Radiocontrast medium induces histamine release in association with upregulation of miR‑19a‑3p and miR‑362‑3p expression
- Authors:
- Published online on: April 19, 2024 https://doi.org/10.3892/br.2024.1780
- Article Number: 93
-
Copyright: © Chang et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
Abstract
Introduction
The development of imaging medicine has seen a steady increase in the use of computerized tomography (CT) technology in the diagnosis of diseases. The application of CT technology requires the use of contrast medium to enhance image quality and output (1). The most commonly used contrast medium in clinical practice is iodinated contrast medium. Although iodinated contrast medium provides a clear image of the body, adverse effects following use of this agent have been reported (2). The most common adverse effect was type I hypersensitivity caused by iodinated contrast medium-induced mast cell activation and histamine release (3). Type I hypersensitivity is often triggered by allergen-specific IgE-mediated immune cell activation, such as mast cell and basophil (4). When mast cells or basophils have been activated, they release histamine and other mediators to cause inflammatory responses such as itching, urticaria and skin rash, and anaphylaxis in severe cases (5). This allergic reaction is regulated by the FcεRI downstream signaling pathway (6). Allergens are recognized by the IgE Fab site and signal transduction is initiated, which first involves activation of tyrosine kinase, providing a docking site for syk kinase. After syk kinase is phosphorylated, downstream adaptors, such as LAT1 and Grb2, are subsequently phosphorylated by syk kinase, leading to type I hypersensitivity (7). As well as cell membrane-localized signal transduction, intracellular signaling pathways, such as PI3K, ERK, JNK, and Akt play a role in histamine release and expression of proinflammatory cytokines by mast cells (8).
In recent years, microRNAs (miRNAs/miRs) have been reported to play an important role in mast cell activation (9). miRNAs are short, single-stranded RNAs consisting of only 19-25 nucleotides, which can bind to target RNA and serve as a negative or positive regulator. For example, when mast cells have been activated, intracellular signal pathways are activated, leading to the release of proinflammatory cytokines and histamines and miR126 has been demonstrated to increase the degranulation of mast cells by enhancing PI3K, Akt, and Ca++ influx (10). miR 302e can reduce mast cell activation by binding to the 3'-UTR of RelA mRNA (11). However, the role of miRNA in contrast-medium-induced mast cell activation remains unclear and needs further clarification.
The present study aimed to investigate the expression of miRNA in mast cells following stimulation with contrast medium in clinical practice. The results showed that miR 19a-3p and miR 362-3p were associated with contrast medium-induced mast cell activation and histamine release.
Materials and methods
Cell culture
The mouse mast cell cell line, P815 (Elabscience Biotechnology, Inc.), was cultured in RPMI1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Cytiva) and 1% penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37˚C incubator with 5% CO2.
Cell viability
P815 cells were incubated at 37˚C with different doses of ultravist (Bayer AG) for 24 h and cell viability was measured by Cell Counting Kit 8 (MilliporeSigma) according to the manufacturer's protocol. Cells were seeded in 96-well culture plates at a density of 1x104 cells per well for 24 h and then incubated with different concentrations of ultravist for 24 h. Then, 10 µl of CCK-8 solution was added to each well and incubated for 1 h at 37˚C. Absorbance was measured at 450 nm by a multi-well plate reader (Sunrise; Tecan Group Ltd.).
β-hexosaminidase measurement
P815 cells were incubated at 37˚C with different doses of ultravist (Bayer AG) for 24 h and cells were lysed by RIPA buffer. Cell lysates were centrifuged at 12,902 x g for 10 min at 4˚C and supernatant was collected for measuring the level of β-hexosaminidase. Briefly, the concentration of β-hexosaminidase was detected by the sandwich ELISA method according to the manufacturer's protocol (Fine Test; Wuhan Fine Biotech Co., Ltd.). Optical density at 450 nm was detected by ELISA reader (Sunrise; Tecan Group Ltd.).
Histamine measurement
P815 cells were incubated at 37˚C with different doses of ultravist (Bayer AG) for 24 h and culture supernatant was centrifuged at 193 x g for 5 min at 4˚C to remove cells and then the concentration of histamine was measured. All procedures of histamine measurement were conducted according to the manufacturer's protocol (Abcam) Briefly, samples were incubated on an antibody-coated ELISA plate for 1 h at room temperature. The plate was washed 3 times and incubated with HRP-conjugated detection antibody for 30 min at room temperature. The ELISA plate was washed 3 times and incubated with TMB substrate for 30 min at room temperature. H2SO4 was used to stop the reaction and optical density at 450 nm was detected by an ELISA reader (Sunrise; Tecan Group Ltd.).
RNA extraction, library preparation and sequencing
P815 cells were stimulated with ultravist (Bayer AG) at 37˚C for 24 h and total RNA was extracted using a Genezol TriRNA pure kit (Geneaid Biotech Ltd.) according to the manufacturer's protocol. A total of 100 ng total RNA was used as input material for the small RNA sample preparations. Sequencing libraries were generated using QIAseq® miRNA Library Kit (Qiagen GmbH) following the manufacturer's recommendations. Briefly, 3' and 5' adaptors were directly and specifically ligated to the 3' and 5' ends of small RNA, respectively. Then first-strand cDNA was synthesized using QIAseq miRNA NGS RT Enzyme and RT primer (Qiagen GmbH). After PCR amplification, the library was size-selected with 170-200 bp by QIAseq beads. The quality of purified libraries was assessed on the Qsep400 system (Qiagen GmbH). The qualified libraries were then sequenced on Illumina NovaSeq 6000 platform (Illumina, Inc.) with trimmed 75 bp single-end reads generated by Genomics, BioSci & Tech Co., Ltd.
Bioinformatics analysis
The adapter sequences in raw sequenced data were removed using TrimGalorev0.6.6 (https://github.com/FelixKrueger/TrimGalore) and mapped to reference genome, mature and hairpin miRNAs (12) (miRBase v.22.1) with Bowtie (13) (v1.3.0) to obtain proper miRNA reads. After the alignment step, bam files were processed using samtools (14) (v1.12) and the expression profile of miRNAs was calculated and normalized using edgeR (15) (v3.26.5). All of the differentially expressed miRNAs (DEmiRNAs) were identified using DEGSeq (16) (v1.48.0) and used for target prediction with three different tools, miRanda (17), Targets can (18), and PITA (19). The functional enrichment analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways among DEmiRNA targets predicted by all three tools was implemented in an R package called clusterProfiler (20) (v3.14.0). The miRNAs of interest were analyzed using mirPATHDB 2.0 to identify significant pathways. Subsequently, the identified KEGG pathways were visualized using Pathview (21). For the intersection analysis, mirNET 2.0(22) was used to input the two miRNAs and to identify the intersecting target genes common to both. The mirNET network analysis feature was then utilized to construct a network diagram representing these connections.
miRNA extraction and reverse transcription-quantitative (RT-q) PCR
A total of 2.5 x105 P815 cells were stimulated with ultravist (Bayer AG) at 37˚C for 24 h and miRNA was extracted from cells using the miTotal RNA extraction miniprep system, according to the manufacturer's protocol (Viogene-Biotek Corporation). The complementary DNA (cDNA) transcripts were amplified using miScript II RT Kit (Qiagen GmbH) according to the manufacturer's protocol. qPCR was performed using a TopQ miRNA probe qPCR assay kit (Topgen Biotechnology Co., Ltd.) according to the manufacturer's protocol and detected by StepOne plus (Thermo Fisher Scientific, Inc.). The method of quantification was used the 2(-Delta Delta C(T)) method (23). The thermocycling conditions of qPCR was initiation at 95˚C for 1 min, followed by 40 cycles at 95˚C for 3 sec and 60˚C for 40 sec. The primer and probe sequences of miR-362-3p and miR-19a-3p are shown in Table I. U6 was used as the internal control and for normalization. The experiments were replicated three times.
Statistical analysis
Data obtained from cell viability, level of histamine, β-hexosaminidase, and miR-362-3p, miR-19a-3p expression were analyzed by one-way ANOVA, followed by Tukey's post hoc test, and analyses were performed using GraphPad Prism 6 (GraphPad; Dotmatics). The results are presented as the mean ± SEM. P<0.05 was considered to indicate a statistically significant difference.
Results
Contrast medium could induce mast cell activation
The cytotoxicity of ultravist was detected to determine the experimental condition for the following experiments. P815 cells were incubated with different doses of ultravist for 24 h and cell viability was measured by CCK-8 test. The results showed that over 150 mg/ml of ultravist could induce cell death (Fig. 1). Next, P815 cells were stimulated with 25, 50 and 100 mg/ml ultravist for 24 h and the level of histamine in the supernatant and intracellular β-hexosaminidase concentration detected. The results showed that histamine levels were increased dose-dependently with ultravist concentration. To further investigate how Ultravist regulated the response of mast cells, the concentration of intracellular β-hexosaminidase was measured. The result showed that expression of intracellular β-hexosaminidase was decreased in a dose-dependent manner with increasing ultravist concentrations (Fig. 2). These results indicated ultravist could induce mast cell activation and histamine release.
miRNA plays a role in mast cell activation induced by contrast medium through the regulation of the mTOR signaling pathway
In the present study, 50 mg/ml of ultravist was identified as a threshold concentration capable of activating mast cells without inducing cytotoxicity. Then, miRNA sequencing was employed to examine changes in miRNA expression following stimulation with 50 mg/ml of ultravist for 24 h. The miRNA expression profile was analyzed and the analysis revealed a total of 20 miRNAs that were differentially upregulated or downregulated (Fig. 3). This result suggested that contrast medium-induced mast cell activation might be associated with miRNA modulation. For further investigation into the correlation between miRNA expression and mast cell activation induced by contrast medium, the present study focused on the mTOR signaling pathway. The mTOR signaling pathway plays a pivotal role in mast cell activation. In the analysis of P815 cell samples, two miRNAs were identified, miR-19a-3p and miR-362-3p, that were upregulated upon stimulation with contrast medium and associated with the activation of the mTOR signaling pathway. The examination of miRNA expression data revealed a significant upregulation of several key components within this pathway, including mTOR, AMP-activated protein kinase, PTEN and protein kinase C, as indicated on the KEGG pathway map (Fig. 4A and B). These changes suggested an enhancement of mTOR activity in samples stimulated with contrast medium compared with the control group.
Validating expression of miR-19a-3p and miR-362-3p following contrast medium stimulation
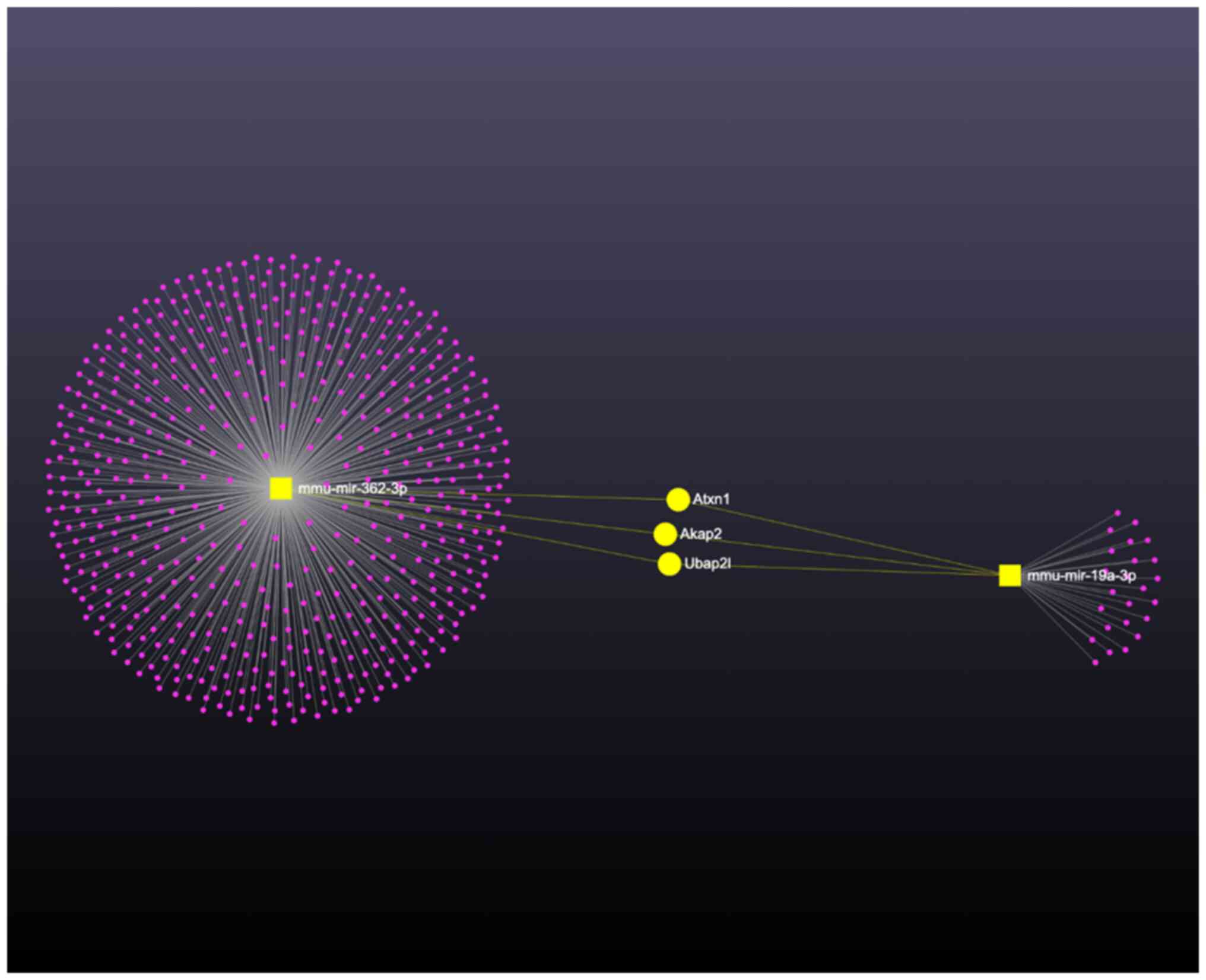
In recent years, miR-19a has been reported to be associated with allergy-related cytokine expression (24) and the role of miR-362-3p in mast cell activation remains unclear. Therefore, these two miRNAs were selected to further validate the effect of contrast medium-induced miRNA overexpression. The results showed that after stimulating with different doses of ultravist for 24 h, statistical analysis confirmed a significant dose-dependent increase in the expressions of miR-19a-3p and miR-362-3p with increasing doses of ultravist (P<0.05; Fig. 5). The present study further analyzed the role of miR-19a-3p and miR-362-3p by KEGG analysis. In its comprehensive analysis of miRNA-mediated regulation in mast cell activation, the present study focused on the co-regulatory effects of miR-362-3p and miR-19a-3p. The network graph in Fig. 6 illustrates the complex interaction landscape, with miR-362-3p and miR-19a-3p at the center of the network. Through the bioinformatic analysis, a subset of genes that are potentially co-regulated by both miR-362-3p and miR-19a-3p were identified. Notably, genes such as Atxn1, Akap2, and Uba2pl emerged as significant targets, indicated by their direct links in the network (highlighted in yellow). These target genes are implicated in key processes within the mTOR signaling pathway, suggesting a synergistic effect of miR-362-3p and miR-19a-3p in modulating the pathway's activity. The identification of these co-regulated genes provides new insights into the molecular mechanisms by which miRNAs may influence mast cell response to contrast medium stimulation.
Discussion
Contrast medium-induced mast cell activation has always been an important issue in radiology. To the best of the authors' knowledge, this is the first report to investigate the correlation between miRNA expression and mast cell activation following stimulation with the contrast medium, ultravist. The present study demonstrated that ultravist could induce mast cell activation through histamine release and decrease the level of β-hexosaminidase. The miRNA profile following stimulation with ultravist in mast cells was also investigated and this further confirmed the role of miR-19a-3p and miR-362-3p in contrast medium-induced mast cell activation.
Histamine release is a landmark feature of mast cell activation. Contrast medium-induced adverse effects, such as skin rash, itching and flushing, are all associated with mast cell overactivation (25). When mast cells are activated, intracellular mediators, such as histamine and β-hexosaminidase, are released into the tissue. This further induces vascular dilation and increases vaso permeability, thus causing skin rash or vaso edema (26). In serious cases, it can lead to anaphylactic shock (27). β-hexosaminidase, a lysosomal enzyme, is commonly used as a marker for mast cell degranulation. The aim of the present study was not only to evaluate mast cell degranulation but also to investigate the overall cellular response to radiocontrast media, including alterations in cellular enzyme levels. Consequently, the level of β-hexosaminidase in the cell lysate was measured. The present study showed that the contrast medium ultravist increased secreted histamine level and decreased intracellular β-hexosaminidase concentration. This finding indicated that ultravist could induce mast cell activation. However, the present study used only a single mast cell line to demonstrate that contrast medium could increase histamine release. It is suggested that multiple cell lines and animal model be used in future research to validate these findings.
In recent years, miRNA has been reported to play a role in mast cell activation (9). In a dinitrophenyl-induced mouse bone marrow-derived mast cell model, a total of 17 miRNA expressions were changed compared with the resident mast cell. Among these results, expressions of miR-21a and miR-3113 showed the highest increase and decrease respectively in the activated mast cells compared with the resident mast cells (28). These miRNA expressions could regulate downstream signaling, such as PI3K and PKC, to inhibit intracellular mediator synthesis and degranulation (29). The present study used a high-throughput method to screen contrast medium-induced miRNA expression and found that the expression of more than 40 miRNA was changed. These findings are consistent with previous reports on mast cell activation.
For the validation of miRNA sequencing data, two miRNAs, miR-19a-3p and miR-362-3p, were selected and in vitro experiment used to confirm the results. Previous reports on the physiological role of miR-19a-3p have shown associations with tumorigenesis and suppression of M1 macrophage differentiation (30,31). The possible mechanisms involve regulation of THBS1 protein expression or inhibition of the STAT1/IRF1 signaling pathway. Furthermore, miR-19 family members have been associated with inflammatory responses in sepsis and induced neuropathic pain. The mechanism is through regulating the expression of KLF7 and miR-19b-3p (32,33). Most reports on miR-362-3p are related to tumor suppressor genes modulation. For example, miR-362-3p can target MCM5 protein, SERBP1 gene and JMJD2A enzyme expression to suppress cervical adenocarcinoma, ovarian cancer progression and nasopharyngeal tumor progression (34-36). In addition to cancer-related studies, miR-362-3p has also been reported to play an important role in the pathogenesis of inflammatory bowel disease (37). However, to the best of the authors' knowledge, there are no data in the literature on the two aforementioned miRNAs regarding their roles in mast cell activation, and thus, the present study is the first to report on the potential roles of miR-19a-3p and miR-362-3p in contrast medium-induced mast cell activation. Furthermore, according to the KEGG pathway analysis, miR-19a-3p and miR-362-3p are also associated with other cell physiology functions. Further study is needed to elucidate the mechanism and downstream signaling pathway.
The present study demonstrated that contrast medium could induce mast cell activation and miRNA overactivation. However, there were some limitations in the present study. First, although contrast medium-induced histamine release and decrease in β-hexosaminidase level were observed, the underlying mechanisms and downstream signaling pathway are unclear and hence further research including multiple cell lines and animal model are warranted. Second, the miRNA array data showed expressions were changed in 40 miRNAs, but only two of them were selected for further evaluation. Furthermore, among the three attempts for the validation, one experiment failed due to the absence of signaling, prompting the analysis of only two of the three datasets to ascertain the effect of the contrast medium on miRNA expression. This decision was made to ensure the accuracy and reliability of the findings. The potential implications of excluding the failed attempt was thoroughly considered and this approach maintained the integrity of the results of the present study. Even so, there was insufficient evidence to prove which one played the more important role in the contrast medium-induced mast cell activation. miRNA inhibitors should be used in future experiments to clarify the effects of selected miRNAs on contrast medium-induced mast cell activation. Third, the present study has a lack of clinical validation in blood samples from individuals with hypersensitivity compared with individuals with no allergic reaction to contrast medium. Further experimentation will be necessary in the future.
In conclusion, the present study showed that contrast medium could induce mast cell activation and miRNA expression. It also reported high throughput data of miRNA expressions after mast cell stimulation with contrast medium. These data demonstrated that miRNAs are involved in contrast medium-induced mast cell activation and that these miRNAs may have potential as a target to prevent contrast medium-induced adverse effects.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Zuoying Branch of Kaohsiung Armed Forces General Hospital, Kaohsiung, Taiwan (grant nos. KAFGH-ZY-A-110020 and KAFGH-ZY_A_111016) and by MacKay Medical College, New Taipei City, Taiwan (grant nos. MMC-RD-112-1C-P004 and MMC-RD-112-1B-P014).
Availability of data and materials
The data generated in the present study may be found in the BioProject database with the accession number PRJNA1085472, which can be accessed via the following URL: http://www.ncbi.nlm.nih.gov/bioproject/1085472.
Authors' contributions
WFC, PWH, ECL and SJY designed the experiments. CLL, HSH, TYC and SJY performed the experiments. HSH and TYC acquired and analyzed the results. WFC, PWH, CLL, ECL and SJY drafted the initial manuscript. CLL and SJY confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
Bae KT: Intravenous contrast medium administration and scan timing at CT: Considerations and approaches. Radiology. 256:32–61. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Bottinor W, Polkampally P and Jovin I: Adverse reactions to iodinated contrast media. Int J Angiol. 22:149–154. 2013.PubMed/NCBI View Article : Google Scholar | |
|
Zhang B, Dong Y, Liang L, Lian Z, Liu J, Luo X, Chen W, Li X, Liang C and Zhang S: The incidence, classification, and management of acute adverse reactions to the low-osmolar iodinated contrast media isovue and ultravist in contrast-enhanced computed tomography scanning. Medicine (Baltimore). 95(e3170)2016.PubMed/NCBI View Article : Google Scholar | |
|
Bannon GA: Hypersensitivity: Anaphylactic (Type I). In Van Nostrand's Scientific Encyclopedia. G.D. Considine (ed.). Wiley & Sons, Inc., Hoboken NJ, 2007. https://doi.org/10.1002/9780471743989.vse9966. | |
|
Nakamura T: The roles of lipid mediators in type I hypersensitivity. J Pharmacol Sci. 147:126–131. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Nagata Y and Suzuki R: FcεRI: A master regulator of mast cell functions. Cells. 11(622)2022.PubMed/NCBI View Article : Google Scholar | |
|
Kanagaratham C, El Ansari YS, Lewis OL and Oettgen HC: IgE and IgG antibodies as regulators of mast cell and basophil functions in food allergy. Front Immunol. 11(603050)2020.PubMed/NCBI View Article : Google Scholar | |
|
Gilfillan AM and Rivera J: The tyrosine kinase network regulating mast cell activation. Immunol Rev. 228:149–169. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Shefler I, Salamon P and Mekori YA: MicroRNA involvement in allergic and non-allergic mast cell activation. Int J Mol Sci. 20(2145)2019.PubMed/NCBI View Article : Google Scholar | |
|
Bao Y, Wang S, Gao Y, Zhang W, Jin H, Yang Y and Li J: MicroRNA-126 accelerates IgE-mediated mast cell degranulation associated with the PI3K/Akt signaling pathway by promoting Ca(2+) influx. Exp Ther Med. 16:2763–2769. 2018.PubMed/NCBI View Article : Google Scholar | |
|
Xiao L, Jiang L, Hu Q and Li Y: MiR-302e attenuates allergic inflammation in vitro model by targeting RelA. Biosci Rep. 38:2018.PubMed/NCBI View Article : Google Scholar | |
|
Griffiths-Jones S: miRBase: The microRNA sequence database. Methods Mol Biol. 342:129–138. 2006.PubMed/NCBI View Article : Google Scholar | |
|
Langmead B and Salzberg SL: Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 2009.PubMed/NCBI View Article : Google Scholar | |
|
Robinson MD, McCarthy DJ and Smyth GK: edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Wang L, Feng Z, Wang X, Wang X and Zhang X: DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 26:136–138. 2010.PubMed/NCBI View Article : Google Scholar | |
|
Enright AJ, John B, Gaul U, Tuschl T, Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol. 5(R1)2003.PubMed/NCBI View Article : Google Scholar | |
|
Agarwal V, Bell GW, Nam JW and Bartel DP: Predicting effective microRNA target sites in mammalian mRNAs. Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar | |
|
Kertesz M, Iovino N, Unnerstall U, Gaul U and Segal E: The role of site accessibility in microRNA target recognition. Nat Genet. 39:1278–1284. 2007.PubMed/NCBI View Article : Google Scholar | |
|
Yu G, Wang LG, Han Y and He QY: ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar | |
|
Luo W, Pant G, Bhavnasi YK, Blanchard Jr SG and Brouwer C: Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 45(W1):W501–W508. 2017.PubMed/NCBI View Article : Google Scholar | |
|
Chang L, Zhou G, Soufan O and Xia J: miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48(W1):W244–W251. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Livak KJ and Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar | |
|
Gil-Martínez M, Lorente-Sorolla C, Naharro S, Rodrigo-Muñoz JM and del Pozo V: Advances and Highlights of miRNAs in Asthma: Biomarkers for diagnosis and treatment. Int J Mol Sci. 24(1628)2023.PubMed/NCBI View Article : Google Scholar | |
|
Kulkarni KN, Hegde RG, Balani A and Joshi AR: Transient angioedema of small bowel secondary to intravenous iodinated contrast medium. Indian J Radiol Imaging. 24:303–305. 2014.PubMed/NCBI View Article : Google Scholar | |
|
Zou W, Yang S, Chen L, Hu S, Hao G and Hu C: Iodixanol activation of mast cells: Implications in the pathogenesis of iodixanol-induced delayed cutaneous adverse reactions. Toxicology. 465(153034)2022.PubMed/NCBI View Article : Google Scholar | |
|
Lee SH, Park JW and Hwang BM: Anaphylactic shock following nonionic contrast medium during caudal epidural injection. Korean J Pain. 28:280–283. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Li Y, Liu J, Zhang J, Zhang W and Wu Z: Characterization of microRNA profile in IgE-mediated mouse BMMCs degranulation. J Microbiol Immunol Infect. 53:550–560. 2020.PubMed/NCBI View Article : Google Scholar | |
|
Teng Y, Zhang R, Yu H, Wang H, Hong Z, Zhuang W and Huang Y: Altered MicroRNA expression profiles in activated mast cells following IgE-FcεRI cross-linking with antigen. Cell Physiol Biochem. 35:2098–2110. 2015.PubMed/NCBI View Article : Google Scholar | |
|
Zhu X, Guo Q, Zou J, Wang B, Zhang Z, Wei R, Zhao L, Zhang Y, Chu C, Fu X and Li X: MiR-19a-3p Suppresses M1 macrophage polarization by inhibiting STAT1/IRF1 pathway. Front Pharmacol. 12(614044)2021.PubMed/NCBI View Article : Google Scholar | |
|
Xu G, Li J and Yu L: miR-19a-3p promotes tumor-relevant behaviors in bladder urothelial carcinoma via targeting THBS1. Comput Math Methods Med. 2021(2710231)2021.PubMed/NCBI View Article : Google Scholar | |
|
Jiang L, Wang M, Sun R, Lin Z, Liu R, Cai H, Tang Z and Zhang R: Methylation of miR-19b-3p promoter exacerbates inflammatory responses in sepsis-induced ALI via targeting KLF7. Cell Biol Int. 45:1666–1675. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Ye L, Morse LR, Falci SP, Olson JK, Shrivastava M, Nguyen N, Linnman C, Troy KL and Battaglino RA: hsa-MiR-19a-3p and hsa-MiR-19b-3p Are Associated with Spinal Cord Injury-Induced Neuropathic Pain: Findings from a Genome-Wide MicroRNA Expression Profiling Screen. Neurotrauma Rep. 2:424–439. 2021.PubMed/NCBI View Article : Google Scholar | |
|
Wang D, Wang H, Li Y and Li Q: MiR-362-3p functions as a tumor suppressor through targeting MCM5 in cervical adenocarcinoma. Biosci Rep. 38(BSR20180668)2018.PubMed/NCBI View Article : Google Scholar | |
|
Cao S, Li N and Liao X: miR-362-3p acts as a tumor suppressor by targeting SERBP1 in ovarian cancer. J Ovarian Res. 14(23)2021.PubMed/NCBI View Article : Google Scholar | |
|
Wang X and Chen P: Aberrant miR-362-3p is Associated with EBV-Infection and prognosis in nasopharyngeal carcinoma and involved in tumor progression by targeting JMJD2A. Onco Targets Ther. 15:121–131. 2022.PubMed/NCBI View Article : Google Scholar | |
|
Omidbakhsh A, Saeedi M, Khoshnia M, Marjani A and Hakimi S: Micro-RNAs-106a and -362-3p in peripheral blood of inflammatory bowel disease patients. Open Biochem J. 12:78–86. 2018.PubMed/NCBI View Article : Google Scholar |