Biochem Biophys Res Commun.
487:90–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu Y, Tang L, Zhang Z, Li S, Liang S, Ji
L, Yang B, Liu Y and Wei W: Long noncoding RNA TUG1/miR-29c axis
affects cell proliferation, invasion, and migration in human
pancreatic cancer. Dis Markers. 22:68570422018.
|
|
14
|
Fang R, Huang Y, Xie J, Zhang J and Ji X:
Downregulation of miR-29c-3p is associated with a poor prognosis in
patients with laryngeal squamous cell carcinoma. Diagn Pathol.
14:1092019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Yu T, Li W, Li M, Zuo Q, Zou Q and
Xiao B: The miR-29c-KIAA1199 axis regulates gastric cancer
migration by binding with WBP11 and PTP4A3. Oncogene. 38:3134–3150.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang S, Jin J, Tian X and Wu L:
sa-miR-29c-3p regulates biological function of colorectal cancer by
targeting SPARC. Oncotarget. 8:104508–104524. 2017.PubMed/NCBI
|
|
17
|
Wu H, Zhang W, Wu Z, Liu Y, Shi Y, Gong J,
Shen W and Liu C: miR-29c-3p regulates DNMT3B and LATS1 methylation
to inhibit tumor progression in hepatocellular carcinoma. Cell
Death Dis. 18:482019. View Article : Google Scholar
|
|
18
|
Lopes CB, Magalhães LL, Teófilo CR, Alves
APNN, Montenegro RC, Negrini M and Ribeiro-Dos-Santos Â:
Differential expression of hsa-miR-221, hsa-miR-21, hsa-miR-135b,
and hsa-miR-29c suggests a field effect in oral cancer. BMC Cancer.
18:7212018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li W, Yi J, Zheng X, Liu S, Fu W, Ren L,
Li L, Hoon DSB, Wang J and Du G: miR-29c plays a suppressive role
in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Clin
Epigenetics. 10:642018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chhabra R: miRNA and methylation: A
multifaceted liaison. Chembiochem. 16:195–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang
L, Zhang H, Chen X, Yang Y and Liu G: miRNA-29c suppresses lung
cancer cell adhesion to extracellular matrix and metastasis by
targeting integrin beta1 and matrix metalloproteinase2 (MMP2). PLoS
One. 8:e701922013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Shou H, Wang Q and Liu S:
Investigation of the potential theranostic role of KDM5B/miR-29c
signaling axis in paclitaxel resistant endometrial carcinoma. Gene.
694:76–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morita S, Horii T, Kimura M, Ochiya T,
Tajima S and Hatada I: miR-29 represses the activities of DNA
methyltransferases and DNA demethylases. Int J Mol Sci.
14:14647–14658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu L, Bi N, Wu L, Ding X, Men Y, Zhou W,
Li L, Zhang W, Shi S, Song Y and Wang L: MicroRNA-29c functions as
a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol
Cancer. 16:502017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reymond A, Meroni G, Fantozzi A, Merla G,
Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et
al: The tripartite motif family identifies cell compartments. EMBO
J. 20:2140–2151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song H, Liu B, Huai W, Yu Z, Wang W, Zhao
J, Han L, Jiang G, Zhang L, Gao C and Zhao W: The E3 ubiquitin
ligase TRIM31 attenuates NLRP3 inflammasome activation by promoting
proteasomal degradation of NLRP3. Nat Commun. 7:137272016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu C, Chen S, Guo Y and Sun C: Oncogenic
TRIM31 confers gemcitabine resistance in pancreatic cancer via
activating the NF-κB signaling pathway. Theranostics. 8:3224–3236.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Zhang Y, Hai J, Wang J, Zhao B, Du L
and Geng X: Knockdown of TRIM31 suppresses proliferation and
invasion of gallbladder cancer cells by down-regulating MMP2/9
through the PI3K/Akt signaling pathway. Biomed Pharmacother.
103:1272–1278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ra EA, Lee TA, Won Kim S, Park A, Choi HJ,
Jang I, Kang S, Hee Cheon J, Cho JW, Eun Lee J, et al: TRIM31
promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat
Commun. 7:117262016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scott DC, Sviderskiy VO, Monda JK, Lydeard
JR, Cho SE, Harper JW and Schulman BA: Structure of a RING E3
trapped in action reveals ligation mechanism for the ubiquitin-like
protein NEDD8. Cell. 157:1671–1684. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sato T, Takahashi H, Hatakeyama S, Iguchi
A and Ariga T: The TRIM-FLMN protein TRIM45 directly interacts with
RACK1 and negatively regulates PKC-mediated signaling pathway.
Oncogene. 34:1280–1291. 2015. View Article : Google Scholar : PubMed/NCBI
|