Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common cancer worldwide. Despite ongoing improvements of
established treatment modalities, the long-term survival rate of
patients with HNSCC has improved only marginally over the past
several decades. More than 60% of patients with advanced tumors or
localized lymph node metastases succumb to the disease within five
years after diagnosis (1).
MicroRNAs (miRs) are small non-coding RNAs that
regulate the translation of many genes. Mature miRs are
single-stranded, non-coding RNAs that are frequently dysregulated
in cancer. miRs play key roles in various cellular processes, such
as differentiation, cell growth, angiogenesis,
epithelial-to-mesenchymal transition (EMT) and invasion (2–4).
Accumulating evidence suggests that a correlation between specific
tumors and differential miR expression profiles exists (5–7),
indicating specific molecular pathways activated in cancer cells
during carcinogenesis and tumor progression. This insight
stimulated an increasing number of studies analyzing miR expression
profiles in different squamous carcinomas, including their
potential clinical relevance (8,9). Both
messenger and non-coding RNAs can be detected in blood and studies
indicate that miRs are particularly stable and abundant (10,11).
Circulating miRs could be derived from passive leakage from
apoptotic or necrotic cancer cells yet also from normal tissue due
to damage (e.g. trauma) or chronic inflammation (12,13).
In addition, both cancer and non-malignant cells, including immune
cells, can actively release miRNAs, either microvesicle-associated,
free or in a selective manner (14).
miRs not only regulate normal cellular development,
they also play important roles in cancer development and
progression. Several studies reported that miRs can act either as
oncogenes or tumor suppressors (15,16).
Others illustrated the potential of manipulating miR expression in
cancer therapy. It is believed that miR-based therapeutic
regulation of miRs has the potential to contribute to curing
cancer, as they regulate whole programs of gene expression by
suppressing hundreds of genes simultaneously (17).
For a few miRs, a functional relevance has already
been demonstrated in HNSCC. miR-200c negatively modulates the
expression of BMI1 and inhibits epithelial-mesenchymal transitions
in malignant HNSCC (18). Ectopic
transfection of miR-138 suppressed cell invasion and led to cell
cycle arrest and apoptosis. Knockdown of miR-138 enhanced cell
invasion and suppressed apoptosis, suggesting that miR-138 acts as
a tumor suppressor and may serve as a therapeutic target for HNSCC
patients at risk of metastasis (19). miR-34 coordinates with other miRs
regulating signaling pathways, including the TGF-Wnt pathway, G1-S
cell cycle progression, VEGF signaling pathway, apoptosis and
survival pathways in nasopharyngeal carcinoma (20). In esophageal squamous cell
carcinoma, miR-25 was upregulated significantly and correlated with
the status of lymph node metastasis and TNM-classification.
Overexpression of miR-25 markedly promoted migration and invasion
of esophageal squamous cell carcinoma (21). miRNA-21 (miR-21), a well-known
oncogenic miRNA, was found to be overexpressed in different types
of human cancer, and it has been implicated in multiple
malignancy-related processes including cell proliferation,
apoptosis, invasion and metastasis (22,23).
The present study clarified the role of a panel of
miRs as potential drivers of biological aggressiveness in HNSCC.
Exploring the expression patterns of these miRs, we noted in
vitro a significant upregulation of miR-21 expression in HNSCC
cell lines UM-SCC11B and UM-SCC9 which was different from the five
other miRs tested (miR-200c, −138-1, −138-2, −25 and −34). Using
software specifically designed to identify miR target genes
(miRanda and TargetScan), we found that miR-21 contains a consensus
sequence to programmed cell death 4 (PDCD4), and identified PDCD4
as a potential target gene of miR-21. Therefore, we explored the
biologic effects of miR-21 on PDCD4 in two distinct HNSCC cell
lines.
PDCD4 is a recently-characterized tumor suppressor
gene involved in the apoptotic machinery and in cell transformation
and invasion, and tumor progression (24). PDCD4 protein expression is
consistently downregulated in human cancer and cancer cell lines
(24–27). Several mechanisms are involved in
PDCD4 dysregulation; among others, the oncogenic miR-21 has been
shown to specifically target the PDCD4 3′-untranslated region
(3′-UTR), which negatively regulates PDCD4 expression.
The purpose of the present study was to explore a
possible presence of the interdependence of miR-21 and PDCD4 in
HNSCC. Consequently, we investigated and manipulated miR-21 and
PDCD4-levels in two exemplary HNSCC lines.
Materials and methods
Cell culture
The HNSCC cell line panel was composed of UD-SCC-1
and −2 (provided by Dr Henning Bier, University of Düsseldorf),
UM-SCC-9, -11B, -47, -104 (provided by Dr Tom Carey, University of
Michigan), of which UM-SCC-47, -104 and UD-SCC-2 are HPV16-related.
Normal primary epidermal keratinocytes (PCS-200-011) were purchased
from American Type Culture Collection (ATCC, Manassas, VA, USA).
All cells were cultured in DMEM or RPMI-1640 supplemented with 10%
fetal bovine serum (both from HyClone, Logan, UT, USA) and 1%
penicillin and streptomycin in a 5% CO2 atmosphere at
37°C.
Quantitative real-time RT-PCR (qRT-PCR)
detection of miRNA expression
qRT-PCR detection of miRs (miR-200c, −21, −138-1,
−138-2, −25 and −34) was performed mostly as previously described
(28). Total RNA was obtained from
cell lines using the mirVana miRNA Isolation kit (Applied
Biosystems, Foster City, CA, USA), according to the manufacturer’s
instructions. The expression of mature miR was determined by TaqMan
real-time RT-PCR using the TaqMan miR assay (Applied Biosystems)
and normalized using the 2−ΔΔCT method relative to
U6-small nuclear RNA. All PCRs were carried out in triplicate.
Transfection with antisense
oligonucleotides
The stability-enhanced miR precursor that mimics
miR-21 and the control non-specific miR precursor [pre-miR
precursor, negative control (NC)] and the anti-miR-21 (miR-21
inhibitor) were purchased from Ambion (Ambion-Life Technologies,
Darmstadt, Germany). The sequence for the miR-21 inhibitor was:
5′-UCAACAUCAGUCUGAUAAGCUA-3′; the sequences for miR-21 mimics were:
5′-UAGCUUAUCAGACUGAUGU UGA-3′ and 5′-AACAUCAGUCUGAUAAGCUAUU-3′. The
sequence for the NC miR inhibitor was: 5′-CAGUACUUUU
GUGUAGUACAA-3′. Cells were trypsinized, counted and seeded onto
6-well plates the day prior to transfection to ensure 50% cell
confluence on the day of transfection. Transfection of miR
precursor/inhibitor into cells was performed using Lipofectamine
2000 (Invitrogen, Carlsbad, CA, USA) in accordance with the
manufacturer’s advised procedure. The miR precursor/inhibitor was
used at a final concentration of 100 nM. Post-transfection,
real-time RT-PCR, western blotting and cell proliferation analysis
were performed. The transfection efficiency was assessed by
fluorescence microscope (nuclei were stained with DAPI). Nearly all
cells exhibited Cy3 staining, indicating that the miR-21
precursor/inhibitor and controls were effectively transfected into
UM-SCC9 cells.
Western blot analysis
The transfected cells and untreated control cells
were isolated 72 h after transfection and proteins were extracted
in a solution of RIPA and Halt™ Protease Inhibitor Cocktail (Thermo
Scientific, Waltham, MA, USA) from cells and subjected to
SDS-polyacrylamide gel electrophoresis. Quantification of total
protein was carried out by bicinchoninic acid (BCA) (Sigma, St.
Louis, MO, USA). The proteins (100 μg) were subjected to 12%
SDS-polyacrylamide gel electrophoresis. Separated proteins were
electrophoretically transferred to nitrocellulose (NC) membrane
(Bio-Rad, Hercules, CA, USA) and immunoblotted with anti-human
PDCD4 polyclonal (1:1,000; Covance, Princeton Township, NJ, USA),
anti-GAPDH (1:5,000; Sigma). Immunoreactive proteins were
visualized using the Odyssey Infrared Imaging System (LI-COR,
Lincoln, NE, USA), as described by the manufacturer.
Cell proliferation assay
A diphenyltetrazolium bromide (MTT) assay was
performed to determine the cell proliferation. Five thousand cells
were seeded to each well of a 96-well plate and grown for 24, 48,
72 and 96 h. Next, the medium was removed and cells were washed
with phosphate-buffered saline (PBS). Then 5 g/l of thiazolyl
tetrazolium (Ameresco, Indianapolis, IN, USA) was added to each
well. After an additional 4 h of incubation, MTT was removed and
150 μl of dimethyl sulfoxide (Sigma) was added. The viability of
the cells was calculated from the absorption at 570/630 nm with an
enzyme-linked immunosorbent assay reader. The experiment was
repeated three times.
Cell cycle analysis
At 48 h post-transfection with the miR-21
precursor/inhibitor or control precursor (100 nM), cells, including
untreated and mock controls, were collected by trypsinization and
washed with PBS. For cell cycle analysis, the cells were fixed with
75% ethanol and stored at 4°C overnight. The following day, fixed
cells were washed with PBS, treated with RNase A (50 μg/ml), and
stained with propidium iodide (PI) (50 μg/ml) for 30 min in the
dark. The stained cells were analyzed by flow cytometry
(FACSCalibur; Becton-Dickinson, Franklin Lakes, NJ, USA). Cellular
debris and fixation artifacts were removed by an exclusion gate and
the cell populations in G0/G1, S and G2/M phases were quantified
using FlowJo 7.6.2 software (Tree Star, Ashland, OR, USA). At least
10,000 cells for each condition were analyzed to obtain a reliable
signal.
Luciferase assays
The full-length 3′-UTR of PDCD4 mRNA containing the
miR-21 binding site was amplified by PCR primers:
5′-ggggagctcatataagaactcttgcagtct-3′ (forward) and
5′-gggaagcttggtgtacattcttctagaac-3′ (reverse), and cloned into the
SacI-HindIII site of the pMIR-REPORT kit (Applied
Biosystems) and termed Luc-PDCD4-Wt. To generate miR-21 binding
site deletion mutants, the seed sequences were deleted using the
QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, La
Jolla, CA, USA), with Luc-PDCD4-Wt as a template. The resulting
mutant was termed Luc-PDCD4-d. For the reporter assays, the cells
were transiently transfected with the luciferase vector as control
vector, and either anti-miR-21 oligonucleotide or negative control
using Lipofectamine 2000. Reporter assays were performed 24-h post
transfection using the Luciferase Assay kit (Promega, Madison, WI,
USA). β-galactosidase activity was used for normalizing the
transfection efficiency.
Statistical analysis
Data are shown as means ± SD and subjected to
one-way analysis of variance with factors of treatment using the
SPSS 13.0 for Windows (SPSS, Chicago, IL, USA). Comparisons between
two groups were performed by an unpaired Student’s t-test.
P<0.05 (one-tailed) was considered to indicate a statistically
significant result.
Results
The expression of miR-21 is upregulated
in HNSCC lines
To explore the role of miRNAs in HNSCC, detecting
expression levels of different miRNAs was a primary consideration.
Expression of miR-21, −200c, −138-1, −138-2, −25 and −34 were
assessed by qRT-PCR detection in 6 HNSCC lines and compared to
normal primary epidermal keratinocytes as control. The cell lines
chosen were derived from human HNSCC, from patients exhibiting an
aggressive clinical course who developed local or regional
recurrence, and who had died within two years of diagnosis
(29). Moreover, with regard to the
etiology of HNSCC, three HPV-related (UM-SCC-47, −104 and UD-SCC-2)
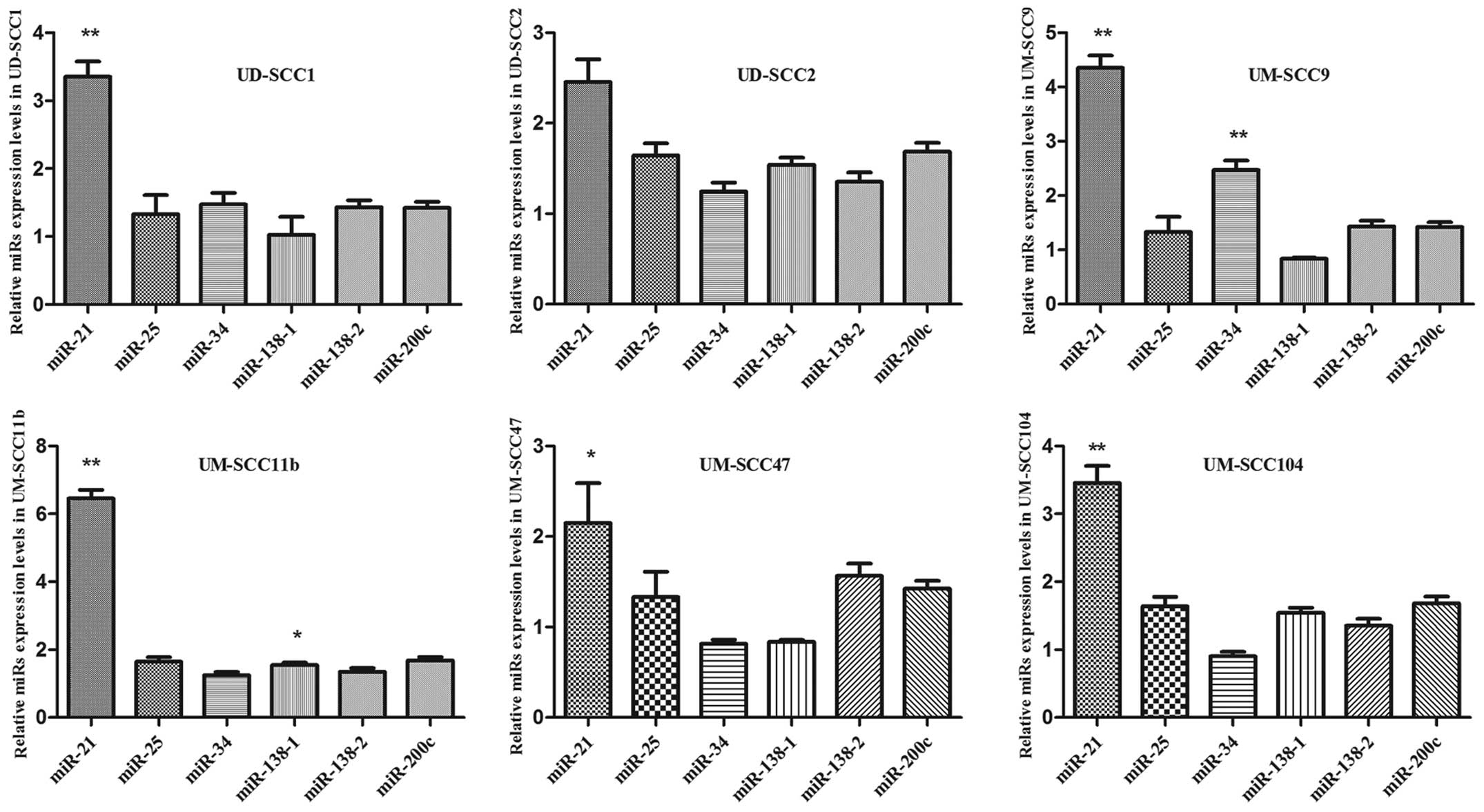
cell lines were included. qRT-PCR revealed different levels of
expression of the tested miRs in those HNSCC cells (Fig. 1). miR-21 exhibited the highest
expression in the tested HNSCC cell lines, with respect to normal
primary epidermal keratinocytes used for control purposes. This
observation has not been previously described. From the other
tested miRs, only miR-34 showed a significant upregulation in
UM-SCC9 and miR-138-1 in UM-SCC11B. Thus, further investigations
were carried out to determine the role of miR-21 in HNSCC.
miR-21 regulates the growth of HNSCC
cells
Considering the marked upregulation of miR-21 in the
HNSCC lines, it was hypothesized that it may function as a tumor
promoter. This hypothesis was supported by results from
computational analyses indicating interactions between miR-21 and
the tumor-suppressor PDCD4 as described below. Consequently, we
focused on proof of principle by testing the effect of miR-21 on
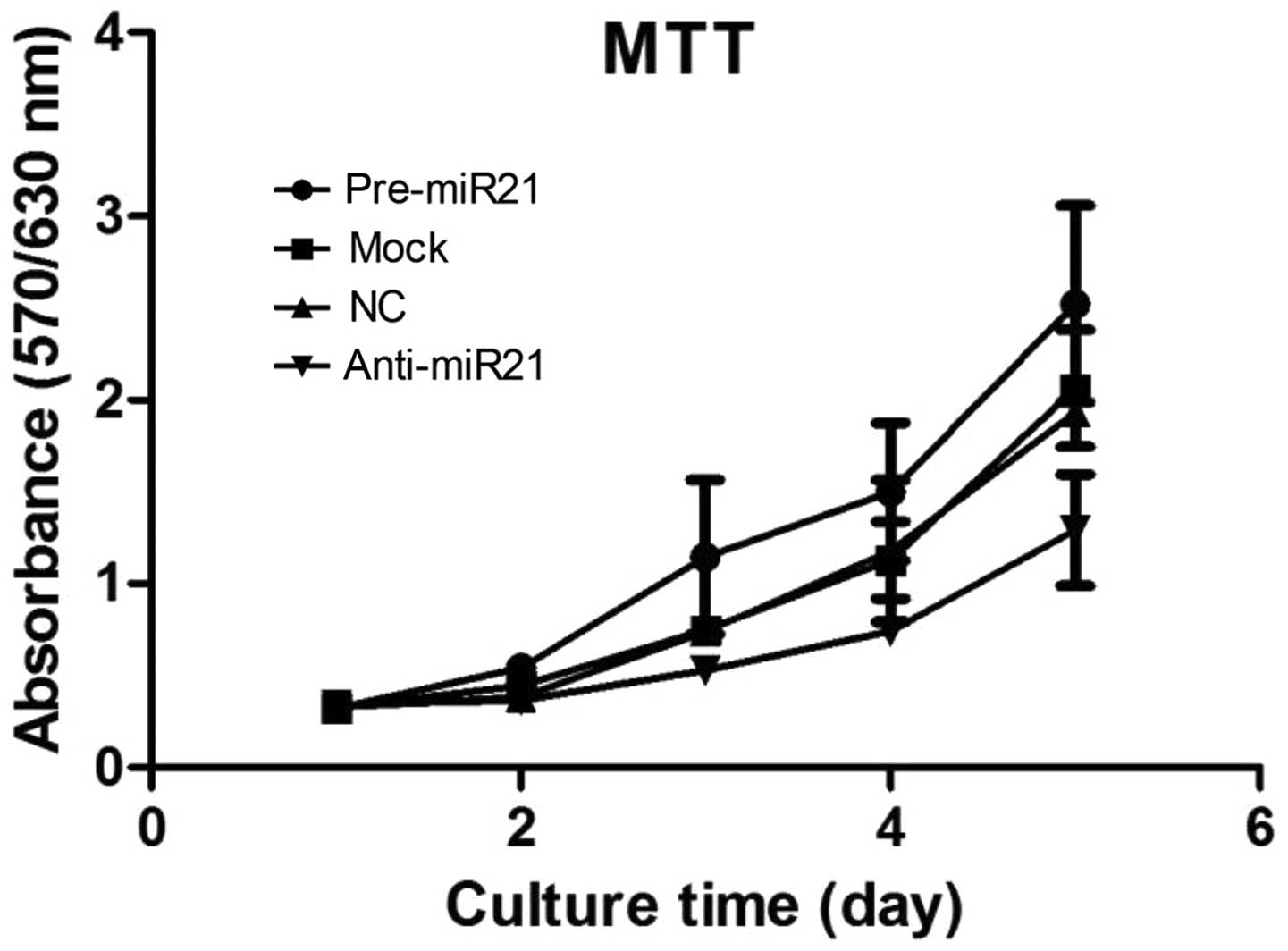
the growth of one exemplary cell line (UM-SCC9). In an MTT
proliferation assay, cells transfected with a miR-21 precursor grew
more rapidly than the mock control (Fig. 2A and B). The difference in the
proliferative activity of transfected cells indicated that
overexpression of miR-21 promotes growth activity in both HNSCC
lines. Nevertheless, inhibition of miR-21 by anti-miR-21 resulted
in growth delay (Fig. 2A and B).
These results suggest that miR-21 promotes cell growth, and
disturbance of miR-21 can effectively restrain the proliferation of
HNSCC cells in vitro.
Downregulation of miR-21 expression
induces cell cycle arrest in G1/S phase
To elucidate the mechanism of miR-21-mediated cell
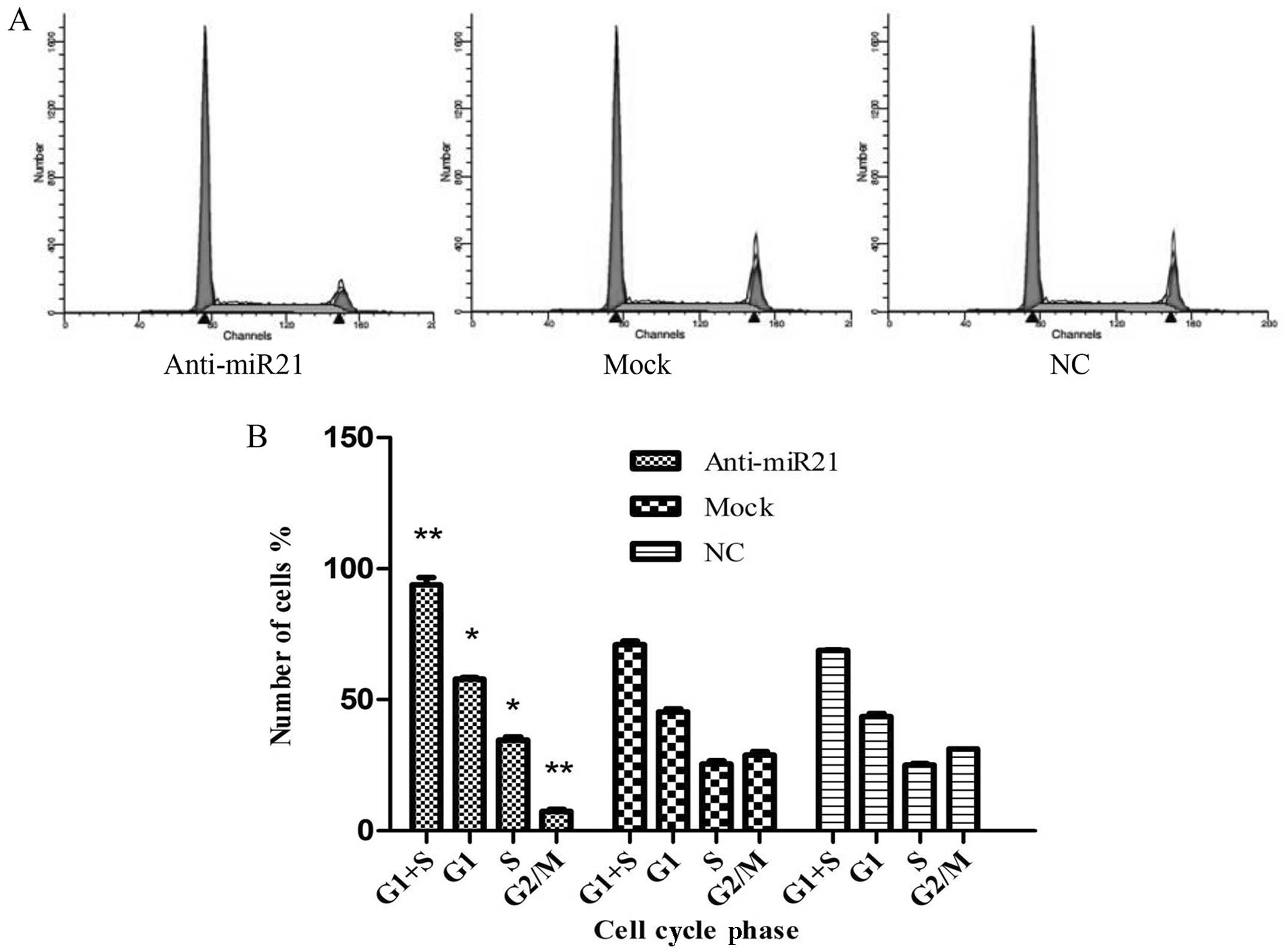
growth of HNSCC cells (UM-SCC9), cell cycle analysis of anti-miR-21
cells was performed (Fig. 3A). The
results demonstrated that, when compared with the mock and normal
control group, the percentage of anti-miR-21-transfected UM-SCC9
cells in G1/S phase increased from 71.03±1.26 to 93.93±2.67%
(P<0.05), whereas the percentage of cells in G2/M phase
decreased from 28.96±1.26 to 7.4±0.79% (P<0.05) (Fig. 3B). These results indicate that
downregulation of miR-21 expression induces G1/S phase arrest,
thereby confirming the stimulating role of miR-21 for cell
division.
miR-21 PDCD4 expression by targeting the
PDCD4-3′-UTR
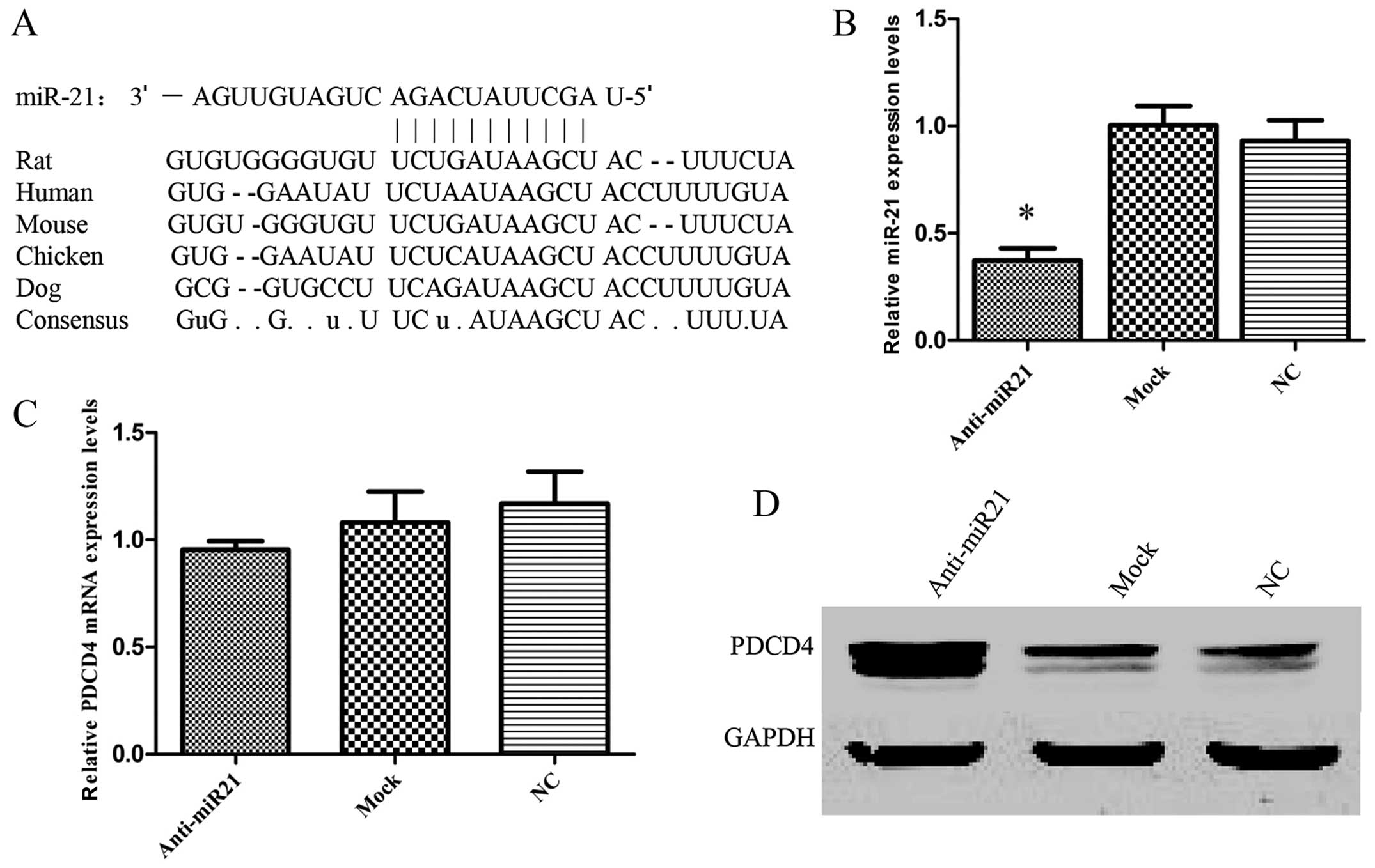
Computational analysis with specifically designed
software indicated that PDCD4 is a potential target gene of miR-21
(Fig. 4A). The sequences for the
binding sites in the 3′UTRs of PDCD4 are highly conserved among
different species. Previous reports demonstrated that there was a
significant inverse correlation between miR-21 expression and PDCD4
protein levels (30,31), and low expression levels of PDCD4 in
primary HNSCC were also found (32). If PDCD4 was regulated by miR-21 as
hypothesized, a miR-21 inhibitor should increase its expression in
HNSCC. To test this hypothesis, UM-SCC9 cells were used to
determine in vitro whether the suppression of miR-21 also
affected PDCD4 expression in HNSCC cells. The downregulation of
endogenous miR-21 with anti-miR-21 (Fig. 4B) led to a significant increase in
PDCD4 protein (Fig. 4D) without any
change in PDCD4 mRNA levels (Fig.
4C). Thus, the results suggested that PDCD4 is a potential
miR-21 target gene. Next, to further confirm that miR-21 is able to
directly bind to PDCD4 and inhibit PDCD4 expression and to
determine whether the 3′-UTR of PDCD4 mRNA is a functional target
of miR-21, a reporter plasmid driven by the SV40 basal promoter,
harboring the full-length 3′-UTR of PDCD4 mRNA at the 3′ position
of the luciferase reporter gene, was cloned (Fig. 4E). The transient transfection of
UM-SCC9 cells with the reporter plasmid and anti-miR-21 inhibitor
led to a significant increase of reporter activity in comparison
with the negative control (Fig.
4F). However, the activity of the reporter construct deleted at
the seed sequences of miR-21 target site was unaffected by a
simultaneous transfection with anti-miR-21 (Fig. 4F). These results indicate that
miR-21 regulates PDCD4 expression at the post-transcriptional level
by targeting the PDCD4-3′-UTR.
Discussion
In the present study, we are first to show that
miR-21 regulates cellular proliferation in HNSCC lines. We
demonstrated that the tumor suppressor PDCD4 is negatively
regulated by miR-21 at the post-transcriptional level via binding
to the 3′-untranslated region (3′-UTR) of PDCD4 mRNA.
miRs regulate a variety of cellular pathways through
the regulation of the expression of multiple target genes (33). In this regard, miR-21 has been
suggested to function as an oncogene, since it is overexpressed in
many types of solid malignancy (6),
particularly in malignancies such as breast cancer (34), glioblastoma (35), prostate (36), ovarian (37), pancreatic (38), colon (39) and gastric cancer (40), cholangiocarcinoma (41), hepatocellular cancer (42), HNSCC (43) and esophageal cancer (44). Furthermore, an association between
miR-21 expression and prognosis has been proposed in pancreatic
cancer and colon adenocarcinoma (45,46).
In the present study, miR-21 expression in HNSCC was significantly
higher than that of matched normal epithelium, and PDCD4 protein
correlated inversely with miR-21 level as shown in previous studies
(43,47). Whether it may serve as a prognostic
factor in HNSCC as observed in other malignancies remains, to date,
unknown (35,42,48–51).
In the present study, we sought to elucidate its function as a
regulator. The tumor suppressor gene PDCD4 was originally
characterized as an inhibitor of cellular transformation in a mouse
cell culture model (52). PDCD4
expression is downregulated or lost in several tumor types
(53) and ectopic expression of
PDCD4 reduces tumor formation in a mouse skin cancer model
(54). On a molecular level, PDCD4
binds and inhibits the translation initiation factor eukaryotic
initiation factor 4a, thereby impacting protein translation
(55). In addition, PDCD4 has been
found to inhibit activator protein-mediated transactivation
(56) and to induce the expression
of the cyclin-dependent kinase inhibitor p21 (57). As a result, the loss of PDCD4
confers growth advantages to the cells by several means, thereby
facilitating the development and promotion of cancer. In recent
studies, PDCD4 was reported as a functional target of miR-21 in
various aspects of tumor progression; cell proliferation, invasion,
metastasis and neoplastic transformation in breast cancer (58) and invasion, intravasation, and
metastasis in colon cancer (47).
In the present study, a high expression of miR-21 was found in the
6 tested HNSCC lines of which two were HPV-associated. Furthermore,
suppression of miR-21 in vitro led to reduction of cellular
proliferation. Therefore, we hypothesized that PDCD4 was also an
important target of miR-21 in HNSCC. As shown in Fig. 4D, anti-miR-21-transfected cells
showed a significant increase in PDCD4 protein without any change
in PDCD4 mRNA. Transient transfection of cells with a reporter
plasmid containing the 3′-UTR of PDCD4 mRNA and anti-miR-21
inhibitor led to a significant increase of reporter activity. These
findings suggest that the PDCD4 is negatively regulated by miR-21
at the post-translational level via binding to the 3′-UTR of PDCD4
mRNA.
In the present study, we also found that
downregulation of miR-21 expression induced cell cycle arrest in
G1/S phase. The actions of some cytotoxic drugs are cell
cycle-specific. The S phase is where DNA synthesis takes place.
Many cell cycle-specific drugs act only on cells that are in the S
phase. These drugs interfere with DNA synthesis in some way,
therefore miR-21 may play a role in increasing the sensitivity of
HNSCC lines to these cell cycle-specific cytotoxic drugs.
In summary, miR-21 was overexpressed in all tested
HNSCC cell lines, including two HPV-associated lines, and
anti-miR-21 inhibited cellular proliferation in vitro. These
effects are possibly due to downregulation of the tumor suppressor
PDCD4 by miR-21. These findings raise the possibility that
anti-miR-21 may have potential therapeutic value in HNSCC patients
and may also play a role as predictor in healthy individuals. It
has been shown that anti-miR oligonucleotides could stay for a
relatively long period of time in animals (59,60).
Therefore, miRs, in particular miR-21, may serve as a potentially
useful target for cancer therapy.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Cheng ZX, Sun B, Wang SJ, et al: Nuclear
factor-κB-dependent epithelial to mesenchymal transition induced by
HIF-1α activation in pancreatic cancer cells under hypoxic
conditions. PLoS One. 6:e237522011.
|
|
3
|
Semenza GL: HIF-1 and tumor progression:
pathophysiology and therapeutics. Trends Mol Med. 8(Suppl 4):
S62–S67. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Imai T, Horiuchi A, Wang C, et al: Hypoxia
attenuates the expression of E-cadherin via up-regulation of SNAIL
in ovarian carcinoma cells. Am J Pathol. 163:1437–1447. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosenfeld N, Aharonov R, Meiri E, et al:
MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol.
26:462–469. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu Y, Zhu H, Lv L, Zhou Y and Huo J:
MiRNAs in oesophageal squamous cancer. Neth J Med. 71:69–75.
2013.PubMed/NCBI
|
|
9
|
Janiszewska J, Szaumkessel M and Szyfter
K: microRNAs are important players in head and neck carcinoma: a
review. Crit Rev Oncol Hematol. 88:716–728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Q, Wang C, Lu Z, Guo L and Ge Q:
Analysis of serum genome-wide microRNAs for breast cancer
detection. Clin Chim Acta. 413:1058–1065. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang H, Gong F, Zhang S, Zhang CY, Zen K
and Chen X: The origin, function, and diagnostic potential of
extracellular microRNAs in human body fluids. Wiley Interdiscip Rev
RNA. 5:285–300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Endo K, Weng H, Kito N, Fukushima Y and
Iwai N: miR-216a and miR-216b as markers for acute phased
pancreatic injury. Biomed Res. 34:179–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olivieri F, Rippo MR, Procopio AD and
Fazioli F: Circulating inflamma-miRs in aging and
age-related diseases. Front Genet. 4:1212013.
|
|
14
|
de Yébenes VG, Bartolomé-Izquierdo N and
Ramiro AR: Regulation of B-cell development and function by
microRNAs. Immunol Rev. 253:25–39. 2013.PubMed/NCBI
|
|
15
|
Stefani G: Roles of microRNAs and their
targets in cancer. Expert Opin Biol Ther. 7:1833–1840. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang J, Hao Y and Xi JJ: Therapeutic
application of microRNAs against human cancers. J Lab Autom.
18:30–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar
|
|
18
|
Lo WL, Yu CC, Chiou GY, et al:
MicroRNA-200c attenuates tumour growth and metastasis of
presumptive head and neck squamous cell carcinoma stem cells. J
Pathol. 223:482–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Jiang L, Wang A, Yu J, Shi F and
Zhou X: MicroRNA-138 suppresses invasion and promotes apoptosis in
head and neck squamous cell carcinoma cell lines. Cancer Lett.
286:217–222. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen HC, Chen GH, Chen YH, et al: MicroRNA
deregulation and pathway alterations in nasopharyngeal carcinoma.
Br J Cancer. 100:1002–1011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu X, Chen Z, Zhao X, et al: MicroRNA-25
promotes cell migration and invasion in esophageal squamous cell
carcinoma. Biochem Biophys Res Commun. 421:640–645. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao W, Xu J, Liu L, Shen H, Zeng H and Shu
Y: A systematic-analysis of predicted miR-21 targets identifies a
signature for lung cancer. Biomed Pharmacother. 66:21–28. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schee K, Boye K, Abrahamsen TW, Fodstad Ø
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12:5052012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Young MR, Santhanam AN, Yoshikawa N and
Colburn NH: Have tumor suppressor PDCD4 and its counteragent
oncogenic miR-21 gone rogue? Mol Interv. 10:76–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Allgayer H: Pdcd4, a colon cancer
prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol.
73:185–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fassan M, Pizzi M, Battaglia G, et al:
Programmed cell death 4 (PDCD4) expression during multistep
Barrett’s carcinogenesis. J Clin Pathol. 63:692–696.
2010.PubMed/NCBI
|
|
27
|
Fassan M, Pizzi M, Giacomelli L, et al:
PDCD4 nuclear loss inversely correlates with miR-21 levels in colon
carcinogenesis. Virchows Arch. 458:413–419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wolf JS, Chen Z, Dong G, et al: IL
(interleukin)-1α promotes nuclear factor-κB and AP-1-induced IL-8
expression, cell survival, and proliferation in head and neck
squamous cell carcinomas. Clin Cancer Res. 7:1812–1820. 2001.
|
|
30
|
Itani S, Kunisada T, Morimoto Y, et al:
MicroRNA-21 correlates with tumorigenesis in malignant peripheral
nerve sheath tumor (MPNST) via programmed cell death protein 4
(PDCD4). J Cancer Res Clin Oncol. 138:1501–1509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horiuchi A, Iinuma H, Akahane T, Shimada R
and Watanabe T: Prognostic significance of PDCD4 expression and
association with microRNA-21 in each Dukes’ stage of colorectal
cancer patients. Oncol Rep. 27:1384–1392. 2012.PubMed/NCBI
|
|
32
|
Wang J and Zhang Y: Expression of
programmed cell death 4 protein is closely correlated with
laryngeal squamous cell carcinomas. Lin Chung Er Bi Yan Hou Tou
Jing Wai Ke Za Zhi. 25:539–541. 2011.(In Chinese).
|
|
33
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi GH, Ye DW, Yao XD, et al: Involvement
of microRNA-21 in mediating chemo-resistance to docetaxel in
androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sin.
31:867–873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu YZ, Xi QH, Ge WL and Zhang XQ:
Identification of serum microRNA-21 as a biomarker for early
detection and prognosis in human epithelial ovarian cancer. Asian
Pac J Cancer Prev. 14:1057–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giovannetti E, Funel N, Peters GJ, et al:
MicroRNA-21 in pancreatic cancer: correlation with clinical outcome
and pharmacologic aspects underlying its role in the modulation of
gemcitabine activity. Cancer Res. 70:4528–4538. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Feng YH, Wu CL, Tsao CJ, et al:
Deregulated expression of sprouty2 and microRNA-21 in human colon
cancer: correlation with the clinical stage of the disease. Cancer
Biol Ther. 11:111–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026.
2012.PubMed/NCBI
|
|
41
|
Selaru FM, Olaru AV, Kan T, et al:
MicroRNA-21 is overexpressed in human cholangiocarcinoma and
regulates programmed cell death 4 and tissue inhibitor of
metalloproteinase 3. Hepatology. 49:1595–1601. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tran N, McLean T, Zhang X, et al: MicroRNA
expression profiles in head and neck cancer cell lines. Biochem
Biophys Res Commun. 358:12–17. 2007. View Article : Google Scholar
|
|
44
|
Huang S, Li XQ, Chen X, Che SM, Chen W and
Zhang XZ: Inhibition of microRNA-21 increases radiosensitivity of
esophageal cancer cells through phosphatase and tensin homolog
deleted on chromosome 10 activation. Dis Esophagus. 26:823–831.
2013. View Article : Google Scholar
|
|
45
|
Schetter AJ, Leung SY, Sohn JJ, et al:
MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dillhoff M, Liu J, Frankel W, Croce C and
Bloomston M: MicroRNA-21 is overexpressed in pancreatic cancer and
a potential predictor of survival. J Gastrointest Surg.
12:2171–2176. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Feber A, Xi L, Luketich JD, et al:
MicroRNA expression profiles of esophageal cancer. J Thorac
Cardiovasc Surg. 135:255–260. 2008. View Article : Google Scholar
|
|
49
|
Fulci V, Chiaretti S, Goldoni M, et al:
Quantitative technologies establish a novel microRNA profile of
chronic lymphocytic leukemia. Blood. 109:4944–4951. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Markou A, Tsaroucha EG, Kaklamanis L,
Fotinou M, Georgoulias V and Lianidou ES: Prognostic value of
mature microRNA-21 and microRNA-205 overexpression in non-small
cell lung cancer by quantitative real-time RT-PCR. Clin Chem.
54:1696–1704. 2008. View Article : Google Scholar
|
|
52
|
Yang HS, Jansen AP, Nair R, et al: A novel
transformation suppressor, Pdcd4, inhibits AP-1 transactivation but
not NF-κB or ODC transactivation. Oncogene. 20:669–676.
2001.PubMed/NCBI
|
|
53
|
Jansen AP, Camalier CE, Stark C and
Colburn NH: Characterization of programmed cell death 4 in multiple
human cancers reveals a novel enhancer of drug sensitivity. Mol
Cancer Ther. 3:103–110. 2004.PubMed/NCBI
|
|
54
|
Jansen AP, Camalier CE and Colburn NH:
Epidermal expression of the translation inhibitor programmed cell
death 4 suppresses tumorigenesis. Cancer Res. 65:6034–6041. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Göke A, Göke R, Knolle A, et al: DUG is a
novel homologue of translation initiation factor 4G that binds
eIF4A. Biochem Biophys Res Commun. 297:78–82. 2002.PubMed/NCBI
|
|
56
|
Yang HS, Jansen AP, Komar AA, et al: The
transformation suppressor Pdcd4 is a novel eukaryotic translation
initiation factor 4A binding protein that inhibits translation. Mol
Cell Biol. 23:26–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Göke R, Barth P, Schmidt A, Samans B and
Lankat-Buttgereit B: Programmed cell death protein 4 suppresses
CDK1/cdc2 via induction of p21Waf1/Cip1. Am J Physiol
Cell Physiol. 287:C1541–C1546. 2004.PubMed/NCBI
|
|
58
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in
breast cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xu L, Dai WQ, Xu XF, Wang F, He L and Guo
CY: Effects of multiple-target anti-microRNA antisense
oligodeoxyribonucleotides on proliferation and migration of gastric
cancer cells. Asian Pac J Cancer Prev. 13:3203–3207. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lennox KA and Behlke MA: A direct
comparison of anti-microRNA oligonucleotide potency. Pharm Res.
27:1788–1799. 2010. View Article : Google Scholar : PubMed/NCBI
|