1. Introduction
MicroRNAs (miRNAs) are short single-stranded RNA
molecules approximately 22 nucleotides in length. They are involved
in the post-transcriptional regulation of gene expression, through
binding to complementary sequences in the 3′ untranslated regions
(3′ UTRs) of target mRNA transcripts (1). miRNAs exist in three forms during
their synthesis: primary miRNAs (pri-miRNAs), precursor miRNAs
(pre-miRNAs) and mature miRNAs. Pri-miRNA contains a 5′ cap and
poly(A) tail and is first transcribed by RNA polymerase II as a
long transcript. Following the processing of pri-miRNA by the RNase
III enzyme Drosha in the nucleus, pre-miRNA is released, exported
to the cytoplasm by exportin-5 and processed by the RNase III
enzyme Dicer to produce a mature miRNA-miRNA duplex (2). Single-stranded mature miRNA is
incorporated from the duplex into an RNA-induced silencing complex
(RISC) where it mediates the silencing of target genes (3,4).
In 2000, Reinhart et al identified a novel
miRNA, let-7, which was able to implement and change the nematode
phenotype during the later development of Caenorhabditis
elegans (5). Numerous members
of the let-7 family have been identified in various species and
different members are found in different species (6). At present, 10 mature subtypes of the
let-7 family have been identified in humans, including let-7a,
let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98 and
miR-202, in which mature let-7a and let-7f were produced by
precursor sequences (let-7a-1, let-7a-2, let-7a-3; let-7f-1,
let-7f-2). In normal physiology, let-7 is primarily involved in
development, muscle formation, cell adhesion and gene regulation.
The regulation of let-7 synthesis has been the subject of
increasing attention and numerous proteins and factors have been
shown to be correlated with let-7 family expression, including
certain complete regulatory loops. Progress has also been made in
understanding the mechanisms of action of let-7 miRNA. A number of
studies have reported that let-7 is downregulated in numerous types
of cancer, including lung cancer (7), gastric tumors (8), colon cancer (9) and Burkitt's lymphoma (10). Low levels of let-7 expression have
been associated with the clinical postoperative survival of
patients with lung cancer (7). The
let-7 miRNA family is involved in the proliferation, apoptosis and
invasion of cancer cells. Oncogenes and their signaling pathways
are a potential connection between the level of expression of let-7
and the biological characteristics of tumor cells. This review
explored the regulation of let-7 expression and its mechanisms of
action, as well as its correlation with oncogenic pathways in
tumors.
2. Regulation of let-7 expression
Let-7 is found in Caenorhabditis elegans and
appears to act as a key regulator of temporal patterning,
coordinating developmental timing across tissues (11). As a conserved sequence, its
expression and function as a regulator of cell differentiation and
proliferation have been confirmed in animal and human cells
(12–15). In addition to regulating the
differentiation and proliferation of normal cells, let-7 has also
been found to be involved in inhibiting the growth of tumor cells
(16,17). The regulation of let-7 expression
has become increasingly important in light of its involvement in
tumor suppression. The expression of let-7 is regulated at various
stages of its biogenesis and the regulation process involves
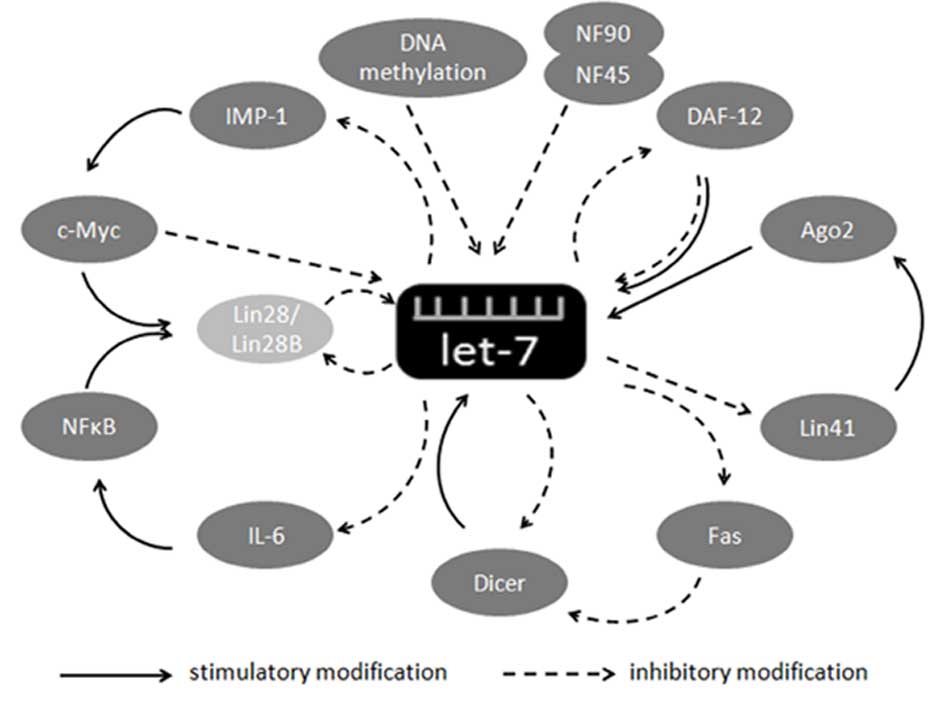
numerous factors and regulatory circuits (Fig. 1). However, although some regulatory
circuits have been demonstrated, others remain theoretical.
Lin28 and Lin28B
Reinhart et al found that the activity of
let-7 was affected by mutations in Lin28 (5). Lin28 and Lin28B have been shown to act
as post-transcriptional repressors of let-7 biogenesis, by binding
to the loop portion of the pri-let-7 hairpin and the stem of
pre-let-7 to inhibit the binding of Drosha or Dicer, thereby
inhibiting its processing (18,19).
The mechanism of Lin28 binding to the terminal loop region of
let-7g has been elucidated (20)
and the cold-shock and zinc-finger domains in Lin28 were shown to
be required for pre-let-7 binding, resulting in >90% inhibition
of let-7g processing upon the upregulation of Lin28. Lin28 is also
considered to be necessary and sufficient to block the
microprocessor-mediated cleavage of pri-let-7 miRNAs (18), as the ectopic expression of Lin28
completely prevented the processing of pri-let-7a and pri-let-7g,
whereas the processing of pri-miR-15a and pri-miR-122 were largely
unaffected. In addition, the transfection of Lin28 cDNA decreased
the endogenous levels of mature let-7 family members, but had no
effect on the levels of endogenous mature miR-21 (18).
A novel mechanism whereby Lin28/Lin28B blocks let-7
processing, other than by Drosha/Dicer inhibition, has been
identified. Lin28 was shown to be able to directly regulate let-7
synthesis by targeting pre-let-7 and affecting its terminal
uridylation (21); Lin28 mediates
the terminal uridylation of pre-let-7, thus diverting Dicer
processing and irreversibly re-routing pre-let-7 to a degradation
pathway. Three enzymes have been reported to be involved in the
terminal uridylation progress. Zcchc11 is a terminal uridylyl
transferase (TUTase) and is required by Lin28 for the mediation of
pre-let-7 uridylation and the subsequent blockade of let-7
processing in mouse embryonic stem cells (22). TUT4 is recruited by Lin28 to
pre-let-7 via the recognition of a tetranucleotide sequence motif
(GGAG) in the terminal loop. TUT4 in turn adds an oligouridine tail
to pre-let-7 to block Dicer processing (23). PUP-2 interacts directly with Lin28
to regulate the stability of Lin28-blockaded let-7 pre-miRNA to
suppress the action of Dicer and contribute to the Lin28-stimulated
uridylation of let-7 pre-miRNA, preventing its maturation in
Caenorhabditis elegans (24).
DNA methylation
It has been suggested that DNA methylation is
correlated with human tumorigenesis. The hyper- or hypomethylation
of CpG islands, often observed in tumor cells, results in the
silencing of genes and is significant in tumor development
(25–27). The results of certain studies
suggest that some miRNA genes are methylated, with consequent
effects on miRNA expression (28–30).
The expression of let-7 has been reported to be regulated by DNA
methylation. The human let-7a-3 gene is located on chromosome
22q13.31 and is associated with a CpG island, which is methylated
by the DNA methyltransferases DNMT1 and DNMT3B. Let-7a-3 is
methylated in normal lung samples, with a similar methylation
pattern to that observed in other human tissues. The
hypomethylation of let-7a-3 was found to increase the level of
expression of miRNA and inhibit the growth of tumor cells in lung
adenocarcinomas (31).
Hypermethylation resulted in the downregulation of let-7a-3 in
epithelial ovarian cancer and this low level of expression of
let-7a-3 in epithelial ovarian cancer was associated with a poor
prognosis (32).
Nuclear factor 90 and nuclear factor
45
Nuclear factor (NF) 90 and NF45 belong to the larger
Drosha complex family, which is needed for the production of
pre-miRNA from pri-miRNA. The overexpression of NF90 and NF45 has
been shown to be associated with the level of pri-miRNA. The
NF90-NF45 complex, rather than either factor alone, binds the
majority of pri-miRNAs with a higher affinity for pri-let-7a-1 than
the DGCR8-Drosha complex, which also binds to pri-miRNAs. The
NF90-NF45 complex may thus inhibit the processing of pri-miRNA by
the DGCR8-Drosha complex. NF90-NF45 also showed a higher binding
affinity for pri-let-7a-1 than for pri-miR-21 (33).
Regulatory circuits involved in let-7
expression
The regulation of let-7 biogenesis is complex and
involves numerous regulatory factors, including a number of factors
that control let-7 expression via regulatory loops. These loops may
be divided into two categories: Lin28-dependent and
Lin28-independent. The regulatory feedback loops that depend on
Lin28 are all positive and include the NFκB-Lin28-let-7-interleukin
(IL)-6-NFκB, Lin28-let-7-insulin-like growth factor II mRNA-binding
protein 1 (IMP1)-c-Myc-Lin28 and Lin28-let-7-Lin28 loops.
Inflammation is clinically linked to cancer and
certain inflammation-related molecules, including NFκB, have been
demonstrated to be key in this association. A regulatory feedback
loop controlling let-7 expression and including NFκB has been
identified and may aid the elucidation of the mechanistic link
between inflammation and cancer. NFκB directly activates Lin28
transcription and rapidly reduces let-7 levels. Let-7 may then
directly inhibit IL-6 expression, which may activate NFκB, thereby
completing a positive feedback loop (34).
As a target oncogene of let-7, c-Myc may be directly
regulated by IMP1, which has been shown to be directly and
negatively regulated by let-7 (35,36).
In addition, c-Myc was shown to be able to directly transactivate
Lin28B, a Lin28 homolog, to inhibit let-7 expression. The
activation of Lin28B was observed to be necessary and sufficient
for Myc-mediated let-7 expression (37,38).
It is also possible, however, that Myc regulates let-7 directly
(39).
Besides indirectly regulating the expression of
Lin28, let-7 may also affect Lin28 directly; the binding of let-7
to the 3′ UTR of Lin28/Lin28B transcripts represses the expression
of Lin28/Lin28B (38). As
discussed, Lin28 is a classical direct inhibitor of let-7, creating
a double-negative regulatory loop for let-7.
Certain loops, however, do not require Lin28,
including the DAF-12-let-7-DAF-12, Ago2-let-7-Lin41-Ago2,
Dicer-let-7-Dicer and Fas-Dicer-let-7-Fas loops.
The DAF-12-let-7-DAF-12 loop consists only of DAF-12
and let-7. DAF-12 is able to directly regulate the transcription of
let-7 either positively (as liganded DAF-12) or negatively (as
unliganded DAF-12). At the same time, let-7 can mediate the
expression of DAF-12 by binding its 3′ UTR (40).
Ago2, as the RISC slicer providing the RNase
activity required to cleave target mRNAs for miRNA, may also
directly bind let-7a or generate an additional miRNA precursor to
stabilize and enhance mature let-7a (41). Lin41, a target gene of let-7, has
been reported to negatively regulate Ago2 levels (42).
Dicer is a classical enzyme which is essential for
all miRNA synthesis processes. Let-7 has been reported to directly
target the miRNA-processing enzyme Dicer within its coding
sequence, thus establishing another autoregulatory negative
feedback loop for let-7 (43).
However, let-7b is downregulated when Dicer is overexpressed in
oral cancer cells (44), indicating
a double-negative regulatory loop for let-7b and Dicer in oral
cancer cells.
Fas (also termed APO-1 or CD95) was first identified
by Yonehara et al in 1989 and was considered to be a novel
cell surface protein antigen that differed from the tumor necrosis
factor (TNF) receptor, but was able to mediate the cytolytic
activity of TNF (45). Fas was
found to induce apoptosis by binding to specific ligands, including
Fas ligand (FasL), TNF-α and Fas-specific monoclonal antibody CH11
(mAb CH11) in sensitive cells (46,47).
The apoptosis-inducing ability of Fas has since been used to
inhibit the growth of tumor cells (48). Fas has been reported to be regulated
by let-7/miR-98 in T cells (49)
and this observation had been confirmed in human colon carcinoma.
Activated Fas is able to inhibit Dicer activation, reducing the
levels of mature let-7 miRNA (50).
These regulatory circuits maintain the balance of
the let-7 levels in a normal organism. The level of expression of
let-7 is affected when factors which take part in the regulatory
circuits are altered. Changes in the level of expression of let-7
may induce normal and abnormal responses. Therefore, the single
nucleotide polymorphisms (SNPs) of tumor suppressor miRNA
biogenesis genes are considered to be high-risk factors for cancer
(51,52). An SNP of the Lin28 gene, rs3811463,
located near the let-7 binding site, resulted in the weakened
repression of Lin28 mRNA induced by let-7 and the downregulation of
let-7 expression via the let-7-Lin28 double negative feedback loop.
This SNP was therefore considered to raise the risk of
tumorigenesis in breast cancer (53).
3. Mechanisms of action
The classical mechanism of let-7 action involves its
binding to the 3′ UTR of target mRNAs to regulate their expression.
However, in addition to this classical mechanism, let-7 may act in
other ways.
Let-7a caused identical responses when it was
targeted to the 5′ or 3′ UTRs of mRNAs containing internal ribosome
entry sites (54), indicating that
let-7 is able to act through binding to sites other than the 3′ UTR
of target mRNAs. Furthermore, let-7 binds not only to untranslated
or non-coding regions, but is also able to bind directly to coding
regions to restrict target mRNAs (43).
The mechanisms of let-7 action on mRNAs remain under
debate. Let-7a may repress the translation of target mRNAs by
binding to and inhibiting translating polyribosomes (55). Deadenylation is another route
whereby let-7 may inhibit translation and contribute to mRNA decay,
with the translational repression function being more prominent
than mRNA decay. However, deadenylation alone is not sufficient to
effect full mRNA repression (56).
4. Target oncogenic signaling pathways of
let-7
c-Myc, ras, high-mobility group A (HMGA), Janus
protein tyrosine kinase (JAK), signal transducer and activator of
transcription 3 (STAT3) and NIRF are oncogenes that are critical in
tumorigenesis, proliferation and invasion and which are targeted by
let-7. These oncogenes activate or upregulate the expression of
their downstream target proteins, which directly regulate the cell
cycle, apoptosis and cell adhesion. Let-7 may thus act as a tumor
suppressor through the inhibition of these oncogenic signaling
pathways.
Regulation of the ras oncogene
Ras is associated with numerous downstream signaling
pathways, including Ras/Raf/mitogen-activated protein kinase,
Ras/phosphoinositide 3-kinase/Akt and Ras/Rho/Ral. Following
activation by complex signaling cascades, Ras is able to bind to
numerous effectors to trigger further signaling cascades, which in
turn modulate cell processes including growth, survival, migration,
differentiation and death. Let-7 is expressed in normal adult lung
tissues (35), but has a lower
level of expression in lung tumors that express high levels of Ras
protein (7). This inverse
correlation between let-7 and Ras suggests a causal relationship.
The three human ras genes (H-ras, K-ras and N-ras) have
let-7-complementary sites in their 3′ UTRs (57). SNPs in the let-7 complementary sites
in the 3′ UTR of K-ras resulted in increased levels of K-ras and
poor prognosis of patients with lung and breast cancer (58–60).
Chin et al found that tumors which contained a variant
allele had lower let-7 levels than those without a variant allele
and hypothesized that SNPs in the let-7 binding site of the K-ras
3′ UTR reduce the levels of let-7 through a feedback loop (59). The overexpression of let-7 resulted
in a lower level of expression of the Ras protein, a reduction in
cell proliferation and migration in glioma and lung cancer cells
and a decrease in the size of gliomas and lung tumors in nude mice
(61,62). These results suggest that let-7
directly and negatively mediates the expression of the Ras protein
(H-Ras, K-Ras and N-Ras) by binding to 3′ UTR complementary sites.
The downstream proteins in Ras signaling pathways should thus be
downregulated when let-7 is overexpressed in cancer cells,
resulting in changes in cell function.
HMGA as a target oncogene of let-7
HMGA proteins (HMGA1a, HMGA1b and HMGA2) are
polypeptides of ~100 amino acid residues characterized by a modular
sequence organization, with three highly positively-charged
regions, termed AT hooks, that bind the minor groove of AT-rich DNA
stretches. These proteins regulate gene expression by altering the
structure of chromatin or by direct protein-protein interactions
with transcription factors (63).
As an oncogene, the overexpression of the HMGA2 protein has been
observed in a number of tumors (64–68).
It has been suggested that HMGA2 expression is controlled by
negatively-acting regulatory elements within the 3′ UTR. The
overexpression of HMGA2 in tumors may thus occur due to this
regulatory element being unable to bind effectively to the 3′ UTR
of HMGA2, which is reduced or absent in tumor cells (69). The level of let-7 expression is
significantly higher in gastric cancer patients with a high HMGA2
expression than in those with low a HMGA2 expression (70). Furthermore, HMGA2 expression was
found to be downregulated when let-7 was overexpressed (71). These results demonstrate that let-7
negatively mediates the expression of the oncogene HMGA2 by binding
to its 3′ UTR and directly suppressing its expression (72).
Myc oncogene and pathway
The proto-oncogene Myc is frequently activated in
tumors (73). Activated Myc
increases cell growth, division and survival by increasing the
synthesis of its target proteins, some of which are involved in the
regulation of the cell cycle and apoptosis (74). The overexpression of let-7a was
found to downregulate Myc mRNA and protein (9,10) and
it has been suggested that let-7 regulates Myc expression directly
by binding to its 3′ UTR. However, let-7 also affects the cell
cycle by directly mediating certain downstream proteins [cyclin D1
and cyclin-dependent kinase (CDK) 6] in the Myc oncogene signaling
pathway (75,76).
JAK-STAT3 pathway
JAK is a member of the protein tyrosine kinase
family. Activated JAK activates STATs, which are commonly expressed
in all tissues and cells in the human body. STAT activation is
involved in the genesis, differentiation and apoptosis of cells and
inflammation, cancer and the immune response (77). STAT3 is a member of the STAT family.
The JAK-STAT3 pathway is activated in a number of types of tumors
and has been confirmed to be correlated with cancer (78–80).
STAT3 was found to be a target of let-7a, mediating cell
proliferation in HepG2 cells (81).
Let-7 may thus also regulate the activity of tumor cells by
targeting the JAK-STAT3 pathway.
NIRF oncogene and let-7
The nucleic protein NIRF is encoded by the UHRF2
gene, which is a member of the UHRF oncogene family. NIRF contains
a ubiquitin-like domain and a ring-finger structural domain which
are essential for UHRF E3 ligase activity to regulate DNA
methylation (82,83). DNA damage may activate a
p53-dependent checkpoint pathway resulting in the induction of
p21WAF1 expression and subsequent cell cycle arrest at the G1/S
phase, through the inhibition of CDK activity and DNA replication.
A reduction in the levels of UHRF family members has been reported
to correlate with p21WAF1 expression (84). Let-7a was found to suppress NIRF
expression via binding to the 3′ UTR of NIRF mRNA and enhance
p21WAF1 expression in A549 lung cancer cells (85), indicating that let-7a may regulate
the cell cycle via a NIRF/p53/p21/CDK signaling pathway.
5. Conclusions
Let-7 miRNA has been the subject of increasing
interest as a potential therapeutic target in cancer. Let-7 is able
to suppress tumor proliferative activities and survival by
negatively mediating a number of oncogenes and by affecting key
regulators of the cell cycle, cell differentiation and apoptotic
pathways. The upregulation of let-7 is expected to provide an
effective cancer therapy. Therefore, although significant progress
has been made in understanding the regulation of let-7 synthesis
and its role in oncogenic signaling pathways, its regulation in
normal and malignant cells and the mechanisms whereby it controls
cell proliferation and survival require further elucidation.
Additional investigations are needed to enable the clinical
application of let-7 regulation to cancer suppression.
Acknowledgements
This study was supported by the China Natural
Science Foundation (81072078, 81101901 and 81172389), the Natural
Science Foundation of Jiangsu Province (201123808 and 2010580),
Jiangsu Province's Key Provincial Talents Program (RC2011051),
Jiangsu Province's Key Discipline of Medicine (XK201117), Program
for Development of Innovative Research Team in the First Affiliated
Hospital of NJMU, a project funded by the Priority Academic Program
Development of Jiangsu Higher Education Institutions, and National
High Technology Research and Development Program 863
(2012AA02A508).
Abbreviations:
|
miRNA
|
microRNA
|
|
3′ UTRs
|
3′ untranslated regions
|
|
NF
|
nuclear factor
|
|
SNP
|
single nucleotide polymorphism
|
|
RNase
|
ribonuclease
|
References
|
1
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 23:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hutvágner G, McLachlan J, Pasquinelli AE,
Bálint E, Tuschl T and Zamore PD: A cellular function for the
RNA-interference enzyme Dicer in the maturation of the let-7 small
temporal RNA. Science. 293:834–838. 2001.PubMed/NCBI
|
|
3
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roush S and Slack FJ: The let-7 family of
microRNAs. Trends Cell Biol. 18:505–516. 2008. View Article : Google Scholar
|
|
7
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang HH, Wang XJ, Li GX, Yang E and Yang
NM: Detection of let-7a microRNA by real-time PCR in gastric
carcinoma. World J Gastroenterol. 13:2883–2888. 2007.PubMed/NCBI
|
|
9
|
Akao Y, Nakagawa Y and Naoe T: Let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grosshans H, Johnson T, Reinert KL,
Gerstein M and Slack FJ: The temporal patterning microRNA let-7
regulates several transcription factors at the larval to adult
transition in C. elegans. Dev Cell. 8:321–330. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sokol NS, Xu P, Jan YN and Ambros V:
Drosophila let-7 microRNA is required for remodeling of the
neuromusculature during metamorphosis. Genes Dev. 22:1591–1596.
2008. View Article : Google Scholar
|
|
13
|
Caygill EE and Johnston LA: Temporal
regulation of metamorphic processes in Drosophila by the
let-7 and miR-125 heterochronic microRNAs. Curr Biol. 18:943–950.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thomson JM, Parker J, Perou CM and Hammond
SM: A custom microarray platform for analysis of microRNA gene
expression. Nat Methods. 1:47–53. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
17
|
Chang TC and Mendell JT: MicroRNAs in
vertebrate physiology and human disease. Annu Rev Genomics Hum
Genet. 8:215–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Viswanathan SR, Daley GQ and Gregory RI:
Selective blockade of microRNA processing by Lin28. Science.
320:97–100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Newman MA, Thomson JM and Hammond SM:
Lin-28 interaction with the let-7 precursor loop mediates regulated
microRNA processing. RNA. 14:1539–1549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Piskounova E, Viswanathan SR, Janas M,
LaPierre RJ, Daley GQ, Sliz P and Gregory RI: Determinants of
microRNA processing inhibition by the developmentally regulated
RNA-binding protein Lin28. J Biol Chem. 283:21310–21314. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heo I, Joo C, Cho J, Ha M, Han J and Kim
VN: Lin28 mediates the terminal uridylation of let-7 precursor
microRNA. Mol Cell. 32:276–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hagan JP, Piskounova E and Gregory RI:
Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in
mouse embryonic stem cells. Nat Struct Mol Biol. 16:1021–1025.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho
J, Yeom KH, Han J and Kim VN: TUT4 in concert with Lin28 suppresses
microRNA biogenesis through pre-microRNA uridylation. Cell.
138:696–708. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lehrbach NJ, Armisen J, Lightfoot HL,
Murfitt KJ, Bugaut A, Balasubramanian S and Miska EA: LIN-28 and
the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in
Caenorhabditis elegans. Nat Struct Mol Biol. 16:1016–1020.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
26
|
Ehrlich M: DNA methylation in cancer: too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Esteller M: Relevance of DNA methylation
in the management of cancer. Lancet Oncol. 4:351–358. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lujambio A, Ropero S, Ballestar E, Fraga
MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A,
Sanchez-Cespedes M, Git A, et al: Genetic unmasking of an
epigenetically silenced microRNA in human cancer cells. Cancer Res.
67:1424–1429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoshitomi T, Kawakami K, Enokida H,
Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama
K, Seki N and Nakagawa M: Restoration of miR-517a expression
induces cell apoptosis in bladder cancer cell lines. Oncol Rep.
25:1661–1668. 2011.PubMed/NCBI
|
|
31
|
Brueckner B, Stresemann C, Kuner R, Mund
C, Musch T, Meister M, Sültmann H and Lyko F: The human let-7a-3
locus contains an epigenetically regulated microRNA gene with
oncogenic function. Cancer Res. 67:1419–1423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu L, Katsaros D, de la Longrais IA,
Sochirca O and Yu H: Hypermethylation of let-7a-3 in epithelial
ovarian cancer is associated with low insulin-like growth factor-II
expression and favorable prognosis. Cancer Res. 67:10117–10122.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sakamoto S, Aoki K, Higuchi T, Todaka H,
Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T and Agata
Y: The NF90-NF45 complex functions as a negative regulator in the
micro RNA processing pathway. Mol Cell Biol. 29:3754–3769. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iliopoulos D, Hirsch HA and Struhl K: An
epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and
IL6 links inflammation to cell transformation. Cell. 139:693–706.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Boyerinas B, Park SM, Shomron N, Hedegaard
MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB and Peter ME:
Identification of let-7-regulated oncofetal genes. Cancer Res.
68:2587–2591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ioannidis P, Mahaira LG, Perez SA,
Gritzapis AD, Sotiropoulou PA, Kavalakis GJ, Antsaklis AI,
Baxevanis CN and Papamichail M: CRD-BP/IMP1 expression
characterizes cord blood CD34+ stem cells and affects c-myc and
IGF-II expression in MCF-7 cancer cells. J Biol Chem.
280:20086–20093. 2005.PubMed/NCBI
|
|
37
|
Chang TC, Zeitels LR, Hwang HW, Chivukula
RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al:
Lin-28B transactivation is necessary for Myc-mediated let-7
repression and proliferation. Proc Natl Acad Sci USA.
106:3384–3389. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dangi-Garimella S, Yun J, Eves EM, Newman
M, Erkeland SJ, Hammond SM, Minn AJ and Rosner MR: Raf kinase
inhibitory protein suppresses a metastasis signalling cascade
involving LIN28 and let-7. EMBO J. 28:347–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chang TC, Yu D, Lee YS, Wentzel EA, Arking
DE, West KM, Dang CV, Thomas-Tikhonenko A and Mendell JT:
Widespread microRNA repression by Myc contributes to tumorigenesis.
Nat Genet. 40:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hammell CM, Karp X and Ambros V: A
feedback circuit involving let-7-family miRNAs and DAF-12
integrates environmental signals and developmental timing in
Caenorhabditis elegans. Proc Natl Acad Sci USA.
106:18668–18673. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diederichs S and Haber DA: Dual role for
argonautes in microRNA processing and posttranscriptional
regulation of microRNA expression. Cell. 131:1097–1108. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rybak A, Fuchs H, Hadian K, Smirnova L,
Wulczyn EA, Michel G, Nitsch R, Krappmann D and Wulczyn FG: The
let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin
ligase for the miRNA pathway protein Ago2. Nat Cell Biol.
11:1411–1420. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Forman JJ, Legesse-Miller A and Coller HA:
A search for conserved sequences in coding regions reveals that the
let-7 microRNA targets Dicer within its coding sequence. Proc Natl
Acad Sci USA. 105:14879–14884. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jakymiw A, Patel RS, Deming N,
Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM and Chan
EK: Overexpression of dicer as a result of reduced let-7 microRNA
levels contributes to increased cell proliferation of oral cancer
cells. Genes Chromosomes Cancer. 49:549–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yonehara S, Ishii A and Yonehara M: A
cell-killing monoclonal antibody (anti-Fas) to a cell surface
antigen co-downregulated with the receptor of tumor necrosis
factor. J Exp Med. 169:1747–1756. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Peter ME, Budd RC, Desbarats J, Hedrick
SM, Hueber AO, Newell MK, Owen LB, Pope RM, Tschopp J, Wajant H,
Wallach D, et al: The CD95 receptor: apoptosis revisited. Cell.
129:447–450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Barnhart BC, Legembre P, Pietras E, Bubici
C, Franzoso G and Peter ME: CD95 ligand induces motility and
invasiveness of apoptosis-resistant tumor cells. EMBO J.
23:3175–3185. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gordon N and Kleinerman ES: Aerosol
therapy for the treatment of osteosarcoma lung metastases:
targeting the Fas/FasL pathway and rationale for the use of
gemcitabine. J Aerosol Med Pulm Drug Deliv. 23:189–196. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang S, Tang Y, Cui H, Zhao X, Luo X, Pan
W, Huang X and Shen N: Let-7/miR-98 regulate Fas and Fas-mediated
apoptosis. Genes Immun. 12:149–154. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Geng L, Zhu B, Dai BH, Sui CJ, Xu F, Kan
T, Shen WF and Yang JM: A let-7/Fas double-negative feedback loop
regulates human colon carcinoma cells sensitivity to Fas-related
apoptosis. Biochem Biophys Res Commun. 408:494–499. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liang D, Meyer L, Chang DW, Lin J, Pu X,
Ye Y, Gu J, Wu X and Lu K: Genetic variants in microRNA
biosynthesis pathways and binding sites modify ovarian cancer risk,
survival, and treatment response. Cancer Res. 70:9765–9776. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Permuth-Wey J, Kim D, Tsai YY, Lin HY,
Chen YA, Barnholtz-Sloan J, Birrer MJ, Bloom G, Chanock SJ, Chen Z,
et al: LIN28B polymorphisms influence susceptibility to epithelial
ovarian cancer. Cancer Res. 71:3896–3903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen AX, Yu KD, Fan L, Li JY, Yang C,
Huang AJ and Shao ZM: Germline genetic variants disturbing the
Let-7/LIN28 double-negative feedback loop alter breast cancer
susceptibility. PLoS Genet. 7:e10022592011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lytle JR, Yario TA and Steitz JA: Target
mRNAs are repressed as efficiently by microRNA-binding sites in the
5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 104:9667–9672.
2007.PubMed/NCBI
|
|
55
|
Nottrott S, Simard MJ and Richter JD:
Human let-7a miRNA blocks protein production on actively
translating polyribosomes. Nat Struct Mol Biol. 13:1108–1114. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Beilharz TH, Humphreys DT, Clancy JL,
Thermann R, Martin DI, Hentze MW and Preiss T: MicroRNA-mediated
messenger RNA deadenylation contributes to translational repression
in mammalian cells. PLoS One. 4:e67832009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: Ras is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nelson HH, Christensen BC, Plaza SL,
Wiencke JK, Marsit CJ and Kelsey KT: KRAS mutation, KRAS-LCS6
polymorphism, and non-small cell lung cancer. Lung Cancer.
69:51–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chin LJ, Ratner E, Leng S, Zhai R, Nallur
S, Babar I, Muller RU, Straka E, Su L, Burki EA, et al: A SNP in a
let-7 microRNA complementary site in the KRAS 3′ untranslated
region increases non-small cell lung cancer risk. Cancer Res.
68:8535–8540. 2008.PubMed/NCBI
|
|
60
|
Paranjape T, Heneghan H, Lindner R, Keane
FK, Hoffman A, Hollestelle A, Dorairaj J, Geyda K, Pelletier C,
Nallur S, et al: A 3′-untranslated region KRAS variant and
triple-negative breast cancer: a case-control and genetic analysis.
Lancet Oncol. 12:377–386. 2011.
|
|
61
|
Lee ST, Chu K, Oh HJ, Im WS, Lim JY, Kim
SK, Park CK, Jung KH, Lee SK, Kim M and Roh JK: Let-7 microRNA
inhibits the proliferation of human glioblastoma cells. J
Neurooncol. 102:19–24. 2010.PubMed/NCBI
|
|
62
|
He XY, Chen JX, Zhang Z, Li CL, Peng QL
and Peng HM: The let-7a microRNA protects from growth of lung
carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer
Res Clin Oncol. 136:1023–1028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sgarra R, Rustighi A, Tessari MA, Di
Bernardo J, Altamura S, Fusco A, Manfioletti G and Giancotti V:
Nuclear phosphoproteins HMGA and their relationship with chromatin
structure and cancer. FEBS Lett. 574:1–8. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma
E, Sioris T, Salo J, Karjalainen A, Knuutila S and Anttila S:
Increased expression of high mobility group A proteins in lung
cancer. J Pathol. 209:206–212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Meyer B, Loeschke S, Schultze A, Weigel T,
Sandkamp M, Goldmann T, Vollmer E and Bullerdiek J: HMGA2
overexpression in non-small cell lung cancer. Mol Carcinog.
46:503–511. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Abe N, Watanabe T, Suzuki Y, Matsumoto N,
Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A and Atomi Y: An
increased high-mobility group A2 expression level is associated
with malignant phenotype in pancreatic exocrine tissue. Br J
Cancer. 89:2104–2109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Miyazawa J, Mitoro A, Kawashiri S, Chada
KK and Imai K: Expression of mesenchyme-specific gene HMGA2 in
squamous cell carcinomas of the oral cavity. Cancer Res.
64:2024–2029. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Belge G, Meyer A, Klemke M, Burchardt K,
Stern C, Wosniok W, Loeschke S and Bullerdiek J: Upregulation of
HMGA2 in thyroid carcinomas: a novel molecular marker to
distinguish between benign and malignant follicular neoplasias.
Genes Chromosomes Cancer. 47:56–63. 2008. View Article : Google Scholar
|
|
69
|
Borrmann L, Wilkening S and Bullerdiek J:
The expression of HMGA genes is regulated by their 3'UTR. Oncogene.
20:4537–4541. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Motoyama K, Inoue H, Nakamura Y, Uetake H,
Sugihara K and Mori M: Clinical significance of high mobility group
A2 in human gastric cancer and its relationship to let-7 microRNA
family. Clin Cancer Res. 14:2334–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shi G, Perle MA, Mittal K, Chen H, Zou X,
Narita M, Hernando E, Lee P and Wei JJ: Let-7 repression leads to
HMGA2 overexpression in uterine leiomyosarcoma. J Cell Mol Med.
13:3898–3905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Albihn A, Johnsen JI and Henriksson MA:
Myc in oncogenesis and as a target for cancer therapies. Adv Cancer
Res. 107:163–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ruggero D: The role of Myc-induced protein
synthesis in cancer. Cancer Res. 69:8839–8843. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Schultz J, Lorenz P, Gross G, Ibrahim S
and Kunz M: MicroRNA let-7b targets important cell cycle molecules
in malignant melanoma cells and interferes with
anchorage-independent growth. Cell Res. 18:549–557. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Leeman RJ, Lui VW and Grandis JR: STAT3 as
a therapeutic target in head and neck cancer. Expert Opin Biol
Ther. 6:231–241. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Achcar RO, Cagle PT and Jagirdar J:
Expression of activated and latent signal transducer and activator
of transcription 3 in 303 non-small cell lung carcinomas and 44
malignant mesotheliomas: possible role for chemotherapeutic
intervention. Arch Pathol Lab Med. 131:1350–1360. 2007.
|
|
80
|
Ma XT, Wang S, Ye YJ, Du RY, Cui ZR and
Somsouk M: Constitutive activation of Stat3 signaling pathway in
human colorectal carcinoma. World J Gastroenterol. 10:1569–1573.
2004.PubMed/NCBI
|
|
81
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus x protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Bronner C, Achour M, Arima Y, Chataigneau
T, Saya H and Schini-Kerth VB: The UHRF family: oncogenes that are
drugable targets for cancer therapy in the near future? Pharmacol
Ther. 115:419–434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Robinson PA and Ardley HC:
Ubiquitin-protein ligases. J Cell Sci. 117:5191–5194. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Arima Y, Hirota T, Bronner C, Mousli M,
Fujiwara T, Niwa S, Ishikawa H and Saya H: Down-regulation of
nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage
checkpoint signals contributes to cell cycle arrest at G1/S
transition. Genes Cells. 9:131–142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
He X, Duan C, Chen J, Ou-Yang X, Zhang Z,
Li C and Peng H: Let-7a elevates p21(WAF1) levels by targeting of
NIRF and suppresses the growth of A549 lung cancer cells. FEBS
Lett. 583:3501–3507. 2009. View Article : Google Scholar : PubMed/NCBI
|