Introduction
Hepatitis B virus (HBV), a hepadnavirus, is a
well-known cause of acute and chronic hepatitis, liver cirrhosis
and hepatocellular carcinoma (HCC). In 2005, ~400 million
individuals carried the virus worldwide, of which >250 million
resided in Asia (1). The HBV
genome is a 3.2-kb circular, partially double-stranded DNA molecule
with 4 overlapping open reading frames (ORFs): PreC-C, PreS-S, P,
and X-ORF. The expression of core and polymerase polypeptides, the
large surface antigen polypeptide, the middle and major surface
antigen polypeptides and the HBV X protein (HBx) polypeptide are
directed by 4 HBV promoters: Cp, PS1p, Sp and Xp, respectively, and
influenced by 2 HBV enhancers, enhancer I and enhancer II (2,3).
Mammalian hepadnaviruses encode the small regulatory
protein HBx. This is transcribed from the X gene independently from
the other viral transcripts under the control of enhancer I and X
promoter. HBx is a small, conserved viral protein that is 154 aa in
length with a number of amino acid substitutions, including those
at codons 94 (4), 31 (5) and 38 (6). It is a multifunctional regulator that
modulates a variety of host processes through directly or
indirectly interacting with virus and host proteins (7–9). HBx
is a promiscuous transactivator that can activate a variety of
viral and cellular promoters and enhancers (10–12).
No clear counterpart exists in the nononcogenic family of avian
hepatitis viruses. HBx has long been suspected to potentiate
hepatocarcinogenesis as HCC has not been observed in avians
infected with avian hepadnaviruses in which X-ORF is absent. The
X-ORF is partly overlapped by the P ORF at the N-terminal, the PreC
ORF and several cis-elements of the PreC promoter at the
C-terminal. Therefore, mutations in the C-terminal portion may
result in mutations of HBx and the cis-elements at the same
time.
In 1990, Loncarevic et al (13) discovered a small ORF upstream of
X-ORF, which was designated the pre-X region, that can be expressed
in-frame with the X-ORF. Pre-X is 168 nt in length and encodes an
additional 56 aa. This implies that the whole of X (PreX-X) encodes
a 210-aa protein. A previous study demonstrated that the strain
coding the pre-X region is common in China (14). Additionally, a study by Takahashi
et al (15) suggested that
the pre-X region is associated with HCC.
Throughout the course of chronic HBV infection
(CHB), the loss of hepatitis B ‘e’ antigen (HBeAg) and the
appearance of antibodies directed against it (anti-HBe) are
commonly accompanied by the cessation of viral replication. Such a
serological profile may be observed in patients who have precore
(PreC) and basal core promoter (BCP) mutants. HBeAg-negative HBV
results from the common genomic mutation; G1896A, which converts
codon 28 of the precore sequence to a termination codon
(TGG>TAG) and thus prevents the expression of HBeAg (16). Another group of mutations affect
the BCP region and this results in a transcriptional reduction of
precore mRNA, but not pregenomic or core mRNA. These
HBeAg-suppressive strains contain the mutations A1762T and G1764A
in the BCP region and are the predominant quasispecies in patients
with chronic hepatitis (17,18).
These changes are associated with the HBeAg-negative CHB, but
another study demonstrated that these mutants are also found in
certain HBeAg-positive patients, particularly those with CHB. This
study suggests that HBeAg may be a target antigen on HBV-infected
hepatocytes, and the failure to produce HBeAg may be a means of
evading immune clearance. Nevertheless, the significance of
additional mutations in the precore and core promoter (CP) regions
for HBeAg production is unknown (19).
A deletion mutation of the X gene/CP region was
previously observed in one patient undergoing HBeAg seroconversion
spontaneously, with a length of 234 bp (accession numbers:
EF608587-EF608597) (20). In the
present study, sequences bearing the PX-X region were investigated,
in addition to the enhancer II, the CP, and the precore start codon
in order to identify the specific mutations associated with
HBeAg-negative CHB.
Materials and methods
Patients
A total of 21 patients were consecutively analyzed:
7 HBeAg-positive and 14 HBeAg-negative; one HBeAg-negative patient
was also HB ‘s’ antigen-negative. All patient profiles were
maintained by routine testing and confirmed by repeat tests every
3–6 months at least twice. A total of 19 patients were inferred to
suffer from CHB and 2 patients were diagnosed with liver cirrhosis.
Of the patients, 2 were brothers (X441 and X464), 2 were mother and
son (X476 and X475, respectively), and 3 were mother, daughter and
son (X461, X462 and X463, respectively). Patients were excluded if
they exhibited evidence of autoimmune hepatitis or markers of
hepatitis C virus, hepatitis D virus (HDV), or human
immunodeficiency virus. Blood samples were collected and the sera
stored at −80°C until use. The present study was conducted in
accordance with the Declaration of Helsinki (1964). Approval from
the Ethics Committee of Zhongshan Hospital (Xiamen, China), and
written informed consent from all participants, was obtained for
the current study.
Polymerase chain reaction (PCR)
DNA was extracted from 200 μl serum with a QIAamp
DNA Mini kit (Qiagen, Hilden, Germany), according to the
manufacturer’s instructions. The HBV PreX/X genomic region was
amplified by PCR using the sense primer PX1
(5′-CAAGTGTTTGCTGACGCAACC-3′, nt 1176–1198) and antisense primer S2
(5′-ACAGCTTGGCGGCTTGAACAG-3′, nt 1859–1880), which amplifies the
entire HBV PX/X region. PCR assays were performed under the
following conditions: 94°C for 3 min, 35 cycles of 94°C for 40 sec,
58°C for 40 sec and 72°C for 40 sec, followed by a final extension
of 72°C for 10 min. The resulting PCR products (25 μl) were loaded
onto a 1% agarose gel and electrophoresed. The DNA band was excised
and purified and then ligated with the pMD19 T vector (Takara Bio,
Inc., Dalian, China). The recombinant plasmids were transformed
into TOP 10 (Tiangen Biotech, Inc., Beijing, China) and screened on
ampicillin/Xgal-IPTG plates, then identified by restriction enzyme
(Takara Bio, Inc., Dalian, China). digestion. The sequencing
reactions were analyzed using the ABI3730 automated sequencer
(Invitrogen Life Technologies, Shanghai, China). The sequences
reported in the current study have been deposited into GenBank
(http://www.ncbi.nlm.nih.gov/genbank/)
under the following accession numbers: EU043336-EU043352.
Phylogenetic analysis
Nucleotide and amino acid sequences were aligned
using Vector NTI, version 10.0 (InforMax Inc., Bethesda, MD,
USA).
Results
PCR
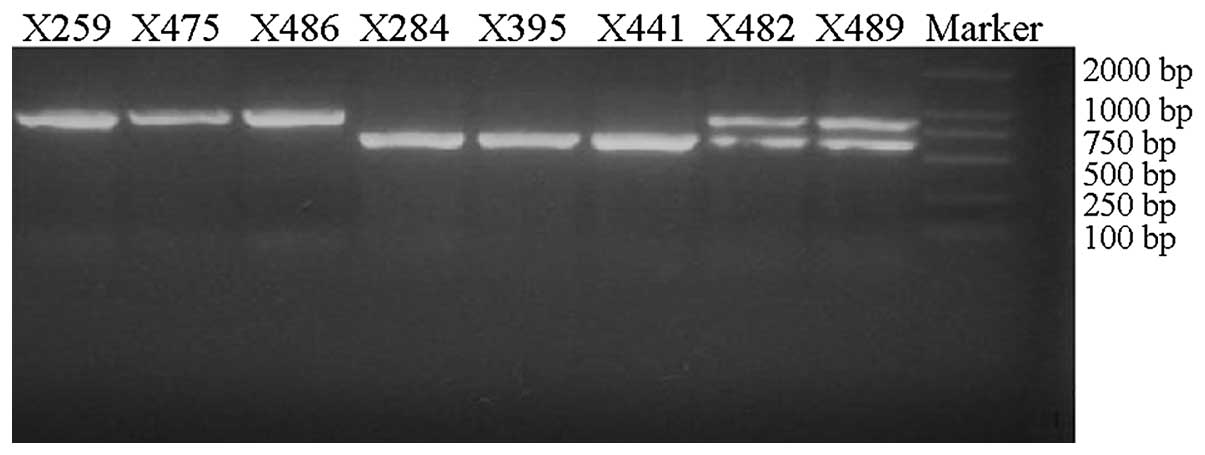
The predicted length of the target region, PreX/X or
the CP region was 704 bp. Electrophoresis of the PCR products
yielded 2 bands following restriction digestion: One that was ~700
bp, and another that was ~500 bp (Fig.
1). This suggests that a 200-bp deletion mutation may reside in
the target region. The map also implies that in different patients
there are 3 types of PreX/X region: Prototype (X259), deletion
mutants (X284) and coexistence of prototype and deletion mutant
(X482).
HBV genomic mutations
Using the electrophoresis results, the PCR products
of various lengths were ligated into the pMD19 T vector. Subsequent
to screening out positive clones, at least 2 clones were selected
out of the recombinant plasmid groups from 1 patient for DNA
sequencing. All sequences were deposited in the GenBank with the
accession numbers EU043336-EU043352. Of these, EU043342, EU043343,
EU043344, EU043345, EU043349 and EU043350 had no deletion
mutations; EU043351 (XX278-3) had a 245-bp deletion mutation,
whilst all others had a 234-bp deletion mutation. None of these
clones code for the pre-X region.
In the present study, 74 clones from 21 patients
were sequenced (Table I). A total
of 54 clones from 19 patients exhibited deletion mutations with a
length of 234 nt, at identical positions (nt 1601–1834). The
deletion region was found downstream of the X gene (nt 1372–1836)
and overlapped the CP (nt 1643–1849). Due to the location of the
deletion site, it was named the core promoter deletion (CPD). In
addition to the CPD, clone XX278-3 (EU043351) also had an 11-nt
deletion, making an overall total of 55 deletion mutations in all
patients. The 2 samples X475 and X486, which were HBeAg-negative,
had no CPD mutants in their recombinant plasmids groups. Notably,
55 clones had 3 point replacement mutations: G/A1515C, G1518C and
A1585T. The sequencing results display that the CPD tandem occurred
with the replacement mutations 1585T, 1518C and 1518C, or
simultaneously.
 | Table ICharacteristics of patients, and
deletion mutations. |
Table I
Characteristics of patients, and
deletion mutations.
| HBeAg-positive | HBeAg-negative |
|---|
| Total patients | 7 | 14 |
| Age (years, mean ±
SD) | 37.4±12.3 | 35.7±16.5 |
| Infection history
(years) | 8.3±11.4 | 9.6±15.4 |
| Family history
(years) | 4 | 7 |
| Sequencing
quantity | 27 | 47 |
| Deletion
mutations | 20 | 35 |
| No deletion
mutation | 7 | 12 |
In addition, sequences of 6 clones from the
brothers, X441 (4) and X464
(2), were identical with CPD. The
sequences of the mother X461 (2),
daughter X462 (3) and the son X463
(2) were also identical with the
CPD. However, there were differences in a number of clones from the
mother (X476) and son (X475); 4 clones from the mother were CPD
mutants, but none of the 4 clones from the son were.
Out of 19 strains without CPD, 2 had a double A1762T
and G1764A nucleotide exchange, XX278-2 and XX278-12, and were from
HBeAg-positive patients. A total of 12 strains from HBeAg-negative
patients had no such mutation.
HBV protein mutations
The 54 clones bearing CPD encoded a truncated HBx
that was only 76 aa in length, so was more like a factor than a
protein. There were 9 different truncated coding patterns of HBx in
these 54 clones (Figs. 2 and
3). A total of 41 strains from 18
patients coded a characteristic amino acid substitution at amino
acids 44 and 45. These CPD strains coded as LL (one is PL), but
those without a CPD coded VV/IV. The replacement mutations were
either a G/A1515C or G1518C. Strain XX278-3, with a CPD that was
245 nt long, coded no HBx-like protein or factor.
Discussion
During CHB, two major HBV core gene variants
frequently occur that affect the expression of HBeAg: i) The PreC
mutants; and ii) the BCP mutants (21,22).
In a previous study, the PreC mutants and the BCP mutants from a
patient undergoing a spontaneous HBeAg seroconversion were
analyzed. In the present study, 20 clones (EF608587-EF608597) with
a long deletion mutation in the CP region that was up to 234 nt in
length were discovered, which was termed CPD. The CPD not only
resulted in deletion of the start codon from enhancer II and preC,
but also produced truncated HBx expression. This is a novel
mutation pattern resulting in HBeAg-negative CHB. In the present
study, samples were collected from 21 patients and the genetic
variability of PreX-X/CP region was analyzed. Of them, 7 were
HBeAg-positive, and 14 were HBeAg-negative. A total of 55 out of 74
(EU043336-EU043352) strains from 19 patients had a CPD. There was 1
strain with a deletion length of 245 nt, whilst the other 54
strains had a deletion length of 234 nt (the location of the latter
deletion was from nt 1601 to 1834) (Fig. 3). In total, 64 strains were
discovered from 20 patients bearing an identical CPD mutation
position in the HBV genome. One deletion mutant was 11 nt longer
than the other 234-nt CPD. CPD prevents the function of the CP, so
the preC start codon is absent, and HBeAg is not expressed. In the
present study, the HBeAg-negative patients did not have a markedly
higher frequency of CPD than that of HBeAg-positive patients.
However, CPD mutants were found in strains from all 7
HBeAg-positive patients, but only in 12 out of 14 HBeAg-negative
patients. Previous studies have suggested that HBV exists as a
quasispecies of wild type and mutant in the HBeAg-positive
patients. A total of 7 strains from 2 HBeAg-negative patients have
no CPD mutation, nor A1762T and/or G1764A replacement mutation. In
the current study, 7 patients from 3 families were included and CPD
mutants were only found in 2 of the families. This implies that the
CPD mutants could be transmitted between family members. In
addition, the CPD mutations occur as 3 point replacement mutations;
G/A1515C, G1518C and A1585T. In view of the data produced in the
current study, we propose that there is an association between the
mutations at nt 1515, 1518 and 1585 and the deletion at nt
1601–1834.
HBx is the only nonstructural regulatory protein of
HBV, and has been demonstrated to be associated with the
development of liver cancer (23).
HBx is a conserved viral protein that is hypothesized, by analogy
to woodchuck hepatitis virus X gene, to be required for HBV
replication in vivo (24).
Notably, in the current study it was discovered that the CPD
mutants did not encode the 154-aa HBx, but a truncated HBx termed X
factor, rather than X protein. Truncated HBx, encoded by CPD
mutants, is 76 aa in length, with characteristic amino acid
substitution at aa 44 and 45, caused by 2 point replacement
mutations at G/A1515C and G1518C. Kumar et al (25) demonstrated that aa 58–140 of HBx is
the minimum domain required for transactivation function. However,
the data from the present study suggests that truncated HBx encoded
by CPD mutants may have no capability to transactivate viral or
host genes as none of the 74 strains encode the preX region.
Therefore, we propose 3 variants of the X-ORF: PreX-X, typical X
and truncated HBx coding regions, but further molecular
epidemiological studies are required to confirm this.
In the current study sequences, including the entire
X region as well as enhancer II, CP and precore/core regions, were
investigated in order to identify specific mutations in
HBeAg-negative HBV. To the best of our knowledge, this is the first
study of CPD mutants in the HBV genome. Using specific PCR primers,
CPD mutants were detected in sera of patients. Electrophoresis
distinguished 3 different PreX-X/CP regions: wild type, CPD mutants
and a mixture of wild type and CPD mutants, which are easily
recognizable by the large CP deletion compared with the wild
type.
Although molecular epidemiological studies have
provided circumstantial evidence for the increased pathogenicity of
CP mutants, observations in patients are complicated by variables
including individual differences in susceptibility to viral
infection or replication, the strength of the immune response, and
the coexistence of viral quasispecies. CPD mutation may be a novel
molecular pattern consistent with HBeAg-negative CHB. However,
neither the CPD mutants nor the PreC or BCP mutants result in the
disappearance of HBeAg. This may be due to the presence of a mixed
infection with the mutant and wild-type viruses. Further studies
are required to identify the prevalence of the CPD mutants in
China, and the biological function of truncated HBx (X factor).
Acknowledgements
The authors thank Dr. Tamara A. Bowman (Center for
Pharmacogenomics, School of Medicine, Washington University in St.
Louis) for a careful language edit. This study was supported by the
Xiamen Municipal Fund for major diseases (WKZ0501) for research on
the relationship between HBV and HCC. This study was presented at
the Asian Pacific Association for the Study of the Liver, 2009.
References
|
1
|
Gish RG: Current treatment and future
directions in the management of chronic hepatitis B viral
infection. Clin Liver Dis. 9:541–565. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ganem D and Varmus HE: The molecular
biology of the hepatitis B viruses. Annu Rev Biochem. 56:651–693.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Seeger C and Mason WS: Hepatitis B virus
biology. Microbiol Mol Biol Rev. 64:51–68. 2000. View Article : Google Scholar
|
|
4
|
Stemler M, Weimer T, Tu ZX, et al: Mapping
of B-cell epitopes of the human hepatitis B virus X protein. J
Virol. 64:2802–2809. 1990.PubMed/NCBI
|
|
5
|
Yeh CT, Shen CH, Tai DI, et al:
Identification and characterization of a prevalent hepatitis B
virus X protein mutant in Taiwanese patients with hepatocellular
carcinoma. Oncogene. 19:5213–5220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Muroyama R, Kato N, Yoshida H, et al:
Nucleotide change of codon 38 in the X gene of hepatitis B virus
genotype C is associated with an increased risk of hepatocellular
carcinoma. J Hepatol. 45:805–812. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tanaka Y, Kanai F, Ichimura T, et al: The
hepatitis B virus X protein enhances AP-1 activation through
interaction with Jab1. Oncogene. 25:633–642. 2006.PubMed/NCBI
|
|
8
|
Feng H, Tan TL, Niu D and Chen WN: HBV X
protein interacts with cytoskeletal signaling proteins through SH3
binding. Front Biosci (Elite Ed). 2:143–150. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pang R, Lee TK, Poon RT, et al: Pin1
interacts with a specific serine-proline motif of hepatitis B virus
X-protein to enhance hepatocarcinogenesis. Gastroenterology.
132:1088–1103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Doria M, Klein N, Lucito R and Schneider
RJ: The hepatitis B virus HBx protein is a dual specificity
cytoplasmic activator of Ras and nuclear activator of transcription
factors. EMBO J. 14:4747–4757. 1995.PubMed/NCBI
|
|
11
|
Qadri I, Conaway JW, Conaway RC, et al:
Hepatitis B virus transactivator protein, HBx, associates with the
components of TFIIH and stimulates the DNA helicase activity of
TFIIH. Proc Natl Acad Sci USA. 93:10578–10583. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qadri I, Fatima K and Abdel-Hafiz H:
Hepatitis B virus X protein impedes the DNA repair via its
association with transcription factor, TFIIH. BMC Microbiol.
11:482011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Loncarevic IF, Zentgraf H and Schröder CH:
Sequence of a replication competent hepatitis B virus genome with a
preX open reading frame. Nucleic Acids Res. 18:49401990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Q, Cheng J, Dong J, et al: Molecular
epidemiological study on pre-X region of hepatitis B virus and
identification of hepatocyte proteins interacting with whole-X
protein by yeast two-hybrid. World J Gastroenterol. 11:3473–3478.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi K, Kishimoto S, Ohori K, et al:
A unique set of mutations in the ‘preX’ region of hepatitis B virus
DNA frequently found in patients but not in asymptomatic carriers:
implication for a novel variant. Int Hepatol Commun. 3:131–138.
1995.
|
|
16
|
Lok AS, Akarca U and Greene S: Mutations
in the pre-core region of hepatitis B virus serve to enhance the
stability of the secondary structure of the pre-genome
encapsidation signal. Proc Natl Acad Sci USA. 91:4077–4081. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buckwold VE, Xu Z, Yen TS and Ou JH:
Effects of a frequent double-nucleotide basal core promoter
mutation and its putative single-nucleotide precursor mutations on
hepatitis B virus gene expression and replication. J Gen Virol.
78:2055–2065. 1997.
|
|
18
|
Kurosaki M, Enomoto N, Asahina Y, et al:
Mutations in the core promoter region of hepatitis B virus in
patients with chronic hepatitis B. J Med Virol. 49:115–123. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kidd-Ljunggren K, Oberg M and Kidd AH:
Hepatitis B virus X gene 1751 to 1764 mutations: implications for
HBeAg status and disease. J Gen Virol. 78:1469–1478.
1997.PubMed/NCBI
|
|
20
|
Zhou F, Mao Q, Lu Y, Chen M, Chen J, Wang
L, Ren J and Dong Z: Hepatitis B virus core promoter deletion
mutations can lead to the expression of HBeAg. Chinese Hepatology.
2008.228–230. 2008.(In Chinese).
|
|
21
|
Baha W, Ennaji MM, Lazar F, et al: HBV
genotypes prevalence, precore and basal core mutants in Morocco.
Infect Genet Evol. 12:1157–1162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Locarnini S, McMillan J and Bartholomeusz
A: The hepatitis B virus and common mutants. Semin Liver Dis.
23:5–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bouchard MJ and Schneider RJ: The
enigmatic X gene of hepatitis B virus. J Virol. 78:12725–12734.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zoulim F, Saputelli J and Seeger C:
Woodchuck hepatitis virus X protein is required for viral infection
in vivo. J Virol. 68:2026–2030. 1994.PubMed/NCBI
|
|
25
|
Kumar V, Jayasuryan N and Kumar R: A
truncated mutant (residues 58-140) of the hepatitis B virus X
protein retains transactivation function. Proc Natl Acad Sci USA.
93:5647–5652. 1996. View Article : Google Scholar : PubMed/NCBI
|