Contents
Introduction
MicroRNA
Polyadenylation
Alternative polyadenylation
MicroRNA targets in alternative 3′ UTRs
MicroRNA targets in COX2 gene: an example
Switch in the use of alternative PAS, a way to
cellular proliferation
Interplay between alternative polyadenylation and
microRNA, a potential cancer link
Future perspectives
Introduction
As is a fact that only a small fraction (∼1.5%) of
the genetic material in the human genome codes for protein, most
genomic DNA is non-coding but involved in the regulation of gene
expression (1,2). Specifically, there are two levels of
regulation for gene expression: transcriptional regulation controls
whether a gene is transcribed or not and to what extent;
post-transcriptional regulation controls the fate of the
transcribed RNA molecules, including their stability, translation
efficiency and subcellular localization (3). Regulation in each level requires
several multi-component cellular machines, each carrying out a
separate step in gene expression pathway. Instead of working as a
simple linear assembly line, gene expression machines couple
extensively to form a complex network to coordinate their
activities, maximizing the efficiency and specificity of each step
in gene expression (4,5). Recent years have witnessed an
increased appreciation for the importance of post-transcriptional
regulation in mammalian organisms. The same primary transcript can
generate a number of different isoforms by processing steps such as
alternative splicing or polyadenylation (6–8). New
classes of non-coding RNA genes have been described, including the
abundant and conserved class of microRNAs (9–13).
Alternative polyadenylation often results in mRNA 3′ UTRs of
different lengths, while microRNAs regulate gene expression by
binding to specific mRNA 3′ UTRs. Earlier studies have formulated
the association between alternative polyadenylation and
nonsense-mediated decay (14–18),
A-U-rich element mediated decay (19–21)
and mRNA surveillance (22), we
have good reason to deduce that there should also be interplay
between alternative polyadenylation and microRNA-mediated decay. In
this review, we address how alternative polyadenylation and
microRNAs will tether to each other for a fine tune of gene
expression and its potential link to disease development, and
cancer in particular.
MicroRNA
MicroRNAs are evolutionarily conserved, endogenous,
single-stranded, non-coding RNA molecules of ∼22 nt in length that
function as post-transcriptional gene regulators (23). It has been commonly maintained that
microRNAs may be able to regulate ≤30% of the protein-coding genes
in the human genome (24), while
later study indicated that >60% of human protein-coding genes
have been under selective pressure to be regulated by microRNAs
(25). The mature microRNA, with
the help of RNA-induced silencing complex, acts by binding to the
3′ UTR of target mRNAs of protein-coding genes and inhibiting their
translation, which has several possible consequences: it can result
in the cleavage of mRNA (26) or
the repression of productive translation (27) or even the destabilization and
reduction in the mRNA concentration by accelerating poly(A) tail
removal (28).
The current consensus is that microRNA target
specificity is determined by both sequence matching and target
accessibility (29). The critical
region for microRNA binding is nucleotides 2–8 from the 5′ end of
the microRNA, called the ‘seed region’, which binds to its target
site on a given mRNA by exact Watson-Crick complementary. Asymmetry
is the general rule for the matching between a microRNA and its
target (30), in that the 5′ end
of the microRNA tends to have more bases complementary to the
target than the 3′ end does. Moreover, because it is an energy
consuming process to free the base pair within mRNA in order to
make the target accessible for microRNA binding, secondary
structures are also required for target recognition to occur. In
truth, microRNA preferentially target 3′ UTR sites that do not have
complex secondary structures and are located in accessible regions
of the RNA based on favorable thermodynamics (31).
Polyadenylation
The architecture of mammalian pre-mRNA consists of
coding regions and non-coding regions. The coding regions are
included within a translational start codon and a stop codon, while
the non-coding regions locate separately at the 5′ and 3′ end of
mRNA (3). The 3′ end of mRNAs is
generated by cleavage followed by polyadenylation (32), which is the last step of the
post-transcriptional processing. Three sequence elements precisely
determine the site of 3′ end cleavage and polyadenylation in
mammalian pre-mRNAs: the poly(A) signal (highly conserved
hexanucleotide AAUAAA) or its close variants, the actual
polyadenylation site (PAS) 11-30 nt downstream of the signal and
the G-U-rich sequence 14–70 nt downstream of the PAS (33). Mainly five protein factors are
involved in the polyadenylation process: cleavage and
polyadenylation specificity factor (CPSF), cleavage stimulatory
factor (CstF), poly(A)polymerase (PAP), cleavage factors I and II
(CFI and CFII) and poly(A)binding protein (PABP) (34). These factors bind as a complex on
the precursor RNA prior to the cleavage and polyadenylation
reactions. The CPSF recognizes the AAUAAA sequence and the CstF
binds to the G-U-rich sequence. The CFI and CFII cleave the RNA and
generate a 3′ end for polyadenylation. The PAP performs
polyadenylation through two stages with the help of PABP, adding
and extending poly(A) tail to the full ∼200 residue length. The 3′
UTR of mature mRNA transcript runs from the stop codon to the PAS,
where pre-mRNAs are polyadenylated (35).
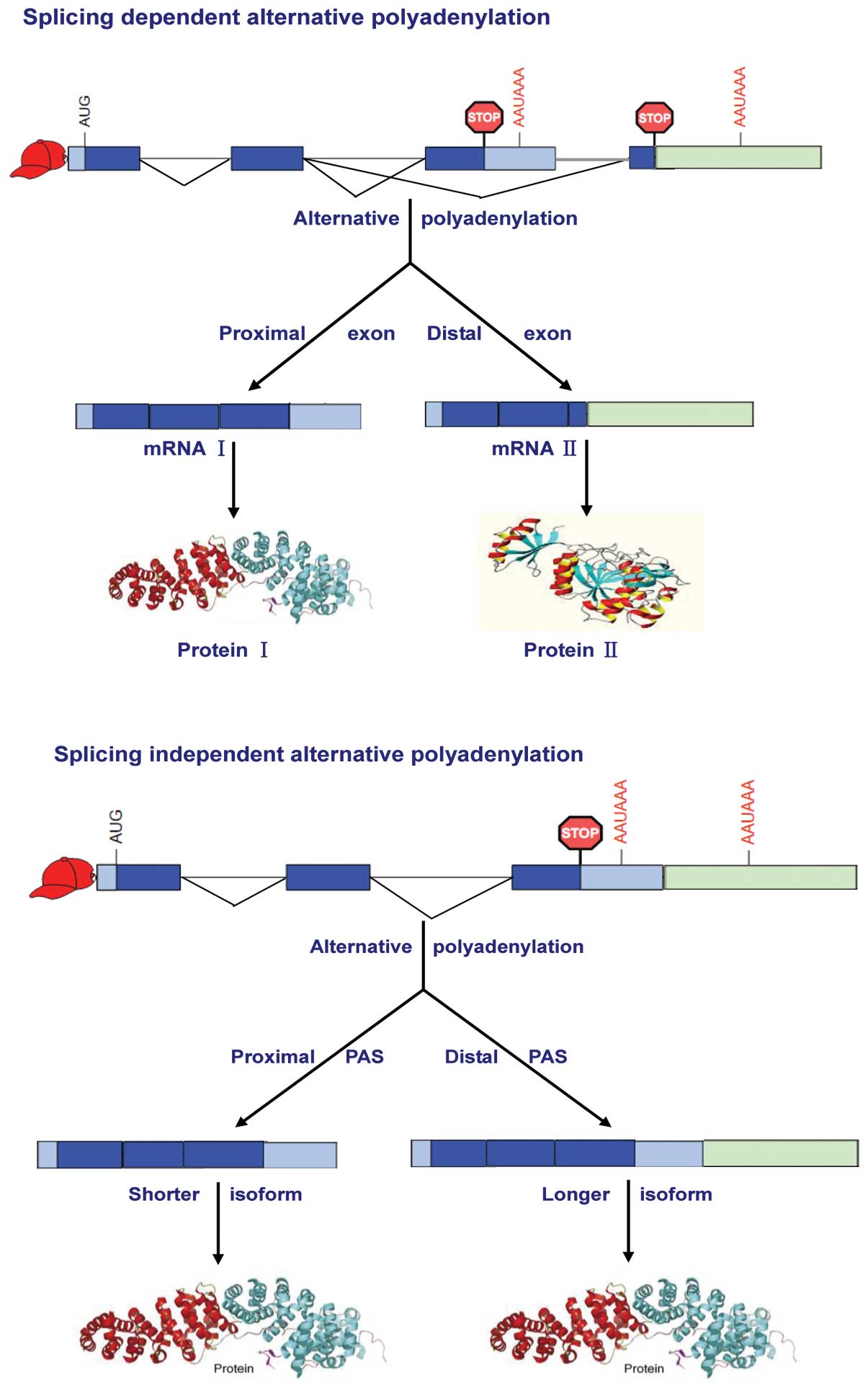
Alternative polyadenylation
Alternative polyadenylation is defined as use of
more than one polyadenylation sites, which is prevalent in at ≥50%
of genes in mammalian genomes, producing mRNAs with different 3′
UTR regions from a single transcript (36–38).
At the end of the past century, Edwalds-Gilbert et al
categorized alternative polyadenylation into three models,
according to their distinct exon arrangement and PAS selection
(35). A large number of genes
have multiple PASs within the 3′ UTR in the terminal exon (39), arranged one behind the other in
tandem and certain regulatory elements may posit between the PASs
which can influence the stability or translatability of the longer
mRNA. Another set of genes have an exon which is a composite of 3′
and 5′ splice sites followed closely by a PAS. Such composite exon
can serve as either internal or terminal and encode different
protein based on circumstances. The genes in the third model have
two or more alternative 3′ terminal exons encoding different
C-termini. They are processed into different mRNAs either by using
the first 3′ terminal exon and its PAS or by skipping over that
exon entirely and using the second 3′ terminal exon with its PAS.
However, later researchers combined the last two models and
classified alternative polyadenylation into two categories, namely
splicing-independent and splicing-dependent (40). In the first case, tandem PASs are
located in the same 3′ exon and the UTR will consist of a
constitutive part followed by an alternative part, producing mRNAs
that differ solely by the length of the 3′ UTR. In the case of
alternative terminal exons, the different 3′ UTR will generally not
share common sequence and the choice of downstream final exon is
likely coupled with suppression of the 3′ splice site of the more
upstream one, producing mRNAs that differ by their coding sequences
(Fig. 1). Although both appeared
as alternative 3′ UTRs, the functional consequences of these two
types of alternative polyadenylation are obviously different. It is
much easier to deduce that splicing dependent alternative
polyadenylation encodes different protein products through the
generation of different mRNAs (41,42);
while splicing independent alternative polyadenylation is more
likely to generate mRNA products with differential stability, which
will ultimately lead to more or less protein product, depending on
enhanced or diminished RNA half-life (19,43–46).
MicroRNA targets in alternative 3′ UTRs
Splicing independent alternative polyadenylation
produces transcript isoforms with 3′ UTRs of different lengths,
containing a constitutive segment and an alternative segment. If a
microRNA target is located within the alternative part of the 3′
UTR, the shorter transcript should be target free and should escape
microRNA-mediated inhibition, while the longer transcript should be
inhibited (47–49). Such variations are very common. It
has been estimated that two thirds of targeted genes have
alternative 3′ UTRs, with 40% of predicted target sites located in
alternative UTR segments. Based on whether the target sites fall
within constitutive and/or alternative UTR segments, Majoros and
Ohler classified microRNA target genes into two broad categories
(50). Constitutive targets
encompass predicted target genes with constitutive UTRs, as well as
genes with alternative UTRs, in which all sites are located within
the constitutive UTR regions. This category accounts for 56.6% of
all targets. Alternatively targeted genes have at least one target
site located in an alternative 3′ UTR segment, which was further,
categorized into on/off targets and modulated targets. On/off
targets are alternative targets in which all target sites fall
exclusively into alternative UTR regions (accounting for 23.7% of
all targets), whereas modulated targets contain alternative targets
with sites in both constitutive and alternative UTR regions
(accounting for 19.6% of all targets). This classification of
target genes perfectly illustrates the functional significance of
the coupling of microRNA regulation and alternative
polyadenylation. MicroRNAs regulate gene expression, while
alternative polyadenylation may decide whether the expression
should be regulated (on/off targets) and to what extent it should
be regulated (modulated targets) (Fig.
2).
MicroRNA targets in COX2 gene: an
example
Cyclooxygenases (COXs) are the key and rate-limiting
enzymes in the production of prostaglandins (51). Two separate COX genes, COX-1
and COX-2, have been identified, divergent strikingly in
their regulation of expression. COX-1 is constitutively expressed,
while COX-2 is strongly induced in response to activation by
hormones, pro-inflammatory cytokines, growth factors, oncogenes,
carcinogens and tumor promoters (52). The COX-2 gene is made up of
10 exons; the 3′ UTR is contained within the last exon. The
COX-2 3′ UTR is larger than average, encompassing ∼2.5 kb,
with several polyadenylation signals. Two of the COX-2
polyadenylation signals are commonly used (53), the proximal one is AUUAAA (604–609
nt from the stop codon) and the distal one is AAUAAA (2485–2490 nt
from the stop codon). The proximal signal was weaker than the
distal signal and the estimated ratio of proximal/distal
polyadenylation signal usage was 1:3. The use of the proximal
polyadenylation signal results in a ∼2.6 kb mRNA, while the use of
the distal polyadenylation signal results in a ∼4.5 kb mRNA. These
two mRNAs were exactly the same in their coding sequences, but
differ in the length of their 3′ UTRs, containing a 0.6-kb
constitutive segment followed by a 1.9-kb alternative segment
(54). Within the 1.9-kb
alternative segment of COX-2 3′ UTR, there is a microRNA
binding site (seed region 1736–1743 nt from the stop codon) for
both miR-101 and miR-199a. In vitro gain- and
loss-of-function experiments show that COX-2 expression is
posttranscriptionally regulated by these two microRNAs (55). Since there is no microRNA target
site confirmed in the constitutive segment of the COX-2 3′
UTR, it should currently be recognized as an on/off target. The
transcription of the longer isoform of COX-2 mRNA will turn
on the target, while the transcription of the shorter isoform will
turn it off.
According to ‘TargetScan’ prediction, there should
be two microRNA target sites within the 3′ UTR of COX-2. The
distal target is exactly that mentioned above, within the
alternative segment, while the proximal target is located in the
constitutive segment (seed region 257-264 nt from the stop codon),
predicted to be targeted by miR-26. However, the proximal
target is not experimentally verified yet. The COX-2 will
change from an on/off target to a modulated target depending on
whether the proximal target is a true target or not. If it is a
true target, then the longer isoform of COX-2 mRNA will have
two target sites and be regulated by two microRNAs, miR-26
and miR-101/miR-199a, the shorter isoform will be targeted
only by miR-26. That is, both the long and the short
isoforms will be regulated, but to different extent. In this sense,
COX-2 gene should modulate the extent of its regulation
through switching from one isoform to the other, or by varying
ratios between the two isoforms.
Switch in the use of alternative PAS, a way
to cellular proliferation
The longer and the shorter isoforms of mRNA have
different regulation efficiencies by microRNAs, resulting in
different levels of protein expression. Will there be any
preferential in the use of the isoforms under certain cellular
circumstances? An interesting study (56) provides evidence that a switch in
the use of alternative PASs is part of a defined program for
cellular proliferation. In this study, Sandberg et al
(56) developed a quantitative
method to compare the expression of the constitutive segment
relative to the alternative segment of the 3′ UTR for genes with
tandem PASs during T cell activation. The expression pattern was
found to differ markedly between resting and stimulated primary T
cells. Specifically, stimulation decreased the use of the
alternative segment of the 3′ UTR in 86% of the genes tested. This
reduction was not associated with significant changes in median
mRNA levels, indicating that the effect is due to a switch in PAS
usage. Since activation of hematopoietic cells is often associated
with a dramatic increase in proliferation, the authors hypothesized
that the decrease in the use of the longer isoform, which leads to
a shorter 3′ UTR, might be associated with cellular proliferation.
Interestingly, Hip2, one of the genes tested by Sandberg
et al, contains in the alternative segment of its 3′ UTR
target sites for miR-21 and miR-155, both of which
are expressed in activated T cells. Overall, levels of Hip2
mRNA were very similar between resting and stimulated T cells.
However, upon T cell activation the relative level of the
alternative segment of 3′ UTR decreased, while protein level
increased substantially. Furthermore, elimination of the target
sequences for both miR-21 and miR-155 from
Hip2 3′ UTR increased expression level similar to that
containing only the constitutive segment of 3′ UTR. These results
show that changes in PAS usage can control the impact of microRNA.
Specifically, the use of the shorter isoform can avoid the
regulation by microRNA and then enhance cellular proliferation
(57).
Although it often escapes microRNA regulation, the
shorter isoform of the alternatively polyadenylated mRNA is not
always upregulated. In their study, Ghosh et al (58) found a microRNA mediated
upregulation of the longer 3′ UTR of the mouse cytoplasmic
β-actin gene. This gene, also named Actb, generates
two alternative transcripts terminated at tandem PAS. The longer
isoform was expressed at a relatively lower level, but it confered
higher translational efficiency to the transcript. In fact, the
longer isoform harbours a conserved binding site for
mmu-miR-34. Sequence specific anti-miRNA molecule, mutations
of the microRNA target site in the 3′ UTR resulted in reduced
expression and the expression could be restored by a mutant
microRNA complementary to the mutated target site. These results
together suggest that miR-34 specifically target the longer
isoform of β-actin transcript and upregulate the target gene
expression, indicating that it is not always the shorter isoform
that contributes to cellular proliferation.
Interplay between alternative
polyadenylation and microRNA, a potential cancer link
This ability has been shown in the above-mentioned
COX2 gene in colorectal cancer cells. Instead of the 4.5-kb
isoform, the 2.6-kb isoform of COX-2 mRNA lacking the
miR-101/miR-199a target of the 3′ UTR was selectively
stabilized upon cell growth to confluence, suggesting that
COX2 mRNA may escape regulation in cancer cells through
alternative PAS usage (6The general association of the use of
shorter 3′ UTR isoforms, avoidance of regulation by microRNAs and
enhanced proliferation leads to the speculation that in some cases
a shift toward expression of proximal PAS isoforms may be required
to evade regulation that would otherwise restrict cell cycle
progression. This is in agreement with the observation that
microRNAs influence many aspects of cellular proliferation and cell
cycle progression (59,60) and accordingly downregulation of
microRNA expression promotes cellular transformation and
tumorigenesis, which is commonly observed in cancer (61,62)
(Fig. 3). Many genes, under
evolutionary selection, have the ability to enrich or deplete their
microRNA binding sites through changes in the lengths of their 3′
UTRs (30). 3). Earlier studies
have shown that increased levels of COX2 protein were present in
both human and animal colorectal tumors, whereas the normal
intestinal mucosa displayed low to undetectable COX2 expression
(64,65). Similarly, elevated COX2 levels
resulting from defects in its regulation was also observed in many
other solid malignancies, including breast, lung, prostate,
pancreas, bladder, stomach, esophagus and head and neck cancers
(66–73). Therefore, it is reasonable to
deduce that interplay between alternative polyadenylation of
COX2 and miR-101/miR-199a regulation may contribute,
at least in part, to the development of cancer.
A cell cycle regulating gene (74), cyclin D1 (CCND1) has two
major spliced transcripts, cyclin D1a and cyclin D1b.
The cyclin D1a has a wild-type isoform of 4.5 kb in length
with a full-length 3′ UTR and another short isoform of 1.5 kb in
length with a truncated 3′ UTR (75). Containing such a long 3′ UTR, the
wild-type cyclin D1a was regulated by a number of microRNAs,
as indicated in certain algorisms. A regulatory feedback loop was
assumed between cyclin D1 and microRNA regulation, in which
cyclin D1 induces the expression of certain microRNAs, in
turn, these microRNAs limit the proliferative function of cyclin
D1, thus regulating cellular tumorigenesis (76). When this feedback loop is broken,
cyclin D1 will be dysregulated and overexpressed, which is
commonly seen in many types of cancer, including breast, lung,
colorectal cancer and lymphomas (77–80).
Many strongly proliferative tumors have exceptionally high
cyclin D1 mRNA levels and preferentially express short
cyclin D1a mRNA isoform. This short isoform lacks many mRNA
destabilization elements present in the wild-type cyclin D1a
mRNA, including a target site for miR-16-1 (81). Thus, truncated cyclin D1a
mRNA deletes miR-16-1 binding sites within the cyclin
D1 mRNA 3′ UTR and alters the microRNA regulatory feedback
loop, leads to increased total cyclin D1 mRNA and increased
cyclin D1 protein expression and then contributes to cancer
development.
Future perspectives
By truncating their 3′ UTRs, certain genes can
escape the regulation of microRNAs, as well as other RNA-binding
proteins and thus proliferate. Just like the gecko, breaking its
tail for escape in time of danger, saving itself from being
injured. This pattern of tail truncation has already been
implicated in the process of disease development, including cancer.
A fairly large proportion of genes have different isoforms of 3′
UTR and there are full of mysteries contained in the alternative
segment of the 3′ UTR, besides nonsense mediated decay, A-U-rich
element mediated decay and mRNA surveillance, microRNA mediated
decay should be an important one. Future studies are needed to
investigate the association of the interplay between alternative
polyadenylation and microRNA regulation in clinical settings, which
will facilitate our understanding of disease etiology and even
helpful in our efforts to work out new ways of disease prevention
and treatment.
References
|
1.
|
Venter JC, Adams MD, Myers EW, et al: The
sequence of the human genome. Science. 291:1304–1351. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Lander ES, Linton LM, Birren B, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Mignone F, Gissi C, Liuni S and Pesole G:
Untranslated regions of mRNAs. Genome Biol. 3:Reviews 0004. 2002.
View Article : Google Scholar
|
|
4.
|
Maniatis T and Reed R: An extensive
network of coupling among gene expression machines. Nature.
416:499–506. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Lackner DH, Beilharz TH, Marguerat S, et
al: A network of multiple regulatory layers shapes gene expression
in fission yeast. Mol Cell. 26:145–155. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
de la Grange P, Dutertre M, Correa M and
Auboeuf D: A new advance in alternative splicing databases: from
catalogue to detailed analysis of regulation of expression and
function of human alternative splicing variants. BMC
Bioinformatics. 8:1802007.PubMed/NCBI
|
|
7.
|
Wang ET, Sandberg R, Luo S, et al:
Alternative isoform regulation in human tissue transcriptomes.
Nature. 456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Tian B, Pan Z and Lee JY: Widespread mRNA
polyadenylation events in introns indicate dynamic interplay
between polyadenylation and splicing. Genome Res. 17:156–165. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Hon LS and Zhang Z: The roles of binding
site arrangement and combinatorial targeting in microRNA repression
of gene expression. Genome Biol. 8:R1662007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Farh KK, Grimson A, Jan C, et al: The
widespread impact of mammalian MicroRNAs on mRNA repression and
evolution. Science. 310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Linsley PS, Schelter J, Burchard J, et al:
Transcripts targeted by the microRNA-16 family cooperatively
regulate cell cycle progression. Mol Cell Biol. 27:2240–2252. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kertesz S, Kerenyi Z, Merai Z, et al: Both
introns and long 3′-UTRs operate as cis-acting elements to trigger
nonsense-mediated decay in plants. Nucleic Acids Res. 34:6147–6157.
2006.
|
|
15.
|
Gilat R and Shweiki D: A novel function
for alternative polyadenylation as a rescue pathway from NMD
surveillance. Biochem Biophys Res Commun. 353:487–492. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Lewis BP, Green RE and Brenner SE:
Evidence for the widespread coupling of alternative splicing and
nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA.
100:189–192. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Durand C, Roeth R, Dweep H, et al:
Alternative splicing and nonsense-mediated RNA decay contribute to
the regulation of SHOX expression. PloS One. 6:e181152011.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Li XL, Andersen JB, Ezelle HJ, Wilson GM
and Hassel BA: Post-transcriptional regulation of RNase-L
expression is mediated by the 3′-untranslated region of its mRNA. J
Biol Chem. 282:7950–7960. 2007.
|
|
20.
|
Rimokh R, Berger F, Bastard C, et al:
Rearrangement of CCND1 (BCL1/PRAD1) 3′ untranslated region in
mantle-cell lymphomas and t(11q13)-associated leukemias. Blood.
83:3689–3696. 1994.
|
|
21.
|
von Roretz C and Gallouzi IE: Decoding
ARE-mediated decay: is microRNA part of the equation? J Cell Biol.
181:189–194. 2008.PubMed/NCBI
|
|
22.
|
Muhlrad D and Parker R: Aberrant mRNAs
with extended 3′ UTRs are substrates for rapid degradation by mRNA
surveillance. RNA. 5:1299–1307. 1999.
|
|
23.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Friedman RC, Farh KK, Burge CB and Bartel
D: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Pillai RS, Bhattacharyya SN, Artus CG, et
al: Inhibition of translational initiation by Let-7 MicroRNA in
human cells. Science. 309:1573–1576. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Wu L, Fan J and Belasco JG: MicroRNAs
direct rapid dead-enylation of mRNA. Proc Natl Acad Sci USA.
103:4034–4039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Didiano D and Hobert O: Molecular
architecture of a miRNA-regulated 3′ UTR. RNA. 14:1297–1317.
2008.PubMed/NCBI
|
|
30.
|
Stark A, Brennecke J, Bushati N, Russell
RB and Cohen SM: Animal MicroRNAs confer robustness to gene
expression and have a significant impact on 3′UTR evolution. Cell.
123:1133–1146. 2005.PubMed/NCBI
|
|
31.
|
Kertesz M, Iovino N, Unnerstall U, Gaul U
and Segal E: The role of site accessibility in microRNA target
recognition. Nat Genet. 39:1278–1284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Lopez F, Granjeaud S, Ara T, Ghattas B and
Gautheret D: The disparate nature of ‘intergenic’ polyadenylation
sites. RNA. 12:1794–1801. 2006.
|
|
33.
|
Legendre M and Gautheret D: Sequence
determinants in human polyadenylation site selection. BMC Genomics.
4:72003. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kubo T, Wada T, Yamaguchi Y, Shimizu A and
Handa H: Knockdown of 25 kDa subunit of cleavage factor Im in Hela
cells alters alternative polyadenylation within 3′-UTRs. Nucleic
Acids Res. 34:6264–6271. 2006.PubMed/NCBI
|
|
35.
|
Edwalds-Gilbert G, Veraldi KL and Milcarek
C: Alternative poly(A) site selection in complex transcription
units: means to an end? Nucleic Acids Res. 25:2547–2561. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Ara T, Lopez F, Ritchie W, Benech P and
Gautheret D: Conservation of alternative polyadenylation patterns
in mammalian genes. BMC Genomics. 7:1892006. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Zhang H, Lee JY and Tian B: Biased
alternative polyadenylation in human tissues. Genome Biol.
6:R1002005. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Feng Z, Wu CF, Zhou X and Kuang J:
Alternative polyadenylation produces two major transcripts of Alix.
Arch Biochem Biophys. 465:328–335. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Moucadel V, Lopez F, Ara T, Benech P and
Gautheret D: Beyond the 3′ end: experimental validation of extended
transcript isoforms. Nucleic Acids Res. 35:1947–1957. 2007.
|
|
40.
|
Lutz CS: Alternative polyadenylation: a
twist on mRNA 3′ end formation. ACS Chem Biol. 3:609–617.
2008.PubMed/NCBI
|
|
41.
|
Heuze-Vourc’h N, Leblond V and Courty Y:
Complex alternative splicing of the hKLK3 gene coding for the tumor
marker PSA (prostate-specific-antigen). Eur J Biochem. 270:706–714.
2003.PubMed/NCBI
|
|
42.
|
Liu H and Johnson EM: Distinct proteins
encoded by alternative transcripts of the PURG gene, located
contrapodal to WRN on chromosome 8, determined by differential
termination/polyadenylation. Nucleic Acids Res. 30:2417–2426. 2002.
View Article : Google Scholar
|
|
43.
|
Yu M, Sha H, Gao Y, Zeng H, Zhu M and Gao
X: Alternative 3′ UTR polyadenylation of Bzw1 transcripts display
differential translation efficiency and tissue-specific expression.
Biochem Biophys Res Commun. 345:479–485. 2006.
|
|
44.
|
Hughes TA: Regulation of gene expression
by alternative untranslated regions. Trends Genet. 22:119–122.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Stojic J, Stohr H and Weber BH: Three
novel ABCC5 splice variants in human retina and their role as
regulators of ABCC5 gene expression. BMC Mol Biol. 8:422007.
View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Hager S, Frame FM, Collins AT, Burns JE
and Maitland NJ: An internal polyadenylation signal substantially
increases expression levels of lentivirus-delivered transgenes but
has the potential to reduce viral titer in a promoter-dependent
manner. Hum Gene Ther. 19:840–850. 2008. View Article : Google Scholar
|
|
47.
|
Legendre M, Ritchie W, Lopez F and
Gautheret D: Differential repression of alternative transcripts: a
screen for miRNA targets. PLoS Comput Biol. 2:e432006. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Tan S, Guo J, Huang Q, et al: Retained
introns increase putative microRNA targets within 3′ UTRs of human
mRNA. FEBS Lett. 581:1081–1086. 2007.PubMed/NCBI
|
|
49.
|
Sivakumaran TA, Resendes BL, Robertson NG,
Giersch AB and Morton CC: Characterization of an abundant COL9A1
transcript in the cochlea with a novel 3′ UTR: expression studies
and detection of miRNA target sequence. J Assoc Res Otolaryngol.
7:160–172. 2006.PubMed/NCBI
|
|
50.
|
Majoros WH and Ohler U: Spatial
preferences of microRNA targets in 3′ untranslated regions. BMC
Genomics. 8:1522007.PubMed/NCBI
|
|
51.
|
Hla T, Bishop-Bailey D, Liu CH, Schaefers
HJ and Trifan OC: Cyclooxygenase-1 and -2 isoenzymes. Int J Biochem
Cell Biol. 31:551–557. 1999. View Article : Google Scholar
|
|
52.
|
Harper KA and Tyson-Capper AJ: Complexity
of COX-2 gene regulation. Biochem Soc Trans. 36:543–545. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Hall-Pogar T, Zhang H, Tian B and Lutz CS:
Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res.
33:2565–2579. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Sawaoka H, Dixon DA, Oates JA and Boutaud
O: Tristetraprolin binds to the 3′-untranslated region of
cyclooxygenase-2 mRNA. A polyadenylation variant in a cancer cell
line lacks the binding site. J Biol Chem. 278:13928–13935.
2003.
|
|
55.
|
Chakrabarty A, Tranguch S, Daikoku T,
Jensen K, Furneaux H and Dey SK: MicroRNA regulation of
cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci
USA. 104:15144–15149. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Sandberg R, Neilson JR, Sarma A, Sharp PA
and Burge CB: Proliferating cells express mRNAs with shortened 3′
untranslated regions and fewer microRNA target sites. Science.
320:1643–1647. 2008.PubMed/NCBI
|
|
57.
|
Zlotorynski E and Agami R: A PASport to
cellular proliferation. Cell. 134:208–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Ghosh T, Soni K, Scaria V, Halimani M,
Bhattacharjee C and Pillai B: MicroRNA-mediated up-regulation of an
alternatively polyadenylated variant of the mouse cytoplasmic
{beta}-actin gene. Nucleic Acids Res. 36:6318–6332. 2008.PubMed/NCBI
|
|
59.
|
Gammell P: MicroRNAs: recently discovered
key regulators of proliferation and apoptosis in animal cells:
identification of miRNAs regulating growth and survival.
Cytotechnology. 53:55–63. 2007. View Article : Google Scholar
|
|
60.
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
O’Rourke JR, Swanson MS and Harfe BD:
MicroRNAs in mammalian development and tumorigenesis. Birth Defects
Res C Embryo Today. 78:172–179. 2006.
|
|
62.
|
Bandres E, Agirre X, Ramirez N, Zarate R
and Garcia-Foncillas J: MicroRNAs as cancer players: potential
clinical and biological effects. DNA Cell Biol. 26:273–282. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Dixon DA: Dysregulated
post-transcriptional control of COX-2 gene expression in cancer.
Curr Pharm Des. 10:635–646. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Eberhart CE, Coffey RJ, Radhika A,
Giardiello FM, Ferrenbach S and DuBois RN: Up-regulation of
cyclooxygenase 2 gene expression in human colorectal adenomas and
adenocarcinomas. Gastroenterology. 107:1183–1188. 1994.PubMed/NCBI
|
|
65.
|
Kutchera W, Jones DA, Matsunami N, et al:
Prostaglandin H synthase 2 is expressed abnormally in human colon
cancer: evidence for a transcriptional effect. Proc Natl Acad Sci
USA. 93:4816–4820. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Ristimaki A, Sivula A, Lundin J, et al:
Prognostic significance of elevated cyclooxygenase-2 expression in
breast cancer. Cancer Res. 62:632–635. 2002.PubMed/NCBI
|
|
67.
|
Wolff H, Saukkonen K, Anttila S,
Karjalainen A, Vainio H and Ristimaki A: Expression of
cyclooxygenase-2 in human lung carcinoma. Cancer Res. 58:4997–5001.
1998.PubMed/NCBI
|
|
68.
|
Yoshimura R, Sano H, Masuda C, et al:
Expression of cyclooxygenase-2 in prostate carcinoma. Cancer.
89:589–596. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Tucker ON, Dannenberg AJ, Yang EK, et al:
Cyclooxygenase-2 expression is up-regulated in human pancreatic
cancer. Cancer Res. 59:987–990. 1999.PubMed/NCBI
|
|
70.
|
Ristimaki A, Nieminen O, Saukkonen K,
Hotakainen K, Nordling S and Haglund C: Expression of
cyclooxygenase-2 in human transitional cell carcinoma of the
urinary bladder. Am J Pathol. 158:849–853. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Ristimaki A, Honkanen N, Jankala H,
Sipponen P and Harkonen M: Expression of cyclooxygenase-2 in human
gastric carcinoma. Cancer Res. 57:1276–1280. 1997.PubMed/NCBI
|
|
72.
|
Ratnasinghe D, Tangrea J, Roth MJ, et al:
Expression of cyclooxygenase-2 in human squamous cell carcinoma of
the esophagus; an immunohistochemical survey. Anticancer Res.
19:171–174. 1999.PubMed/NCBI
|
|
73.
|
Chan G, Boyle JO, Yang EK, et al:
Cyclooxygenase-2 expression is up-regulated in squamous cell
carcinoma of the head and neck. Cancer Res. 59:991–994.
1999.PubMed/NCBI
|
|
74.
|
Sherr CJ: Mammalian G1 cyclins. Cell.
73:1059–1065. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Wiestner A, Tehrani M, Chiorazzi M, et al:
Point mutations and genomic deletions in CCND1 create stable
truncated cyclin D1 mRNAs that are associated with increased
proliferation rate and shorter survival. Blood. 109:4599–4606.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Yu Z, Wang C, Wang M, et al: A cyclin
D1/microRNA 17/20 regulatory feedback loop in control of breast
cancer cell proliferation. J Cell Biol. 182:509–517. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Nosho K, Kawasaki T, Chan AT, et al:
Cyclin D1 is frequently overexpressed in microsatellite unstable
colorectal cancer, independent of CpG island methylator phenotype.
Histopathology. 53:588–598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Elsheikh S, Green AR, Aleskandarany MA, et
al: CCND1 amplification and cyclin D1 expression in breast cancer
and their relation with proteomic subgroups and patient outcome.
Breast Cancer Res Treat. 109:325–335. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
79.
|
Radovic S, Babic M, Doric M, et al:
Non-small cell lung carcinoma: cyclin D1, bcl-2, p53, Ki-67 and
HER-2 proteins expression in resected tumors. Bosn J Basic Med Sci.
7:205–211. 2007.PubMed/NCBI
|
|
80.
|
Shakir R, Ngo N and Naresh KN: Correlation
of cyclin D1 transcript levels, transcript type and protein
expression with proliferation and histology among mantle cell
lymphoma. J Clin Pathol. 61:920–927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
81.
|
Chen RW, Bemis LT, Amato CM, et al:
Truncation in CCND1 mRNA alters miR-16-1 regulation in mantle cell
lymphoma. Blood. 112:822–829. 2008. View Article : Google Scholar : PubMed/NCBI
|