1. Introduction
Matrix-assisted laser desorption/ionization (MALDI)
time-of-flight (TOF) techniques are versatile analytical tools used
in various areas of medical diagnostics and basic research. Mass
spectrometry (MS) has been successfully applied to study
microbiological colonies (1–3),
plants (4), insects (5), vertebrates including whole animals
(6–8), human cells (9,10)
and tissues (11,12).
This method allows the analysis of proteins
(13,14), peptides (15,16),
lipids or phospholipids (6,17–19),
carbohydrates or glycoconjugates (20) and exogenous or endogenous small
molecules, especially molecules involved in drug metabolism
(21–27).
MALDI imaging mass spectrometry (IMS), first
described by Caprioli et al (28), links molecular evaluation of
numerous analytes and the high sensitivity and selectivity of mass
spectrometry with morphological information about spatial
distribution of molecules in cells or tissues (29) and thereby provides unbiased
visualization of the arrangement of biomolecules in tissue
(30).
Since cancer tissue contains all information on
proteomic and genetic changes, it represents the best possible
sample material for any molecular research (29). Thus, pathology is not only a large
field of medical research but also a basis for the diagnosis of
various diseases with direct impact on clinical treatment
decisions. The information contained in tissues cannot be replaced
by investigation of serum or blood (29).
MALDI IMS has been applied to various organ systems
and diseases such as cardiovascular- (31), neurologic- (32), or joint disorders (11,33),
amyloidosis (34) inflammatory
bowel diseases (35), renal
diseases (36–38), the field of reproductive medicine
(39) and various tumor entities
(13,15,40–45).
The following review will focus on MALDI IMS in human tumor tissues
as well as applications that have a potential in pathology
(22).
2. Technical considerations
Despite some technical limitations, mainly in the
area of detection limits and instrument sensitivity at high spatial
resolution, this method has the potential to improve cancer
diagnostics (42). There are
several reviews addressing the complete process of MALDI IMS, in
particular regarding experimental considerations: sample
collection, sample preparation, matrix deposition, ionization
process, instrumentation and data processing (22,30,31,46–54).
IMS experiments can generally be performed in two
related modes, imaging (57) and
profiling (58). Whereas imaging
allows correlation with histology, profiling is generally used when
tissues are completely homogeneous and, therefore, different
compartments do not have to be considered.
Concerning sample collection and sample preparation,
workflow solutions have to be exactly defined beginning from the
handling of various tissues in the operation theatre, followed by
dehydration and paraffin-embedding process as well as deposition of
sections onto conductive slides.
Sample handling and preparation are critical to
achieve good spectral quality and reproducibility. In the past few
years, several groups have worked on strategies to improve the
detection of peptides and proteins (55,56);
however, optimization and standardization in clinical application
to minimize variability of results are still needed.
IMS requires minimal sample preparation for analysis
(59). In the past years, there
has been a significant improvement in image spatial resolution that
is in the range of 20–50 μm and can be improved to up to 5 μm with
special techniques (60,61).
Data processing and statistical analysis are of
tremendous importance for accurate and reliable results. In the
processing step MALDI datasets are typically subjected to
smoothing, baseline correction and calibration. Normalization is
generally the next step intended to minimize the experimental
variation arising from day-to-day instrument fluctuations, or
artifacts related to sample preparation (62). Statistical analysis is then
performed in order to elucidate spectral features that are
consistently associated within a cohort for biomarker discovery or
for disease classification. Most statistical methods can be
performed using a supervised method requiring at least two groups
of spectral data that need to be discriminated or an unsupervised
analysis applied without any prior knowledge of the samples to
generate a classification model. Principal component analysis (PCA)
is a well known method used to describe the variability in the
dataset in a reduced number of dimensions (63). The software for data analysis, such
as ClinProTools and SCiLS Lab (Bruker Daltonik), MassLynx (Waters)
and BioWorks (Thermo Fisher Scientific) is usually commercially
available and provided by the manufacturer of the MALDI
instruments. In addition, BioMap software (Novartis, Basel,
Switzerland) is freely available for calculating basic statistics
of the full dataset. Despite the different data analysis approaches
that have been developed, there is a need for more advanced
biostatistical software in order to be able to handle huge data
files (5,000–50,000 spectra).
A new and emerging application is the
three-dimensional illustration of organ structures. It has been
shown that MALDI IMS can be combined with other imaging modalities
such as positron emission tomography (PET) and magnetic resonance
imaging (MRI), improving the interpretation of the proteomic data
and providing 3D visualization of proteins that may not be apparent
with other imaging methods (64,65).
Another promising development represents MS for
single cell analysis to determine protein and peptide contents in
cells (47).
Combination of MALDI IMS with cytological
preparations of oral brush biopsies has been shown by Remmerbach
et al (9). These authors
could discern non-malignant from malignant cells by MS.
Furthermore, IMS could be applied as a specific diagnostic tool for
routine cervical cytology specimen (10). Thus, IMS as a relatively new
technique is still at an early stage, and technical advances
concerning sample preparation and instrumentation will certainly
improve specificity and sensitivity and enlarge the potential for
further applications.
3. MALDI IMS as an adjunct to
immunohistochemistry
Contrary to antibody-based methods, target
structures do not need to be known when MALDI IMS is applied
(52) which makes it a valuable
discovery tool, since broad range of proteins may be surveyed
simultaneously (67).
Furthermore, MS techniques may have a higher
reproducibility between laboratories than immunohistochemistry
because human bias is abolished (68).
However, MALDI IMS cannot only be an adjunct tool to
immunohistochemistry but also to histochemistry. For examle, it has
been shown that the Berlin-blue reaction for the detection of
hemosiderin can be replaced by MALDI IMS of ferritin chains
(33). Likewise, mucous substances
detected by histochemical reactions could be identified applying MS
(69) as well as soft tissue
constituents (70–72).
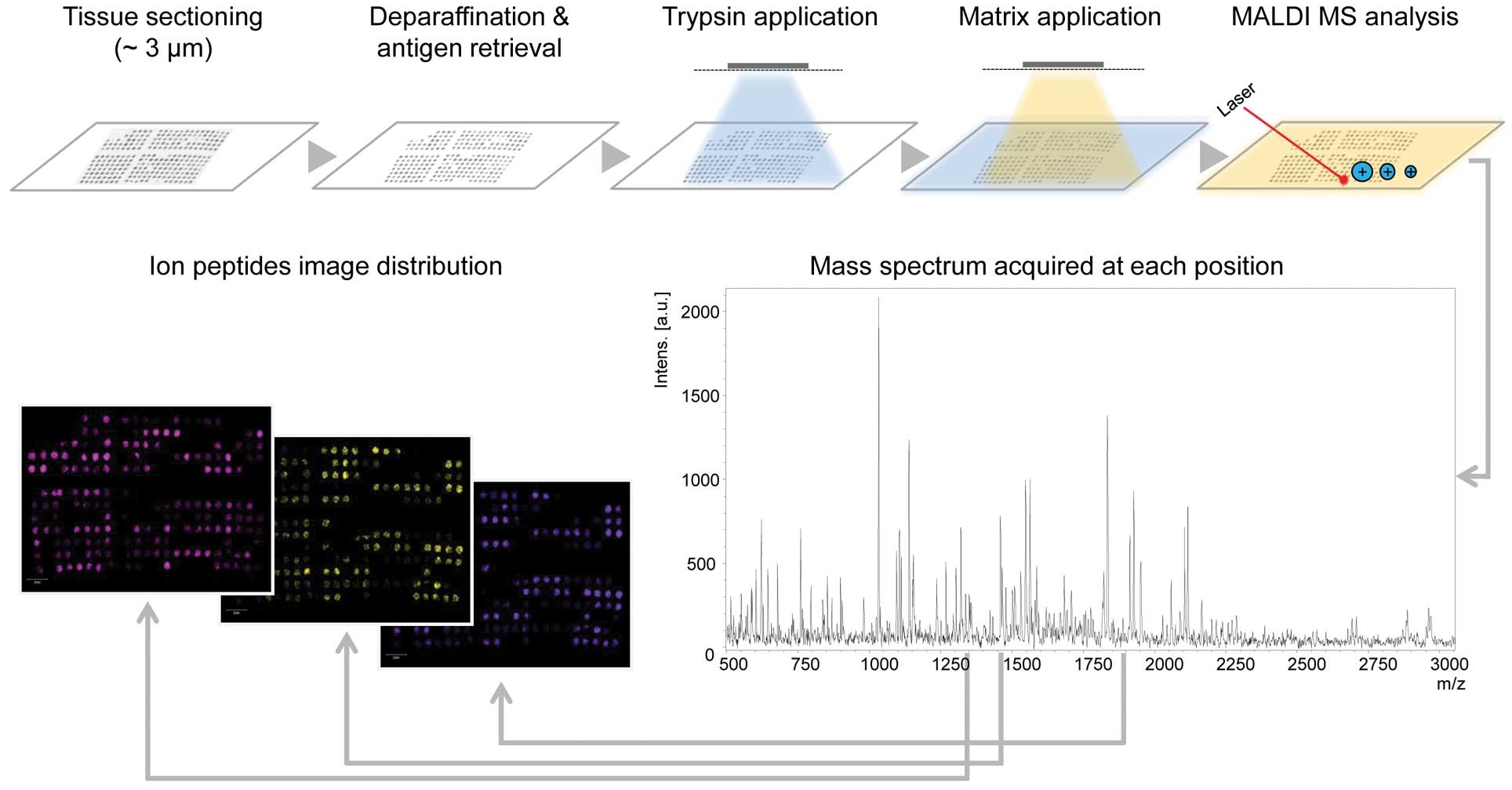
4. Frozen vs. formalin-fixed
paraffin-embedded tissue samples and analysis of tissue
microarrays
The investigation of frozen tissues by MALDI IMS
requires immediate freezing, storage at −80°C and rapid cutting
(48) enabling the study of
proteins, drugs or their metabolites and lipids. Formalin-fixed
paraffin-embedded (FFPE) tissues have to be digested and,
therefore, allow analysis of peptides only (Fig. 1). Although frozen samples are
easier to be analyzed and imaged as they do not require multiple
steps for sample preparation (deparaffination and antigen
retrieval), handling and storage of frozen specimens has
disadvantages, since proteins are degraded during repeated
freeze-thaw cycles and there is a need for expensive freezers.
Despite the difficulties in sample handling,
numerous reports have described the application of MALDI IMS on
whole sections (12,13,43,44,57,73–78)
and even on small biopsy samples of fresh frozen tissue (79).
Millions of FFPE tissues of various diseases
including numerous cancer tissues are available in the archives of
institutes of pathology. These samples are well documented and
include information on patient treatments, treatment response,
disease progression and other relevant clinical data (51).
One of the advantages of using fixed specimens is
that FFPE tissues are well suited for tissue microarray (TMA)
construction, where multiple FFPE core biopsies can be assembled in
a single paraffin block. The direct analysis of FFPE TMA using
MALDI IMS allows the analysis of multiple tissue samples in a
single experiment (67,80). This high-throughput method was used
to investigate gastric (16),
pancreatic (81), non-small cell
lung (67), prostate (82), renal cancer (15) and to discriminate pancreatic from
breast cancer tissues (83). The
correlation of MALDI IMS data with histology can be performed by
two methods: either by staining a serial section with hematoxylin
and eosin (H&E) (67) or by
staining the tissue after the MALDI experiment, since the tissue is
not destroyed during the laser ionization process (47).
5. Identification of peptides and
proteins
There are many techniques used in the study of
proteins, such as western blot analysis, liquid chromatography and
reverse phase protein arrays (84–87).
The most common approach to identify proteins using
MS technique is the bottom-up approach achieved through protein
separation/isolation, proteolytic digestion, sequencing of peptides
by LC-MS/MS, and matching the peptides to genome-based protein
databases (88–90). Another approach is top-down, where
proteins of interest are isolated from a complex protein mixture
and directly fragmented in the mass spectrometer without previous
digestion of proteins. This approach provides direct measurement of
the molecular masses of the intact proteins (91,92).
An alternative method for protein identification is on-tissue
digestion, where tissue samples are in situ digested so that
proteins of interest can be isolated and directly fragmented from
the tissue section using MALDI MS/MS. This method offers the
advantage to identify proteins from a tissue section without
separation or homogenization, while preserving the spatial
integrity of the tissue sample (30,67).
However, protein identification directly from tissue is not always
straightforward due to sample complexity and limited sample per
unit area, unless peptides are present at high abundance in the
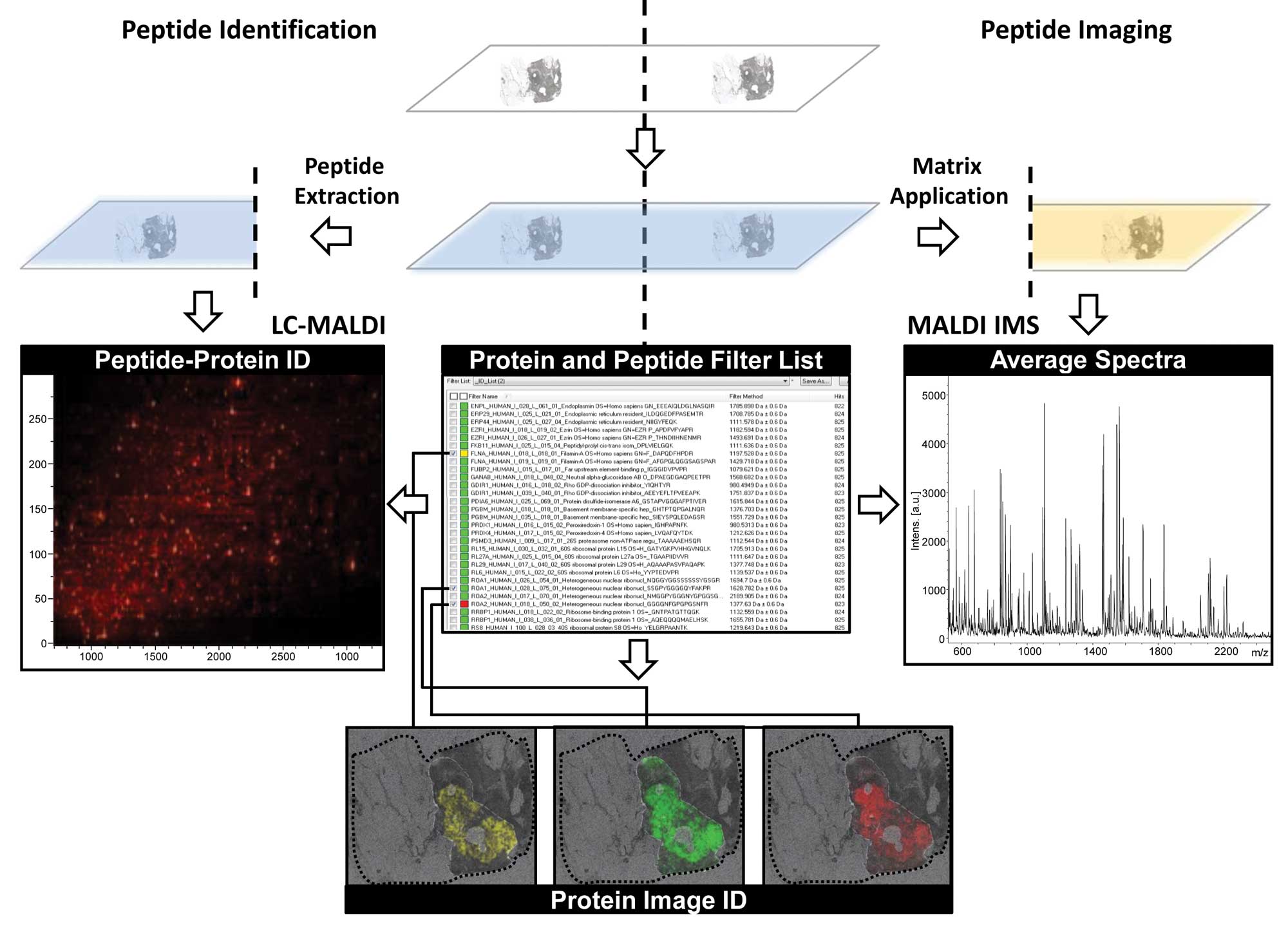
sample. Recently, an imageID approach has been developed where
consecutive tissue sections are processed in parallel to provide
both, protein identification by LC-MS/MS and spatial information by
MALDI IMS simultaneously (83)
(Fig. 2). New public databases for
the identification of proteins such as MSiMass list (93) and MaTisse (94) have been created enabling users to
search either for proteins or for m/z values and assign identities
to the peaks observed in their experiments. Such databases will
improve implementation of MSI in the histopathological and clinical
setting.
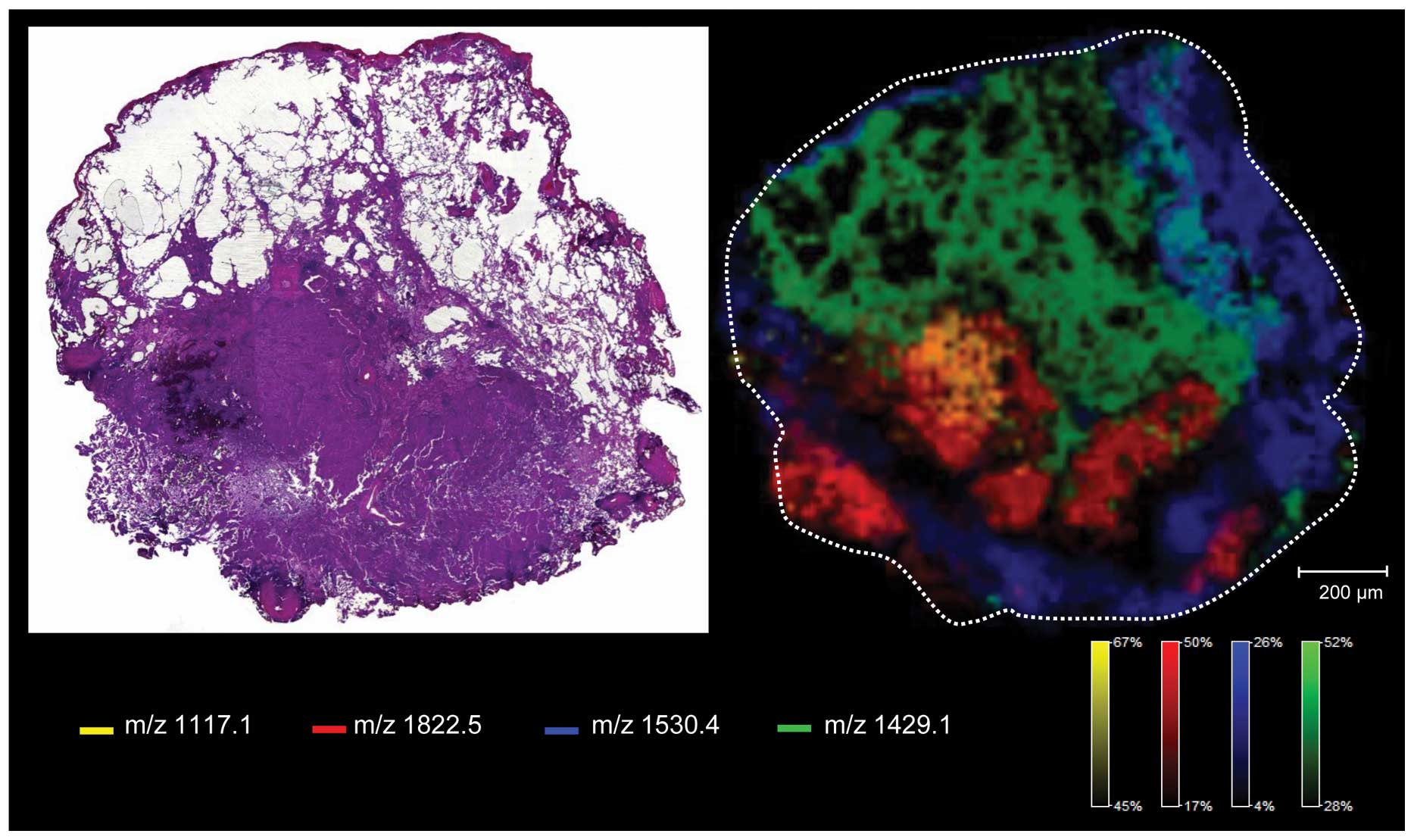
6. Tumor heterogeneity
The biology of a tumor depends not only on tumor
cells but also on the interaction with stroma, blood vessels and
the immune system (e.g. tumor-associated macrophages and
lymphocytes). Homogenization of the tissue sample would lead to
loss of information (29).
Moreover, when considering personalized medicine, demonstration of
tumor heterogeneity (Fig. 3) will
be one of the most challenging issues in the future and MALDI IMS
seems to be the ideal tool to solve this problem (29,40,95).
As an alternative to MALDI IMS in complete tissue sections,
captured cells can be mounted directly onto a MALDI target
following microdissection thereby maintaining the spatial
conformation for IMS (72). DNA-
and RNA-based molecular methods have limitations to show tumor
heterogeneity since spatial integrity is destroyed during the
preparation process of the nucleic acids.
The potential application of MALDI IMS in a clinical
workflow has been suggested by Caprioli (96) who stated in 2005 that this method
may aid in diagnosis, prognosis and therapy of cancer bringing a
new dimension of molecular information into pathological assessment
of tissue specimen.
Today, nearly 10 years later, MALDI IMS is
considered a powerful technique to evaluate tumor margins, and to
acquire information on tumor typing, grading, prognosis and
prediction.
7. Tumor margins
MALDI IMS is an objective method to demonstrate
molecular tumor margins (resection borders). This was first
proposed by Chaurand et al (97). Evidence has shown that adjacent
tissue that appears histologically normal may already be
molecularly compromised. For example, it has been shown in tumors
of the ovary that MALDI analysis could detect two
interface-specific proteins, plastin 2 and peroxiredoxin 1 as
differentially expressed in the interface zones between tumors and
normal tissues, suggesting these proteins as predictive markers for
the presence of residual disease (98). Another study on tumor margins was
conducted on kidney samples by Oppenheimer et al (44). These authors demonstrated that
cells without phenotypic characteristics of tumor cells, have
malignant molecular features. Specifically, mass spectral profiles
were compared among four different regions of tissue consisting of
tumor, margin-tumor, margin-normal and normal. Two statistical
methods, the significance analysis of microarrays (SAM) and
permutation t-test, were used in this work demonstrating the
presence of patterns resembling tumor in the margin-normal
microenvironment outside of the histological tumor border. For
example, proteins involved in the mitochondrial electron transport
system were consistently underexpressed in the tumor as well as in
the histologically normal tissue adjacent to the tumor. Another
application of MALDI MS dealing with surgical tumor margins has
been shown on liver tissue (99).
These investigators detected molecular signatures as classifiers of
non-tumor and tumor regions that could be applied to the interface
between normal liver tissue and hepatocellular carcinoma.
Thus, a potential advantage of IMS for the
evaluation of tumor margins is the ability to identify molecular
changes that are not detectable by histopathological evaluation
alone (47).
8. Tumor typing
Numerous antibodies are often required for the
correct diagnosis of a tumor. In the future, pathologists will be
confronted more frequently with small biopsies since minimally
invasive methods have been emerged in the past. Hence, effective
and tissue sparing molecular techniques are important for correct
classification of tumors. Despite the introduction of
multi-antibody stains in immunohistochemistry, numerous tissue
sections are often required for tumor typing. Since only one tissue
section is needed when MALDI IMS is applied, this technique is
perfect for the evaluation of very small tissue samples, and the
remaining tissue is available for further molecular classification
such as mutation screening (e.g. in lung tumors).
The combination of MALDI IMS for tumor
classification with typing, grading and subsequent application of
MALDI TOF techniques for mutation screening would be a very fast,
sensitive and cost efficient workflow able to reduce human
bias.
Published results indicate that MALDI IMS has the
potential to develop into a valuable diagnostic technique that can
complement established histological and histochemical as well as
immunohistochemical methods. Biomarkers or signal classifiers can
be robustly reproduced in different biomedical centers (73).
MALDI IMS for tumor typing has been demonstrated on
frozen (13) as well as on FFPE
tissue (16,67). It has been shown that
classification of 6 common tumor entities was possible using frozen
tissue sections (13).
Furthermore, squamous carcinoma and adenocarcinoma of the lung
(67), papillary and clear cell
renal cancer (15) and gastric
carcinoma (16) have been
subclassified.
9. Grading and prognosis
MALDI IMS has been successfully used to identify
subsets of markers that correlate with cancer progression in
several cancers including non-small cell lung cancer (76) and pre-invasive bronchial lesions
(43), brain tumors (21,101), breast (58), ovarian (102), renal cancer (15), Barrett’s adenocarcinoma (103), intestinal-type gastric cancer
(78), prostate carcinoma
(104) lymph node metastasis of
melanoma (105), prostate cancer
(82), colonic cancer (13), papillary thyroid carcinoma
(14) and urothelial neoplasia
(106).
The accuracy to discriminate normal, pre-invasive
and invasive lung tissues obtained by MALDI IMS was >90%
(43).
MS studies have also been used to determine tumor
grading. In myxoid sarcomas high-grade tumors could be separated
from low-grade tumors. Likewise, MS/MS technique revealed signals
specific for poorly differentiated gastric cancer (16).
Additionally, proteomic tissue profiling improved
the grading of non-invasive papillary urothelial neoplasia
(106). Variation in the protein
expression pattern of low-grade and high-grade gliomas could also
be demonstrated (107,108).
Besides grading, IMS has been employed to identify
early stage disease (79) as well
as predictive proteomic signatures for lymph node metastasis in
gastric cancer (109). Since
hypoxia is related to poor survival and increased frequency of
metastasis, one study investigated hypoxia associated
protein-patterns and identified around 100 proteins that may serve
as prognostic markers (81).
Notably, one group could also show MALDI IMS as a tool to identify
peptide signals to predict tumor response to molecular targeted
therapy in a breast cancer mouse model (110). All of these applications have a
great potential to find their way into routine diagnostics, as
methods are needed that reduce human bias.
10. Identification of drugs and
metabolites
IMS has been used to study drug localization in
human tissues as well as in animal models (54,111–119). The visualization and quantitative
analysis of the native drug distribution by IMS techniques with
enhanced resolution and sensitivity is desirable for evaluating
drug effects and optimizing drug design as it has been shown for
paclitaxel in tumor-bearing mice (118). Fresh frozen tumor sections from
mice bearing BxPC3 pancreatic cancer xenografts, were analyzed by
MALDI IMS and LC-MS after paclitaxel (PTX)-incorporating micelle
(NK105) administration (50 or 100 mg/kg) at various time-points (15
min, 1, 24, 48 and 72 h). These sections were compared with tumor
sections from mice treated with PTX alone at the same time-points
as well as untreated mice. MALDI IMS demonstrated that NK105
successfully delivered a large amount of the PTX within the tumor
tissue after NK105 injection compared with PTX injection alone.
Quantification of PTX and NK105 were evaluated by LC-MS analysis of
the extracted drugs from serial tissue sections for MALDI IMS. The
content of the PTX released from NK105 was significantly higher and
retained for a longer period of time than the free PTX and control
(untreated), suggesting that the antitumor activity of NK105 is
superior to that of PTX alone. The high sensitivity and selectivity
of IMS combined with visualization of molecular spatial
distribution in tissue provides a valuable platform in targeted
drug and untargeted metabolomic analysis in biological and clinical
research (115). IMS is useful in
the comparison of different ways of drug delivery to organs and
tissues (120).
Another application is the identification of drugs
and their metabolites in the primary or metastatic tumor tissue
(112). Furthermore, IMS allowed
the characterization of the distribution of small molecules in the
microenvironment of tissue compartments of tumors isolated from
lung cancer patients and rat xenograft tissues (111). Three experimental models were
described in the present study for measuring drug localization. In
the ‘concentration gradient model’, an entire solid xenograft tumor
(lung adenocarcinoma A549) tissue was immersed in a solution of the
drug mixture containing tiotropium and erlotinib for 2.5 h. A
passive transfer of the drug mixture into solid tissue samples
displayed a decreasing concentration gradient toward the center of
the tissue. In this model investigators wanted to show the
efficiency of transport of drugs through various cell types. For
example, the signal intensity of drugs from tumor cells was
substantially lower than that from stroma. In the ‘dispersion
model’, 10-μm tissue section of resected lung adenocarcinoma
specimens were immersed into the drug solution, where either
adsorption or adsorption/absorption occurred. In this model, the
distribution of erlotinib localized well to the area of tumor cells
identified by the pathologist. Finally, in the ‘directed dosage
model’ a drug solution was evenly deposited in small spots (~150 μm
in diameter) on the tissue surface of adenocarcinoma tumors using a
microdispenser-based spotter apparatus. As in the previous model,
the localization of erlotinib was found in tumor areas.
As tumor tissue can be resected during the course of
therapy, IMS is suitable for the evaluation of treatment efficacy
and the therapy may be optimized (113).
11. MALDI IMS in various organs or
tissues
To enable readers of the present review a fast
overview on application of MALDI IMS to tumor tissue, Table I provides a brief summary of the
different studies and findings.
 | Table IOutline of MALDI MS-based studies and
findings in the field of cancer research. |
Table I
Outline of MALDI MS-based studies and
findings in the field of cancer research.
| Tissue
type/organ/entity | Content of the
study/findings | Authors (ref.) |
|---|
| Brain | Peptide and protein
patterns of primary glial brain tumors and non-tumor tissue;
discrimination of increasing tumor grade | Schwartz et
al (108) |
| Identification of
protein pattern of gliomas, which could predict short-term and
long-term survival (prognosis) | Schwartz et
al (101) |
| Grading of
meningioma based on different protein pattern | Agar et al
(122) |
| Classification of
brain tumors based on lipid information; discrimination of
meningioma from gliomas | Eberlin et
al (100) |
| Demonstration of
drug transit of small molecules through the blood-brain
barrier | Liu et al
(114) |
| Visualization of
brain and tumor distribution of PI3K-inhibitors in models of
glioblastoma | Salphati et
al (119) |
| Breast | MALDI IMS of normal
breast tissue, ductal carcinoma in situ, invasive breast
cancer and peritumoral stroma | Cornett et
al (58) |
| Optimized tissue
preparation for obtaining peptide mass spectrum profiles from FFPE
tissue of breast cancer | Ronci et al
(123) |
| Visualization of
proteins specific for ductal carcinoma in situ and invasive
ductal carcinoma (different stage of the disease) | Seeley and Caprioli
(34) |
| Determination of
lymph node status of breast carcinoma and prediction of response to
chemotherapy | Seeley and Caprioli
(125) |
| Localization of
choline metabolites and cations in viable and necrotic tumor
regions in a breast cancer xenograft model | Amstalden van Hove
et al (127) |
| Novel approach that
combines MALDI-IMS and principal component analysis for generating
tumor classification | Djidja et al
(63) |
| Protein
identification to predict Her2-status of breast
carcinomas-identified cysteine-rich intestinal protein 1 | Rauser et al
(74) |
| IMS of
acylcarnitines, phosphatidylcholines, and sphingomyelin in breast
tumor models | Chughtai et
al (128) |
| Discrimination of
breast and pancreatic carcinoma based on the peptide profile | Casadonte et
al (83) |
| Comparison of
protein signals of intratumoral and extratumoral stroma in breast
cancer; interlaboratory comparison of spectra | Dekker et al
(73) |
| Gastrointestinal
tract |
| Oral | IMS of biopsy
specimen of oral squamous cell carcinoma with reproducible
images | Patel et al
(129) |
| Esophagus | Detection of
COX7A2, TAGLN2 and S100-A10 as prognostic markers in Barrett’s
adenocarcinoma | Elsner et al
(103) |
| Stomach | Distinguished
gastric cancer from normal mucosa; signals overexpressed in tumor
were defensins and calgranulins; stage of disease could be
predicted | Kim et al
(79) |
| TMA with
classification of signals which could discern between normal
gastric tissue and adenocarcinoma; signal specific to poor
differentiation corresponds to histone H4 | Morita et al
(16) |
| Identification of
prognostic seven protein signature in intestinal-type gastric
cancer | Balluff et
al (78) |
| Identification of
S100A8 and S100A9 as negative regulators for lymph node metastasis
of gastric cancer | Choi et al
(109) |
| Colon | Classification
algorithm of six common tumors; liver metastasis of colonic cancer
was identified | Meding et al
(13) |
| Lipid signatures in
biopsies of liver metastases of colonic cancer | Thomas et al
(130) |
| IMS of
platinum-based metallodrugs in colorectal and ovarian cancer | Bianga et al
(113) |
| Pancreas | Discrimination of
m/z species of normal pancreas, intraepithelial neoplasia, and
intraductal papillary mucinous neoplasms | Grüner et al
(77) |
| TMA peptide
profiles to discriminate pancreatic from breast cancer | Casadonte et
al (83) |
| Imaging of
glucose-regulated protein in pancreatic adenocarcinoma | Djidja et al
(81) |
| Liver | Demonstration of
protein profiles of hepatocellular carcinoma and liver
cirrhosis | Le Faouder et
al (75) |
| Lung | Classification of
protein peaks according to lung cancer histology; distinguished
primary tumor from metastases, discrimination of patients with good
and poor prognosis (frozen tissue) | Yanagisawa et
al (76) |
| Predictive accuracy
of over 90% of normal tissue, preinvasive lesions and invasive lung
cancer based on proteomic profiles of proteins in frozen
sections | Rahman et al
(131) |
| TMA of lung tissues
with distinction of adenocarinoma and squamous cell carcinoma of
the lung by peptide pattern; identification of peptides | Groseclose et
al (67) |
| Non-small cell
cancers, classified according to the MALDI lipid profile | Lee et al
(132) |
| Erlotinib and
gefitinib and tiotropium were localized in their
microenvironment | Vegvari et
al (111) |
| Lymphoma | Discrimination of
Hodgkin’s lymphoma and lymphadenitis with a sensitivity and
specificity of 83.92% and 89.37% by IMS | Schwamborn et
al (134) |
| Skin | Detection of
protein signatures for survival and recurrence in malignant
melanoma | Hardesty et
al (105) |
| Differentiation of
Spitz nevi from Spitzoid malignant melanoma by IMS | Lazova et al
(135) |
| Identification of
vemurafenib in malignant melanoma and provided evidence of
colocalization of drug and target | Sugihara et
al (112) |
| Thyroid | Identification of
S100A10, thioredoxin, and S100-A6 as biomarkers of papillary
thyroid carcinomas with lymph node metastasis | Nipp et al
(14) |
| Detection of
ribosomal protein P2 specific for papillary thyroid cancer | Min et al
(136) |
| Good
reproducibility of IMS data on cytological samples to distinguish
different thyroid lesions; characterization of the proteomic
profile of medullary thyroid carcinoma | Pagni et al
(66) |
| Urogenital
tract |
| Kidney | Analysis of tumor
margins in renal cell carcinoma | Oppenheimer et
al (44) |
| Detection of race
specific profiles in Wilms’ tumors | Axt et al
(139) |
| Discrimination of
papillary and clear cell renal cancer using TMA samples, prediction
of tumor survival | Steurer et
al (140) |
| Bladder | Proteomic tissue
profiling improved grading of non-invasive papillary urothelial
neoplasia | Oezdemir et
al (106) |
| Evaluation of
signals associated with tumor aggressiveness, histologic phenotype
(growth pattern), and cell proliferation in bladder cancer | Steurer et
al (140) |
| Ovary | Investigation of
the interface between serous ovarian carcinoma and normal
tissue | Kang et al
(98) |
| Identification of a
fragment of the 11S proteasome activator complex and Reg α fragment
as biomarkers for ovarian carcinoma | Lemaire et
al (102) |
| Platinum anticancer
imaging in ovarian carcinomas | Bianga et al
(113) |
| Integrated
transcriptomic and lipidomic analysis of sulfatides in ovarian
cancer | Liu et al
(141) |
| Presentation of a
tryptic peptide reference dataset for IMS on FFPE ovarian cancer
tissues | Meding et al
(142) |
| Prostate | Discrimination of
cancerous from non-cancerous prostatic tissue based on protein
profile obtained by IMS | Schwamborn et
al (41) |
| Identification of
cancer and non-cancer tissue on the basis of specific fragment of
mitogen-activated protein kinase/extracellular signal regulated
kinase | Cazares et
al (143) |
| Detection of
signals associated with favorable tumor phenotype, prolonged time
to PSA relapse; multiple signals associated with the ERG-fusion
status and with ERG expression | Steurer et
al (82) |
| Sarcomas | Identification of
proteins and lipids in myxoid sarcomas specific to tumor type and
grade; could show intra-tumor heterogeneity | Willems et
al (40) |
Brain
There are only a few studies applying IMS in
neurobiological science. The reasons for this include limited
access to the technology, relatively low sample throughput and
difficult sample preparation. However, MS techniques have been
applied to a variety of brain tumors including oligodendrogliomas,
astrocytomas, meningiomas and gliomas (100,108,121,122). The technique was used in the
differential diagnosis, defining the tumor margins, the assessment
of the tumor grade and to distinguish different survival groups.
Schwartz et al (101)
developed a peptide-based profile that could sub-classify gliomas
according to different histologic grades on human brain biopsies.
Moreover, a short-long and long-term survival-group could be
distinguished by the investigators, using the weighted flexible
compound covariate method (WFCCM). A proteomic signature of 24
different MS signals discriminated between patients from a
short-term survival group (52 patients with a mean survival of
<15 months) and a long-term survival group (56 patients with a
mean survival of >90 months). These results were also verified
as an independent predictor of patient survival using a
multivariate Cox proportional hazards model.
IMS has great potential in exploring biomolecular
mechanisms underlying disorders. An IMS study developed a
preliminary proteomic algorithm that could discriminate meningioma,
glioma and non-tumor tissue, although the number of patients was
very small (122). In the present
review, investigators used support vector machine models able to
classify meningioma spectra using a training set built from: i) a
single brain tumor class that compared both glioma and meningioma
with non-tumor tissue (including blood); ii) a meningioma brain
tumor class and non-tumor brain class; and iii) a meningioma class,
a glioma class, and a non-tumor brain class. Training sets were
built based on the gold standard of WHO histopathology
classification of tumors of the central nervous system. Another
group was able to discern glioblastoma and oligodendroglioma tumor
biopsies by MS but not by IMS (121).
In conjunction with proteomic approaches to assess
brain tumors, lipidomic signatures could also provide beneficial
information regarding diagnosis, grading and the resection margins
(100). Furthermore,
pharmacokinetics and pharmacodynamics studies have been performed.
Salphati et al (119)
visualized and compared the distribution of PI3K inhibitors in
glioblastoma multiforme in mouse brain by IMS. Drug transit through
the blood-brain barrier (BBB) is essential for therapeutic response
in malignant glioma and was studied by IMS (114). In this review, three examples of
drug transit through the BBB were reported in mouse brain. Drugs
and/or drugs metabolites (BKM120, RAF265 and erlotinib) were imaged
in mouse brain tissue sections together with heme, used as a
molecular marker delineating the lumen of blood vessels in the
brain. In all cases, drugs were found to penetrate the BBB.
Specifically, RAF265 and erlotininb were localized within tumor
regions, whereas BKM120 distributed in the brain parenchyma.
Notably, drugs distributed independently of the heme signal,
indicating the escape of the drug molecules from the tumor
vasculature.
Breast
MALDI IMS has been applied in breast cancer tissue
(58,74,83,123). Additionally, other proteomic mass
spectrometric techniques contributed to new insights into breast
cancer pathogenesis and prognosis (reviewed in ref. 124). Dijda et al (63) used human breast tumor tissue for
the optimization of the protocol for detection of peptides in FFPE
sections.
Cornett et al (58) described an automated workflow
integrating histopathology and breast tissue profiling of various
components in breast cancer tissue. A further IMS analysis of human
breast cancer sections revealed three proteins to be specifically
localized in different regions of the cancer tissue (34). IMS of metastatic breast cancer in a
lymph node found protein signals specifically localized in
histologically defined regions (34). An IMS study comparing breast
primary tumors from patients with or without lymph node involvement
highlighted six differently expressed proteins that could predict
lymph node status (125). Besides
efforts to elucidate prognostic biomarkers, also predictive
profiles have been identified. In a study of breast cancer patients
IMS has been shown as a new tool for HER2/neu receptor analysis
(74). This is of special interest
since at present HER2 amplification status is determined by
immunohistochemistry or in situ hybridization or both. These
methods suffer from subjective interpretation as well as from high
costs and are time-consuming (126).
Dekker et al (73) were able to demonstrate mass signals
for stromal activation in breast carcinoma. Furthermore, IMS has
been utilized to assess the distribution of lipids in breast cancer
showing that heterogeneity is not only restricted to the proteome
(127,128).
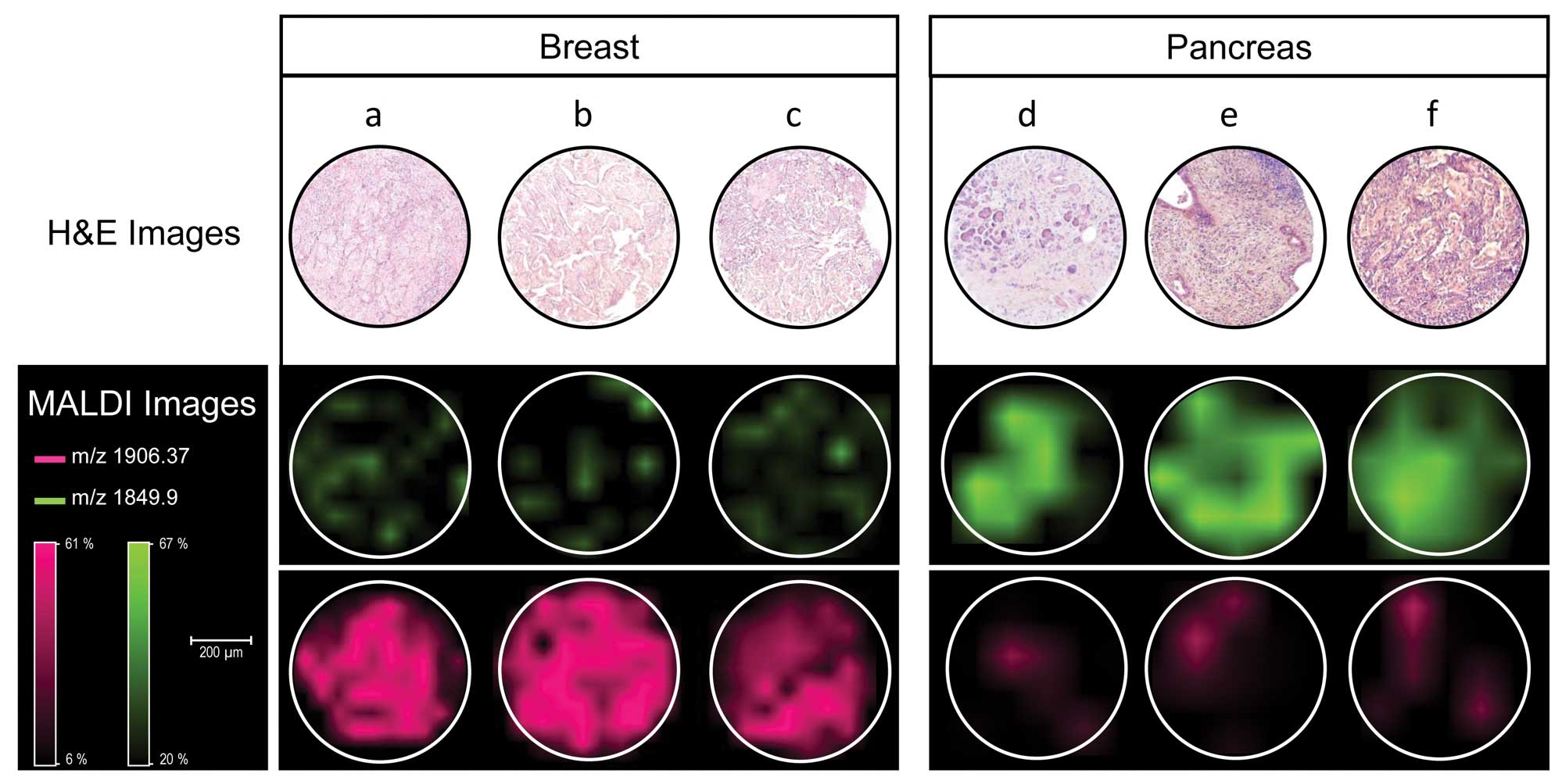
Since most metastases are adenocarcinomas it might
be challenging for the pathologist to determine the origin of the
primary tumor. Casadonte et al (83) developed an algorithm to
discriminate metastases of breast and pancreatic cancer by MALDI
IMS (Fig. 4).
Gastrointestinal tumors
Various studies have addressed MALDI IMS of
gastrointestinal tumors including typing, grading and prognosis
(13,16,35,75,77,79).
Oral
MALDI IMS was applied to human oral biopsies and
revealed a proteomic signature for squamous cell carcinomas
(129).
Esophagus
Several prognostic markers could be identified in
Barrett’s adenocarcinoma (103).
Stomach
IMS was able to discern between the gastric cancer
and the normal gastric mucosa (79). Furthermore, protein profiles from
early stage gastric cancer were significantly different from
advanced stage gastric tumors (79). Also, specific signals for poorly
differentiated gastric cancer were found (16). Choi et al (109) identified proteins as negative
regulators for lymph node metastasis in gastric adenocarcinoma. A
seven-protein signature was associated with unfavorable overall
survival independent of major clinical covariates as demonstrated
by MALDI IMS on frozen tissues (78).
Colon
Thomas et al (130) applied IMS technique to detect
lipids in human colorectal liver metastasis biopsies. With the help
of a tissue based proteomic approach, a large panel of proteins
that are associated with regional lymph node metastasis was
identified (13). The penetration
and distribution of platinum-based metallodrugs was demonstrated in
metastatic tissue of colon cancer by IMS (113).
Pancreas
Djidja et al (81) identified a glucose-regulated
protein as a biomarker within pancreatic tumor tissue
sections. Grüner et al (77) detected several discriminative m/z
species that could distinguish between intraepithelial neoplasia,
intraductal papillary mucinous neoplasm and normal pancreatic
tissue in genetically engineered mouse models. Moreover, metastatic
tissue from breast cancer could be discerned from pancreatic cancer
with an overall accuracy of 83.38%, a sensitivity of 85.95% and a
specificity of 76.96% resulting in a more accurate result than
immunohistochemistry even when several antibodies are applied
(83).
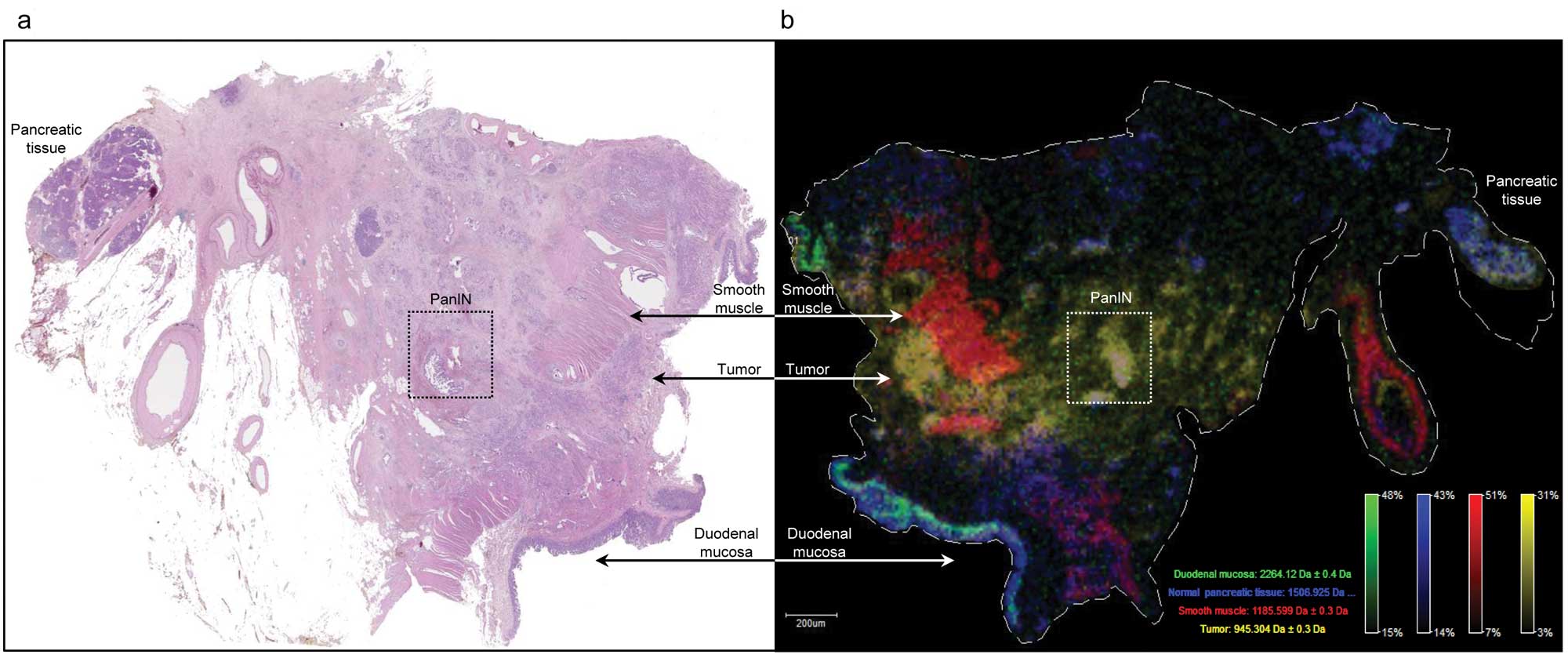
Investigation of large tissue sections of duodenal
mucosa, normal pancreatic tissue and pancreatic carcinoma by MALDI
IMS could clearly discern the various tissue compartments (Fig. 5). This method allows ‘painting’ of
the different tissues by tissue specific peptide signals in tumor
cells (m/z 945), duodenal epithelium (m/z 2264), normal pancreatic
tissue (m/z 1506) and smooth muscle (m/z 1185).
Liver
Le Faouder et al (75) were able to distinguish
hepatocellular carcinoma from cirrhosis by IMS.
Lung
Rahman et al (131) and Yanagisawa et al
(76) reported proteomic patterns
specific for normal alveolar and bronchial tissues, pre-invasive
lesions and invasive lung cancer. Analysis of microarrays of
biopsies from adenocarcinoma and squamous cell carcinoma was
performed by Groseclose et al (67) and could clearly discern both tumor
types without immunohistochemistry. Besides the proteomic profile,
lung cancer could be subclassified also by lipid analysis (132). Proteomic strategies in lung
cancer diagnostics in tissue, blood and serum are described
elsewhere in detail (133).
Notably, EGF-receptor antagonists as target drugs in lung cancer
were characterized in lung cancer tissue (111).
Lymphoma
IMS was applied to Hodgkin’s lymphoma and could
result in pinpointing differentially expressed molecules, which
might represent potential biomarkers. Schwamborn et al
(134) could distinguish between
classical Hodgkin lymphoma and lymphadenitis.
Skin
Skin malignancies, especially melanoma, are another
field where MALDI IMS has been applied successfully. Hardesty et
al (105) found peptides that
discriminate prognostic subgroups in melanoma patients.
Additionally, this group identified signatures associated with
favorable and poor survival. IMS has also been utilized to
differentiate Spitz naevus and melanoma, which is important due to
the completely different therapeutic consequences of both entities
(135). Sugihara et al
(112) introduced IMS to provide
new information on one of the drugs (vemurafenib) currently used in
the treatment of malignant melanoma.
Thyroid
MALDI IMS studies have rarely been performed on
thyroid cancer tissue. IMS techniques have been used to
discriminate thyroid cancer specific peptides from normal thyroid
tissue (136). The authors found
a thyroid cancer specific protein, however, only five patients were
included in the study. Other investigators (14) identified not only a specific
proteomic pattern to discriminate non-metastatic from metastatic
thyroid cancers but showed also a functional relationship between
these proteins and a specific pathway. Pagni et al (66) were able to distinguish between
different thyroid lesions applying IMS to fine needle aspiration
smears. In the present review, unsupervised statistical analysis
and principal component analysis were used to cluster proteomic
spectra between papillary thyroid carcinoma, Hürthle cell adenoma,
medullary thyroid carcinoma and hyperplastic nodules. Different
proteomic signatures were obtained between the histological types
with a receiver operating characteristic (ROC) analysis showing an
area under the curve >0.8, and a Student’s t-test P-value
<0.05. For example, three different proteins detected at 4965,
6278 and 6651 m/z were downregulated in the Hürthle cell adenoma,
medullary thyroid carcinoma and papillary thyroid carcinoma
patients, when compared to patients with hyperplastic nodules.
Nevertheless, by comparing the proteomic profiles obtained from the
Hürthle cell adenoma patients with the medullary thyroid carcinoma
patients, proteins at 7263, 8294 and 8310 m/z were differentially
expressed between the two entities. With these results,
investigators highlighted a potential role of MALDI IMS as a
supporting tool to discriminate malignant from benign thyroid
lesions as well as between different types of thyroid lesions.
Urogenital tract
Kidney
Oppenheimer et al (44) studied tumor margins of renal cell
carcinoma by IMS. Junker et al (137) analyzed stage-related protein
alterations in kidney cancer using MALDI TOF MS/MS but not IMS.
Chinello et al (138) were
able to discriminate patients with renal cell carcinoma from normal
controls by MALDI TOF technique applied to serum. IMS has been used
to identify and map specific peptides that accurately distinguished
malignant from normal renal tissue (72). Axt et al (139) identified spectra from both Wilms’
tumor blastema and stroma that independently classified specimens
according to race with an accuracy of >80%.
Bladder
Oezdemir et al (106) reported improvement of grading of
non-invasive papillary urothelial neoplasia applying IMS. A recent
report utilized IMS for the analysis of a bladder cancer tissue
microarray to evaluate prognostically relevant molecules (140). From the analysis of 697 patients,
numerous m/z signals were found statistically associated with one
or several phenotypes including decrease (12 signals) or increase
(6 signals) of cell proliferation, papillary (3 signals) or solid
(5 signals) growth pattern, and tumor aggressiveness based on
stage, grade and nodal status (30 signals).
Ovary
Lemaire et al (102) not only discriminated between
benign and malignant ovarian tumors, but identified a new candidate
protein biomarker only present in ovarian carcinoma.
Specific proteins in tumor interface zones
associated with ovarian serous cancer tissues, were described by
Kang et al (98). Liu et
al (141) demonstrated that
sulfatides are more abundant in ovarian cancer than control
tissues.
By analysis of tryptic digests of the FFPE tissue
microarray cores by LC-MS/MS, various m/z species observed in MALDI
IMS experiments on ovarian cancer could be correctly identified
(142). Drug distribution of
platinum-based drugs was shown in peritoneal carcinomatosis of
ovarian cancer (113).
Prostate
Studies of prostate cancer have utilized IMS to aid
diagnosis and for the discovery of proteins relevant to the
underlying biology (41,82,143). The first clinical study of human
prostate cancer by IMS was performed by Schwambor et al
(41).
A specific fragment of mitogen activated protein
kinase could discriminate cancer from normal prostatic tissue
(143). Furthermore, a marker for
early recurrence of prostate cancer has been identified by Steurer
et al (82).
Sarcomas
One report utilized IMS for the classification of
myxoid sarcomas. Willems et al (40) could discriminate myxofibrosarcoma
and myxoid liposarcoma that may share a similar histology.
Furthermore, intratumoral heterogeneity was demonstrated in
myxofibrosarcomas.
12. Summary
MALDI IMS is a versatile technique which had been
applied to various organisms including plants, animals and human
tissue. In human tissue and cells, tumor-associated alterations
were studied in various entities. MALDI IMS may improve exact
determination of tumor margins, tumor typing, grading and prognosis
and could have therapeutic implications.
Diagnostic procedures in pathology are based on
histology, histochemistry, immunohistochemistry and molecular
pathology and will include proteomic techniques like MALDI IMS to
improve diagnostic accuracy.
13. Perspectives
MALDI IMS will extend or replace histochemical
techniques such as periodic acid-Schiff- and Prussian blue-reaction
as well as methods that highlight connective tissue compounds as
Masson’s trichrome reaction. Additionally, we foresee that
immunohistochemical methods will be at least partly replaced by
IMS. This is of great importance, especially if molecular testing
is required for diagnostics and only small tissue biopsies are
available for such tests. In this regard, MALDI IMS requires only
one tissue section for the analysis, thus adequate remaining tissue
is available for additional molecular testing. In this scenario,
elaborate molecular testing of DNA and RNA alterations will be
performed either by next generation sequencing or by mass
spectrometric techniques. Applications of IMS in the clinic are
growing as this technology has proved its feasibility and
versatility. The implementation of biocomputational tools combined
with IMS will allow biostatistical evaluation for determination of
molecular signals able to type or grade tumors and will provide
information about prognosis. Furthermore, drug distribution in
different tissue types and cellular components may be monitored by
IMS. To ensure similar sample preparation and processing, robust
standard operating procedures need to be developed for the
application of IMS in routine histopathology. Quality control will
be achieved by interlaboratory validation as well as by sharing
large databases of MALDI signatures of tumors or other
diseases.
References
|
1
|
Patel R: MALDI-TOF mass spectrometry:
transformative proteomics for clinical microbiology. Clin Chem.
59:340–342. 2013. View Article : Google Scholar
|
|
2
|
Clark AE, Kaleta EJ, Arora A and Wolk DM:
Matrix-assisted laser desorption ionization-time of flight mass
spectrometry: a fundamental shift in the routine practice of
clinical microbiology. Clin Microbiol Rev. 26:547–603. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moore JL, Caprioli RM and Skaar EP:
Advanced mass spectrometry technologies for the study of microbial
pathogenesis. Curr Opin Microbiol. 19C:45–51. 2014. View Article : Google Scholar
|
|
4
|
Kueger S, Steinhauser D, Willmitzer L and
Giavalisco P: High-resolution plant metabolomics: from mass
spectral features to metabolites and from whole-cell analysis to
subcellular metabolite distributions. Plant J. 70:39–50. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schoenian I, Spiteller M, Ghaste M, Wirth
R, Herz H and Spiteller D: Chemical basis of the synergism and
antagonism in microbial communities in the nests of leaf-cutting
ants. Proc Natl Acad Sci USA. 108:1955–1960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chaurand P, Cornett DS, Angel PM and
Caprioli RM: From whole-body sections down to cellular level,
multiscale imaging of phospholipids by MALDI mass spectrometry. Mol
Cell Proteomics. 10:O110.004259. 2011. View Article : Google Scholar :
|
|
7
|
Trim PJ, Henson CM, Avery JL, et al:
Matrix-assisted laser desorption/ionization-ion mobility
separation-mass spectrometry imaging of vinblastine in whole body
tissue sections. Anal Chem. 80:8628–8634. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reyzer ML, Chaurand P, Angel PM and
Caprioli RM: Direct molecular analysis of whole-body animal tissue
sections by MALDI imaging mass spectrometry. Methods Mol Biol.
656:285–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Remmerbach TW, Maurer K, Janke S, et al:
Oral brush biopsy analysis by matrix assisted laser
desorption/ionisation-time of flight mass spectrometry profiling: a
pilot study. Oral Oncol. 47:278–281. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schwamborn K, Krieg RC, Uhlig S, Ikenberg
H and Wellmann A: MALDI imaging as a specific diagnostic tool for
routine cervical cytology specimens. Int J Mol Med. 27:417–421.
2011.
|
|
11
|
Kriegsmann M, Seeley EH, Schwarting A, et
al: MALDI MS imaging as a powerful tool for investigating synovial
tissue. Scand J Rheumatol. 41:305–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seeley EH and Caprioli RM: MALDI imaging
mass spectrometry of human tissue: method challenges and clinical
perspectives. Trends Biotechnol. 29:136–143. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meding S, Nitsche U, Balluff B, et al:
Tumor classification of six common cancer types based on proteomic
profiling by MALDI imaging. J Proteome Res. 11:1996–2003. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nipp M, Elsner M, Balluff B, et al:
S100-A10, thioredoxin, and S100-A6 as biomarkers of papillary
thyroid carcinoma with lymph node metastasis identified by MALDI
imaging. J Mol Med. 90:163–174. 2012. View Article : Google Scholar
|
|
15
|
Steurer S, Seddiqi AS, Singer JM, et al:
MALDI imaging on tissue microarrays identifies molecular features
associated with renal cell cancer phenotype. Anticancer Res.
34:2255–2261. 2014.PubMed/NCBI
|
|
16
|
Morita Y, Ikegami K, Goto-Inoue N, et al:
Imaging mass spectrometry of gastric carcinoma in formalin-fixed
paraffin-embedded tissue microarray. Cancer Sci. 101:267–273. 2010.
View Article : Google Scholar
|
|
17
|
Gode D and Volmer DA: Lipid imaging by
mass spectrometry - a review. Analyst. 138:1289–1315. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Horn PJ and Chapman KD: Lipidomics in
situ: insights into plant lipid metabolism from high resolution
spatial maps of metabolites. Prog Lipid Res. 54:32–52. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sparvero LJ, Amoscato AA, Dixon CE, et al:
Mapping of phospholipids by MALDI imaging (MALDI-MSI): realities
and expectations. Chem Phys Lipids. 165:545–562. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harvey DJ: Analysis of carbohydrates and
glycoconjugates by matrix-assisted laser desorption/ionization mass
spectrometry: An update for 2009–2010. Mass Spectrom Rev. May
26–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
21
|
Reyzer ML, Hsieh Y, Ng K, Korfmacher WA
and Caprioli RM: Direct analysis of drug candidates in tissue by
matrix-assisted laser desorption/ionization mass spectrometry. J
Mass Spectrom. 38:1081–1092. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Norris JL and Caprioli RM: Analysis of
tissue specimens by matrix-assisted laser desorption/ionization
imaging mass spectrometry in biological and clinical research. Chem
Rev. 113:2309–2342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Groseclose MR and Castellino S: A mimetic
tissue model for the quantification of drug distributions by MALDI
imaging mass spectrometry. Anal Chem. 85:10099–10106. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Honisch C, Chen Y, Mortimer C, et al:
Automated comparative sequence analysis by base-specific cleavage
and mass spectrometry for nucleic acid-based microbial typing. Proc
Natl Acad Sci USA. 104:10649–10654. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okamoto I, Sakai K, Morita S, et al:
Multiplex genomic profiling of non-small cell lung cancers from the
LETS phase III trial of first-line S-1/carboplatin versus
paclitaxel/carboplatin: results of a West Japan Oncology Group
study. Oncotarget. 5:2293–2304. 2014.PubMed/NCBI
|
|
26
|
Castellino S, Groseclose MR and Wagner D:
MALDI imaging mass spectrometry: bridging biology and chemistry in
drug development. Bioanalysis. 3:2427–2441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rompp A, Guenther S, Takats Z and Spengler
B: Mass spectrometry imaging with high resolution in mass and space
(HR(2) MSI) for reliable investigation of drug compound
distributions on the cellular level. Anal Bioanal Chem. 401:65–73.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caprioli RM, Farmer TB and Gile J:
Molecular imaging of biological samples: localization of peptides
and proteins using MALDI-TOF MS. Anal Chem. 69:4751–4760. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schone C, Hofler H and Walch A: MALDI
imaging mass spectrometry in cancer research: combining proteomic
profiling and histological evaluation. Clin Biochem. 46:539–545.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gessel MM, Norris JL and Caprioli RM:
MALDI imaging mass spectrometry: spatial molecular analysis to
enable a new age of discovery. J Proteomics. 107:71–82. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thomas A, Lenglet S, Chaurand P, et al:
Mass spectrometry for the evaluation of cardiovascular diseases
based on proteomics and lipidomics. Thromb Haemost. 106:20–33.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
McClure RA, Chumbley CW, Reyzer ML, et al:
Identification of promethazine as an amyloid-binding molecule using
a fluorescence high-throughput assay and MALDI imaging mass
spectrometry. Neuroimage Clin. 2:620–629. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kriegsmann M, Casadonte R, Randau T, et
al: MALDI imaging of predictive ferritin, fibrinogen and proteases
in haemophilic arthropathy. Haemophilia. 20:446–453. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seeley EH and Caprioli RM: Molecular
imaging of proteins in tissues by mass spectrometry. Proc Natl Acad
Sci USA. 105:18126–18131. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seeley EH, Washington MK, Caprioli RM and
M’Koma AE: Proteomic patterns of colonic mucosal tissues delineate
Crohn’s colitis and ulcerative colitis. Proteomics Clin Appl.
7:541–549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lalowski M, Magni F, Mainini V, et al:
Imaging mass spectrometry: a new tool for kidney disease
investigations. Nephrol Dial Transplant. 28:1648–1656. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mainini V, Pagni F, Ferrario F, et al:
MALDI imaging mass spectrometry in glomerulonephritis: feasibility
study. Histopathology. 64:901–906. 2014. View Article : Google Scholar
|
|
38
|
Xu BJ, Shyr Y, Liang X, et al: Proteomic
patterns and prediction of glomerulosclerosis and its mechanisms. J
Am Soc Nephrol. 16:2967–2975. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lagarrigue M, Becker M, Lavigne R, et al:
Revisiting rat spermatogenesis with MALDI imaging at 20-microm
resolution. Mol Cell Proteomic. 10:M110.005991. 2011. View Article : Google Scholar
|
|
40
|
Willems SM, van Remoortere A, van Zeijl R,
Deelder AM, McDonnell LA and Hogendoorn PC: Imaging mass
spectrometry of myxoid sarcomas identifies proteins and lipids
specific to tumour type and grade, and reveals biochemical
intratumour heterogeneity. J Pathol. 222:400–409. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schwamborn K, Krieg RC, Reska M, Jakse G,
Knuechel R and Wellmann A: Identifying prostate carcinoma by
MALDI-Imaging. Int J Mol Med. 20:155–159. 2007.PubMed/NCBI
|
|
42
|
Rodrigo MA, Zitka O, Krizkova S, Moulick
A, Adam V and Kizek R: MALDI-TOF MS as evolving cancer diagnostic
tool: a review. J Pharm Biomed Anal. 95:245–255. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rahman SM, Gonzalez AL, Li M, et al: Lung
cancer diagnosis from proteomic analysis of preinvasive lesions.
Cancer Res. 71:3009–3017. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oppenheimer SR, Mi D, Sanders ME and
Caprioli RM: Molecular analysis of tumor margins by MALDI mass
spectrometry in renal carcinoma. J Proteome Res. 9:2182–2190. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jones EE, Powers TW, Neely BA, et al:
MALDI imaging mass spectrometry profiling of proteins and lipids in
clear cell renal cell carcinoma. Proteomics. 14:924–935. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Angel PM and Caprioli RM: Matrix-assisted
laser desorption ionization imaging mass spectrometry: in situ
molecular mapping. Biochemistry. 52:3818–3828. 2013. View Article : Google Scholar
|
|
47
|
Boggio KJ, Obasuyi E, Sugino K, Nelson SB,
Agar NY and Agar JN: Recent advances in single-cell MALDI mass
spectrometry imaging and potential clinical impact. Expert Rev
Proteomics. 8:591–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Eriksson C, Masaki N, Yao I, Hayasaka T
and Setou M: MALDI imaging mass spectrometry-A mini review of
methods and recent developments. Mass Spectrom. 2:S00222013.
View Article : Google Scholar
|
|
49
|
Dreisewerd K: Recent methodological
advances in MALDI mass spectrometry. Anal Bioanal Chem.
406:2261–2278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fujimura Y and Miura D: MALDI mass
spectrometry imaging for visualizing in situ metabolism of
endogenous metabolites and dietary phytochemicals. Metabolites.
4:319–346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gorzolka K and Walch A: MALDI mass
spectrometry imaging of formalin-fixed paraffin-embedded tissues in
clinical research. Histol Histopathol. 29:1365–1376.
2014.PubMed/NCBI
|
|
52
|
Schwamborn K and Caprioli RM: Molecular
imaging by mass spectrometry: looking beyond classical histology.
Nat Rev Cancer. 10:639–646. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chaurand P: Imaging mass spectrometry of
thin tissue sections: a decade of collective efforts. J Proteomics.
75:4883–4892. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rompp A and Spengler B: Mass spectrometry
imaging with high resolution in mass and space. Histochem Cell
Biol. 139:759–783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Goodwin RJ: Sample preparation for mass
spectrometry imaging: small mistakes can lead to big consequences.
J Proteomics. 75:4893–4911. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kaletas BK, van der Wiel IM, Stauber J, et
al: Sample preparation issues for tissue imaging by imaging MS.
Proteomics. 9:2622–2633. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Stoeckli M, Chaurand P, Hallahan DE and
Caprioli RM: Imaging mass spectrometry: a new technology for the
analysis of protein expression in mammalian tissues. Nat Med.
7:493–496. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Cornett DS, Mobley JA, Dias EC, et al: A
novel histology-directed strategy for MALDI-MS tissue profiling
that improves throughput and cellular specificity in human breast
cancer. Mol Cell Proteomics. 5:1975–1983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Seeley EH, Schwamborn K and Caprioli RM:
Imaging of intact tissue sections: moving beyond the microscope. J
Biol Chem. 286:25459–25466. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zavalin A, Yang J and Caprioli R: Laser
beam filtration for high spatial resolution MALDI imaging mass
spectrometry. J Am Soc Mass Spectrom. 24:1153–1156. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Caprioli RM: Imaging mass spectrometry:
molecular microscopy for enabling a new age of discovery.
Proteomics. 14:807–809. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Norris JL, Cornett DS, Mobley JA, et al:
Processing MALDI Mass Spectra to improve mass spectral direct
tissue analysis. Int J Mass Spectrom. 260:212–221. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Djidja MC, Claude E, Snel MF, et al: Novel
molecular tumour classification using MALDI-mass spectrometry
imaging of tissue microarray. Anal Bioanal Chem. 397:587–601. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Andersson M, Groseclose MR, Deutch AY and
Caprioli RM: Imaging mass spectrometry of proteins and peptides: 3D
volume reconstruction. Nat Methods. 5:101–108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Crecelius AC, Cornett DS, Caprioli RM,
Williams B, Dawant BM and Bodenheimer B: Three-dimensional
visualization of protein expression in mouse brain structures using
imaging mass spectrometry. J Am Soc Mass Spectrom. 16:1093–1099.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Pagni F, Mainini V, Garancini M, et al:
Proteomics for the diagnosis of thyroid lesions: preliminary
report. Cytopathology. Jul 20–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
67
|
Groseclose MR, Massion PP, Chaurand P and
Caprioli RM: High-throughput proteomic analysis of formalin-fixed
paraffin-embedded tissue microarrays using MALDI imaging mass
spectrometry. Proteomics. 8:3715–3724. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Gancberg D, Jarvinen T, di Leo A, et al:
Evaluation of HER-2/NEU protein expression in breast cancer by
immunohistochemistry: an interlaboratory study assessing the
reproducibility of HER-2/NEU testing. Breast Cancer Res Treat.
74:113–120. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Thaysen-Andersen M, Wilkinson BL, Payne RJ
and Packer NH: Site-specific characterisation of densely
O-glycosylated mucin-type peptides using electron transfer
dissociation ESI-MS/MS. Electrophoresis. 32:3536–3545. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ronci M, Sharma S, Chataway T, et al:
MALDI-MS-imaging of whole human lens capsule. J Proteome Res.
10:3522–3529. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Bona A, Papai Z, Maasz G, et al: Mass
spectrometric identification of ancient proteins as potential
molecular biomarkers for a 2000-year-old osteogenic sarcoma. PLoS
One. 9:e872152014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Morgan TM, Seeley EH, Fadare O, Caprioli
RM and Clark PE: Imaging the clear cell renal cell carcinoma
proteome. J Urol. 189:1097–1103. 2013. View Article : Google Scholar :
|
|
73
|
Dekker TJ, Balluff BD, Jones EA, et al:
Multicenter Matrix-assisted laser desorption/ionization mass
spectrometry imaging (MALDI MSI) identifies proteomic differences
in breast-cancer-associated stroma. J Proteome Res. May
2–2014.(Epub ahead of print). View Article : Google Scholar
|
|
74
|
Rauser S, Marquardt C, Balluff B, et al:
Classification of HER2 receptor status in breast cancer tissues by
MALDI imaging mass spectrometry. J Proteome Res. 9:1854–1863. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Le Faouder J, Laouirem S, Chapelle M, et
al: Imaging mass spectrometry provides fingerprints for
distinguishing hepatocellular carcinoma from cirrhosis. J Proteome
Res. 10:3755–3765. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yanagisawa K, Shyr Y, Xu BJ, et al:
Proteomic patterns of tumour subsets in non-small-cell lung cancer.
Lancet. 362:433–439. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gruner BM, Hahne H, Mazur PK, et al: MALDI
imaging mass spectrometry for in situ proteomic analysis of
preneoplastic lesions in pancreatic cancer. PLoS One. 7:e394242012.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Balluff B, Rauser S, Meding S, et al:
MALDI imaging identifies prognostic seven-protein signature of
novel tissue markers in intestinal-type gastric cancer. Am J
Pathol. 179:2720–2729. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Kim HK, Reyzer ML, Choi IJ, et al: Gastric
cancer-specific protein profile identified using endoscopic biopsy
samples via MALDI mass spectrometry. J Proteome Res. 9:4123–4130.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Casadonte R and Caprioli RM: Proteomic
analysis of formalin-fixed paraffin-embedded tissue by MALDI
imaging mass spectrometry. Nat Protoc. 6:1695–1709. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Djidja MC, Chang J, Hadjiprocopis A, et
al: Identification of hypoxia-regulated proteins using MALDI-mass
spectrometry imaging combined with quantitative proteomics. J
Proteome Res. 13:2297–2313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Steurer S, Borkowski C, Odinga S, et al:
MALDI mass spectrometric imaging based identification of clinically
relevant signals in prostate cancer using large-scale tissue
microarrays. Int J Cancer. 133:920–928. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Casadonte R, Kriegsmann M, Zweynert F, et
al: Imaging mass spectrometry to discriminate breast from
pancreatic cancer metastasis in formalin-fixed paraffin-embedded
tissues. Proteomics. 14:956–964. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ikeda K, Monden T, Kanoh T, et al:
Extraction and analysis of diagnostically useful proteins from
formalin-fixed, paraffin-embedded tissue sections. J Histochem
Cytochem. 46:397–403. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Becker KF, Schott C, Hipp S, et al:
Quantitative protein analysis from formalin-fixed tissues:
implications for translational clinical research and nanoscale
molecular diagnosis. J Pathol. 211:370–378. 2007. View Article : Google Scholar
|
|
86
|
Chung JY, Lee SJ, Kris Y, Braunschweig T,
Traicoff JL and Hewitt SM: A well-based reverse-phase protein array
applicable to extracts from formalin-fixed paraffin-embedded
tissue. Proteomics Clin Appl. 2:1539–1547. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nirmalan NJ, Harnden P, Selby PJ and Banks
RE: Development and validation of a novel protein extraction
methodology for quantitation of protein expression in
formalin-fixed paraffin-embedded tissues using western blotting. J
Pathol. 217:497–506. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Crockett DK, Lin Z, Vaughn CP, Lim MS and
Elenitoba-Johnson KS: Identification of proteins from
formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest.
85:1405–1415. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hood BL, Darfler MM, Guiel TG, et al:
Proteomic analysis of formalin-fixed prostate cancer tissue. Mol
Cell Proteomics. 4:1741–1753. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Palmer-Toy DE, Krastins B, Sarracino DA,
Nadol JB Jr and Merchant SN: Efficient method for the proteomic
analysis of fixed and embedded tissues. J Proteome Res.
4:2404–2411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Becker KF, Berg D, Malinowsky K, et al:
Update on protein analysis of fixed tissues. Pathologe. 31(Suppl
2): 263–267. 2010. View Article : Google Scholar
|
|
92
|
Shi SR, Liu C, Balgley BM, Lee C and
Taylor CR: Protein extraction from formalin-fixed,
paraffin-embedded tissue sections: quality evaluation by mass
spectrometry. J Histochem Cytochem. 54:739–743. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
McDonnell LA, Walch A, Stoeckli M and
Corthals GL: MSiMass list: a public database of identifications for
protein MALDI MS imaging. J Proteome Res. 13:1138–1142. 2014.
View Article : Google Scholar
|
|
94
|
Maier SK, Hahne H, Gholami AM, et al:
Comprehensive identification of proteins from MALDI imaging. Mol
Cell Proteomics. 12:2901–2910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Deininger SO, Ebert MP, Futterer A,
Gerhard M and Rocken C: MALDI imaging combined with hierarchical
clustering as a new tool for the interpretation of complex human
cancers. J Proteome Res. 7:5230–5236. 2008. View Article : Google Scholar
|
|
96
|
Caprioli RM: Deciphering protein molecular
signatures in cancer tissues to aid in diagnosis, prognosis, and
therapy. Cancer Res. 65:10642–10645. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chaurand P, Sanders ME, Jensen RA and
Caprioli RM: Proteomics in diagnostic pathology: profiling and
imaging proteins directly in tissue sections. Am J Pathol.
165:1057–1068. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Kang S, Shim HS, Lee JS, et al: Molecular
proteomics imaging of tumor interfaces by mass spectrometry. J
Proteome Res. 9:1157–1164. 2010. View Article : Google Scholar
|
|
99
|
Han EC, Lee YS, Liao WS, Liu YC, Liao HY
and Jeng LB: Direct tissue analysis by MALDI-TOF mass spectrometry
in human hepatocellular carcinoma. Clin Chim Acta. 412:230–239.
2011. View Article : Google Scholar
|
|
100
|
Eberlin LS, Norton I, Orringer D, et al:
Ambient mass spectrometry for the intraoperative molecular
diagnosis of human brain tumors. Proc Natl Acad Sci USA.
110:1611–1616. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Schwartz SA, Weil RJ, Thompson RC, et al:
Proteomic-based prognosis of brain tumor patients using
direct-tissue matrix-assisted laser desorption ionization mass
spectrometry. Cancer Res. 65:7674–7681. 2005.PubMed/NCBI
|
|
102
|
Lemaire R, Menguellet SA, Stauber J, et
al: Specific MALDI imaging and profiling for biomarker hunting and
validation: fragment of the 11S proteasome activator complex, Reg
alpha fragment, is a new potential ovary cancer biomarker. J
Proteome Res. 6:4127–4134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Elsner M, Rauser S, Maier S, et al: MALDI
imaging mass spectrometry reveals COX7A2, TAGLN2 and S100-A10 as
novel prognostic markers in Barrett’s adenocarcinoma. J Proteomics.
75:4693–4704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Flatley B, Malone P and Cramer R: MALDI
mass spectrometry in prostate cancer biomarker discovery. Biochim
Biophys Acta. 1844:940–949. 2014. View Article : Google Scholar
|
|
105
|
Hardesty WM, Kelley MC, Mi D, Low RL and
Caprioli RM: Protein signatures for survival and recurrence in
metastatic melanoma. J Proteomics. 74:1002–1014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Oezdemir RF, Gaisa NT, Lindemann-Docter K,
et al: Proteomic tissue profiling for the improvement of grading of
noninvasive papillary urothelial neoplasia. Clin Biochem. 45:7–11.
2012. View Article : Google Scholar
|
|
107
|
Chaurand P, Schwartz SA, Billheimer D, Xu
BJ, Crecelius A and Caprioli RM: Integrating histology and imaging
mass spectrometry. Anal Chem. 76:1145–1155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Schwartz SA, Weil RJ, Johnson MD, Toms SA
and Caprioli RM: Protein profiling in brain tumors using mass
spectrometry: feasibility of a new technique for the analysis of
protein expression. Clin Cancer Res. 10:981–987. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Choi JH, Shin NR, Moon HJ, et al:
Identification of S100A8 and S100A9 as negative regulators for
lymph node metastasis of gastric adenocarcinoma. Histol
Histopathol. 27:1439–1448. 2012.PubMed/NCBI
|
|
110
|
Reyzer ML, Caldwell RL, Dugger TC, et al:
Early changes in protein expression detected by mass spectrometry
predict tumor response to molecular therapeutics. Cancer Res.
64:9093–9100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Vegvari A, Fehniger TE, Rezeli M, et al:
Experimental models to study drug distributions in tissue using
MALDI mass spectrometry imaging. J Proteome Res. 12:5626–5633.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Sugihara Y, Vegvari A, Welinder C, et al:
A new look at drugs targeting malignant melanoma - an application
for mass spectrometry imaging. Proteomics. 14:1963–1970. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bianga J, Bouslimani A, Bec N, et al:
Complementarity of MALDI and LA ICP mass spectrometry for platinum
anticancer imaging in human tumor. Metallomics. 6:1382–1386. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Liu X, Ide JL, Norton I, et al: Molecular
imaging of drug transit through the blood-brain barrier with MALDI
mass spectrometry imaging. Sci Rep. 3:28592013.PubMed/NCBI
|
|
115
|
Buck A and Walch A: In situ drug and
metabolite analyzes in biological and clinical research by MALDI MS
imaging. Bioanalysis. 6:1241–1253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jirasko R, Holcapek M, Kunes M and Svatos
A: Distribution study of atorvastatin and its metabolites in rat
tissues using combined information from UHPLC/MS and
MALDI-Orbitrap-MS imaging. Anal Bioanal Chem. 406:4601–4610. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Huber K, Aichler M, Sun N, et al: A rapid
ex vivo tissue model for optimising drug detection and ionisation
in MALDI imaging studies. Histochem Cell Biol. 142:361–371. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Yasunaga M, Furuta M, Ogata K, et al: The
significance of microscopic mass spectrometry with high resolution
in the visualisation of drug distribution. Sci Rep. 3:30502013.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Salphati L, Shahidi-Latham S, Quiason C,
et al: Distribution of the phosphatidylinositol 3-kinase inhibitors
Pictilisib (GDC-0941) and GNE-317 in U87 and GS2 intracranial
glioblastoma models-assessment by matrix-assisted laser desorption
ionization imaging. Drug Metab Dispos. 42:1110–1116. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Zecchi R, Trevisani M, Pittelli M, et al:
Impact of drug administration route on drug delivery and
distribution into the lung: an imaging mass spectrometry approach.
Eur J Mass Spectrom. 19:475–482. 2013. View Article : Google Scholar
|
|
121
|
Bouamrani A, Ternier J, Ratel D, et al:
Direct-tissue SELDI-TOF mass spectrometry analysis: a new
application for clinical proteomics. Clin Chem. 52:2103–2106. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Agar NY, Malcolm JG, Mohan V, et al:
Imaging of meningioma progression by matrix-assisted laser
desorption ionization time-of-flight mass spectrometry. Anal Chem.
82:2621–2625. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Ronci M, Bonanno E, Colantoni A, et al:
Protein unlocking procedures of formalin-fixed paraffin-embedded
tissues: application to MALDI-TOF imaging MS investigations.
Proteomics. 8:3702–3714. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Qin XJ and Ling BX: Proteomic studies in
breast cancer (Review). Oncol Lett. 3:735–743. 2012.PubMed/NCBI
|
|
125
|
Seeley EH and Caprioli RM: Imaging mass
spectrometry: towards clinical diagnostics. Proteomics Clin Appl.
2:1435–1443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Walch A, Rauser S, Deininger SO and Hofler
H: MALDI imaging mass spectrometry for direct tissue analysis: a
new frontier for molecular histology. Histochem Cell Biol.
130:421–434. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Amstalden van Hove ER, Blackwell TR,
Klinkert I, Eijkel GB, Heeren RM and Glunde K: Multimodal mass
spectrometric imaging of small molecules reveals distinct
spatio-molecular signatures in differentially metastatic breast
tumor models. Cancer Res. 70:9012–9021. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Chughtai K, Jiang L, Greenwood TR, Glunde
K and Heeren RM: Mass spectrometry images acylcarnitines,
phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor
models. J Lipid Res. 54:333–344. 2013. View Article : Google Scholar :
|
|
129
|
Patel SA, Barnes A, Loftus N, et al:
Imaging mass spectrometry using chemical inkjet printing reveals
differential protein expression in human oral squamous cell
carcinoma. Analyst. 134:301–307. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Thomas A, Patterson NH, Marcinkiewicz MM,
Lazaris A, Metrakos P and Chaurand P: Histology-driven data mining
of lipid signatures from multiple imaging mass spectrometry
analyses: application to human colorectal cancer liver metastasis
biopsies. Anal Chem. 85:2860–2866. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Rahman SM, Shyr Y, Yildiz PB, et al:
Proteomic patterns of preinvasive bronchial lesions. Am J Respir
Crit Care Med. 172:1556–1562. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Lee GK, Lee HS, Park YS, et al: Lipid
MALDI profile classifies non-small cell lung cancers according to
the histologic type. Lung Cancer. 76:197–203. 2012. View Article : Google Scholar
|
|
133
|
Hassanein M, Rahman JS, Chaurand P and
Massion PP: Advances in proteomic strategies toward the early
detection of lung cancer. Proc Am Thorac Soc. 8:183–188. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Schwamborn K, Krieg RC, Jirak P, et al:
Application of MALDI imaging for the diagnosis of classical Hodgkin
lymphoma. J Cancer Res Clin Oncol. 136:1651–1655. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Lazova R, Seeley EH, Keenan M, Gueorguieva
R and Caprioli RM: Imaging mass spectrometry - a new and promising
method to differentiate Spitz nevi from Spitzoid malignant
melanomas. Am J Dermatopathol. 34:82–90. 2012. View Article : Google Scholar :
|
|
136
|
Min KW, Bang JY, Kim KP, et al: Imaging
mass spectrometry in papillary thyroid carcinoma for the
identification and validation of biomarker proteins. J Korean Med
Sci. 29:934–940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Junker H, Venz S, Zimmermann U, Thiele A,
Scharf C and Walther R: Stage-related alterations in renal cell
carcinoma-comprehensive quantitative analysis by 2D-DIGE and
protein network analysis. PLoS One. 6:e218672011. View Article : Google Scholar
|
|
138
|
Chinello C, Gianazza E, Zoppis I, et al:
Serum biomarkers of renal cell carcinoma assessed using a protein
profiling approach based on ClinProt technique. Urology.
75:842–847. 2010. View Article : Google Scholar
|
|
139
|
Axt J, Murphy AJ, Seeley EH, et al: Race
disparities in Wilms tumor incidence and biology. J Surg Res.
170:112–119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Steurer S, Singer JM, Rink M, et al: MALDI
imaging-based identification of prognostically relevant signals in
bladder cancer using large-scale tissue microarrays. Urol Oncol.
Aug 14–2014. View Article : Google Scholar : (Epub ahead of
print).
|
|
141
|
Liu Y, Chen Y, Momin A, et al: Elevation
of sulfatides in ovarian cancer: an integrated transcriptomic and
lipidomic analysis including tissue-imaging mass spectrometry. Mol
Cancer. 9:1862010. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Meding S, Martin K, Gustafsson OJ, et al:
Tryptic peptide reference data sets for MALDI imaging mass
spectrometry on formalin-fixed ovarian cancer tissues. J Proteome
Res. 12:308–315. 2013. View Article : Google Scholar
|
|
143
|
Cazares LH, Troyer D, Mendrinos S, et al:
Imaging mass spectrometry of a specific fragment of
mitogen-activated protein kinase/extracellular signal-regulated
kinase kinase kinase 2 discriminates cancer from uninvolved
prostate tissue. Clin Cancer Res. 15:5541–5551. 2009. View Article : Google Scholar : PubMed/NCBI
|