Introduction
Glioma is the most common type of primary malignant
tumor of the central nervous system worldwide. Glioma is associated
with high rates of disability and mortality, and a poor prognosis
(1). According to the World
Health Organization (WHO) pathological classification, grades I and
II represent low-grade glioma (LGG), whereas grades III and IV
represent high-grade glioma (HGG) (1). Among all glioma cases, glioblastoma
(GBM) accounts for ~57% in the US, and its median survival time is
<2 years (2). Although the
survival duration of patients with LGG is longer than HGG, this
type of tumor may eventually progress to HGG after multiple
relapses (3,4). At present, the preferred treatment
for glioma remains surgical resection with maximum safety, with
temozolomide chemotherapy and radiotherapy as auxiliary methods
(5). However, owing to
characteristics such as drug resistance, radiotherapy resistance
and frequent recurrence of glioma, therapeutic efficacy remains
poor (6,7). Therefore, identifying effective
molecular targets (8-11) that help navigate this issue in
glioma treatment is necessary.
Transposons or transposable elements (TEs) are
present in almost all organisms; these mobile genetic elements
account for ~50% of the human genome (12,13). Transposons are usually classified
as RNA-based retrotransposons and 'cut-paste' or 'cut-copy' types
of transposons (14). Transposons
not only serve an active role in the regulation of gene expression,
but can occasionally impose negative effects, as their insertion
into genes may disrupt normal gene function and cause genomic
instability. They maintain genomic diversity and promote adaptive
evolution (15), thus serving as
a crucial factor at different stages of human growth and
development (16). However, owing
to the adverse effects of TE on the genome, they are usually
inactive (17) and the expression
mode of TE is strictly regulated during the human lifespan
(18). Evidence from published
studies has shown that changes in the regulatory mechanisms of TE
may lead to genomic instability, chromosome breakage and carcinogen
activation, triggering the development of various immune,
neurological and genetic diseases (19-21).
PiggyBac transposons are a category of TE. At
present, five complete piggyBac elements have been identified,
namely piggyBac transportable element derived (PGBD)1, 2, 3, 4 and
5 (22). Genomic data have shown
that PGBD5 in humans is the most conserved piggyBac sequence. This
gene can form a 'cut-paste' type of DNA transposon (23). PGBD5 is synthesized in human cells
in response to certain cellular conditions or signals. This
includes during developmental stages, in the presence of
inflammatory signals, and possibly in response to processes that
can lead to cancer. Its DNA transposition must occur in the entire
genome for the transposon to be precisely excised and
preferentially inserted into TTAA sites (24,25). The expression of PGBD5 in humans
and mice has been shown to be primarily limited to some regions of
the early embryonic and adult brains (26,27). In mice, PGBD5 is primarily
expressed in the nucleus, preferentially in specific areas of the
brain and central nervous system, which are rich in granulosa
cells; therefore, PGBD5 may be primarily expressed in granulosa
cells, which are a small population of neurons different from other
nerve cells in terms of morphology and function; some of these
cells can exert effects in adult nerve growth (26). Relevant studies have shown that
the expression of PGBD5 is sufficient to not only promote
carcinogenic genome rearrangement (28,29), but can also induce non-anchored
cell growth in vitro and tumor formation in vivo.
Consequently, the transformation of cells induced by PGBD5 has been
linked to various chromosomal alterations, including deletions,
inversions and translocations (28). In addition, in other studies,
abnormal PGBD5 expression has been reported in neural tissues and
most solid tumors in children, such as medulloblastoma,
rhabdomyoma, neuroblastoma, Ewing's sarcoma and ependymoma
(25,26,28). PGBD5 is frequently upregulated in
solid tumors in children and adults, suggesting its role in cancer
development through inducing DNA rearrangements. This upregulated
pattern offers a plausible mechanism for site-specific genomic
alterations observed in carcinogenesis (28). In a recent study, a survival-risk
prediction model was established using bioinformatics methods.
According to the model, the expression of PGBD5 was negatively
associated with the survival period of GBM, and it served as a
marker of poor prognosis (30).
Peroxisome proliferator-activated receptors (PPARs)
are ligand-induced transcription factors that belong to the nuclear
receptor family (31). When
integrating with ligands, PPARs translocate to the nucleus, bind to
peroxisome promoter response elements on DNA and heterodimerize
with retinoic-acid X receptor. When a ligand binds to a PPAR, it
primarily acts as a transcriptional regulator of specific target
genes (32). The role of PPARs in
lipid glucose metabolism and the regulation of homeostasis has been
studied widely. In mammals (33),
three subtypes of PPAR are present: PPARα, PPARβ/δ and PPARγ, and
their biological distribution patterns and ligand affinity vary
widely in different organs (34).
PPARα is primarily expressed in the liver, heart, kidney,
intestine, brown fat and skeletal muscle, activating fatty acid
catabolism and gluconeogenesis to regulate energy balance. In the
past decade, increasing evidence has shown that PPARα can regulate
one or more pathways to adjust tumorigenesis (35). However, contradictory effects of
PPARα on tumor regulation have been identified. Relevant studies
have shown that a reduction in the transcriptional activity of
PPARα enhances cell migration and invasion in vitro
(36,37), and PPARα agonists can restore
normal cellular behavior in cancer cell lines (38). Another study demonstrated that
PPARα upregulation activates liver cancer stem cells and promotes
the occurrence of early hepatocellular carcinoma (39). PPARβ/δ are commonly expressed in
various tissues; however, their biological functions vary within
these tissues. In addition, the roles of PPARβ/δ in various cancer
cell models differ and remain unclear (40). PPARγ has two subtypes: PPARγ1 and
PPARγ2; the former is dominant and is commonly expressed in various
tissues, whereas the latter is primarily confined to adipose
tissues. The primary functions of PPARγ include regulation of the
glucose-lipid balance, insulin sensitivity, adipogenesis,
inflammation, immune response and tumorigenesis (41).
Prior studies have indicated that PGBD5 is not only
involved in the regulation of gene expression, but is associated
with genomic instability and chromosome rearrangements. Therefore,
the present study aimed to investigate PGBD5 in glioma. Existing
research has suggested that PGBD5 is highly expressed in various
solid tumors, including glioma, in both children and adults
(28). This provides a strong
foundation for exploring its relevance in glioma pathogenesis. The
present study aimed to assess the specific molecular functions and
roles of PGBD5 in glioma, to understand whether its silencing
inhibits the proliferation, migration and invasion of glioma cells.
The present study explored the molecular mechanisms contributing to
glioma formation, with the aim of improving understanding.
Materials and methods
Tissues
A total of five patients (age range, 36-64 years;
mean age, 48.6 years) with glioma were recruited from the
Department of Neurosurgery, The First People's Hospital of Yunnan
Province (Kunming, China) between October and December 2021. The
exclusion criteria included patients with a history of neurological
disorders other than glioma, those who had undergone chemotherapy
or radiation therapy within the last 6 months, and individuals
<18 or >65 years old. Paired tumor tissues and para-cancerous
tissues, located at a specified distance of 2 cm from the tumor
margin, were obtained from the same patients. Only patients from
whom para-cancerous tissues could be successfully harvested during
surgical resection were included in the study. Tissues were
collected at the time of tumor resection to ensure the relevance
and freshness of the samples for subsequent analysis. Among the
five patients, three were male and two were female. Preoperative
diagnosis ruled out other tumors and chronic diseases, and the
patients were yet to undergo any treatment for glioma. The tumor
pathological grades corresponding to the tested specimens were as
follows Two cases of anaplastic astrocytoma (WHO III) and three
cases of GBM (WHO IV). The tissues obtained from each patient
underwent immunohistochemistry analysis to detect specific
proteins, including GFAP and IDH1. The staining intensity was
evaluated by at least two professional pathologists (data not
shown). Residual tumor tissues and adjacent tissues were collected
after pathological examination and were immediately stored in
liquid nitrogen. After the immunohistochemical diagnosis was
confirmed, the tissues were used for further research. The enrolled
patients provided written informed consent, the experimental
contents complied with relevant national regulations, and the
experimental procedures were approved by the Ethics Committee of
the Medical School at Kunming University of Technology (approval
no. KMUST-MEC-092; Kunming, China). The study was conducted in
accordance with The Declaration of Helsinki.
Cell culture
Human glioma cell lines [A172, U251 and U87 MG
(American Type Culture Collection version, GBM cell line of unknown
origin, hereinafter referred to as U87)] and normal human
astrocytes (NHAs; cat. no. CP-H122) were purchased from Procell
Life Science & Technology Co., Ltd. The cells were examined
using short-tandem repeat profiling to confirm authenticity and to
ensure they were free from cross-contamination. A172, U251 and NHA
cells were individually cultured in Dulbecco's modified Eagle
medium (Biological Industries; Sartorius AG) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in
a 5% CO2-humidified atmosphere at 37°C. U87 cells were
cultured in minimum essential medium-non-essential amino acids
(Biological Industries; Sartorius AG) supplemented with 10% FBS in
a 5% CO2-humidified atmosphere at 37°C. Penicillin was
added to all media at a concentration of 100 U/ml.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from glioma tissues and A172,
U251 and U87 cell lines using RNAIso Plus (Takara Biotechnology,
Ltd.; cat. no. 9108) and reverse-transcribed to cDNA using a
HiScript III All-in-one RT SuperMix Perfect for qPCR kit (Vazyme
Biotech Co., Ltd.) according to the manufacturer's protocol. qPCR
was performed using a ChamQ Universal SYBR qPCR Master Mix Kit
(Vazyme Biotech Co., Ltd.) and a LightCycler480II (Roche
Diagnostics), with an initial dena-turation step at 95°C for 5 sec,
followed by a joint annealing and extension step at 60°C for 20
sec. Relative mRNA expres-sion levels were determined using the
2-ΔΔCq method (42),
normalized to β-actin and represented graphically using GraphPad
Prism 9.0 (Dotmatics). The sequences of primers (TsingKe Biological
Technology) are listed in Table
I.
 | Table IList of primers and shRNA sequences
used. |
Table I
List of primers and shRNA sequences
used.
| Name | Sequence |
|---|
| PGBD5-F |
5′-CATGTCCTTGATCTGCTGGTAC-3′ |
| PGBD5-R |
5′-TGATGGCGAACCAGAACACCTG-3′ |
| β-actin-F |
5′-CCTTCCTGGGCATGGAGTC-3′ |
| β-actin-R |
5′-TGATCTTCATTGTGCTGGGTG-3′ |
| sh-PGBD5 #1 |
5′-GCAGAUACGAUGACAAAUATTCTCGAGUAUUUGUCAUCGUAUCUGCTT-3′ |
| sh-NT |
5′-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3′ |
Western blotting
Proteins were extracted from glioma tissues or cells
using the radioimmunoprecipitation assay lysis buffer (Beijing
Solarbio Science & Technology Co., Ltd.) containing a protease
inhibitor (Bimake). Proteins were quantified using a bicinchoninic
acid kit (Beyotime Institute of Biotechnology) and the final
protein system had a concentration of 40 μg/lane. The
proteins were then separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis on 10% gels and
transferred to polyvinylidene fluoride membranes (MilliporeSigma).
After blocking with 5% skimmed milk for 1.5 h at room temperature,
the membranes were incubated with primary antibodies (dilution
1:1,000) overnight at 4°C. The membranes were then washed three
times with TBS-0.1% Tween (TBST) for 10 min at room temperature,
after which, they were incubated with goat anti-rabbit IgG
(dilution 1:5,000) or anti-mouse IgG (dilution 1:5,000) horseradish
peroxidase-conjugated secondary antibodies (both from ABclonal
Biotech Co., Ltd.) for 1 h at room temperature. Subsequently, the
membranes were washed three times with TBST for 10 min. Finally,
exposure and development were performed using a super ECL developer
solution (ABP Biosciences, LLC). Blots were detected using the
ChemiDoc XRS+ Gel imaging system (Bio-Rad Laboratories, Inc.) and
were semi-quantified using Image Lab 6.0.1 (Bio-Rad Laboratories,
Inc.). The list of antibodies used in the present study is included
in Table II.
 | Table IIList of antibodies used. |
Table II
List of antibodies used.
| Antibody | Supplier (cat.
no.) |
|---|
| PGBD5 Mouse
mAb | Thermo Fisher
Scientific, Inc. (cat. no. MA5-32886) |
| β-actin Mouse
mAb | ABclonal Biotech
Co., Ltd. (cat. no. AC004) |
| CDK1 Rabbit
mAb | ABclonal Biotech
Co., Ltd. (cat. no. A11420) |
| Cyclin B1 Rabbit
pAb | ABclonal Biotech
Co., Ltd. (cat. no. A16800) |
| GAPDH Mouse
mAb | ABclonal Biotech
Co., Ltd. (cat. no. AC002) |
| HRP Goat
Anti-Rabbit IgG | ABclonal Biotech
Co., Ltd. (cat. no. AS014) |
| HRP Goat Anti-Mouse
IgG | ABclonal Biotech
Co., Ltd. (cat. no. AS003) |
| Bax Rabbit
Antibody | Cell Signaling
Technology, Inc. (cat. no. 2772) |
| Bcl-2 Rabbit
mAb | Cell Signaling
Technology, Inc. (cat. no. 3498) |
| Caspase-3 Rabbit
Antibody | Cell Signaling
Technology, Inc. (cat. no. 14220) |
| Ki-67 Rabbit
Antibody | MXB Biotechnologies
(cat. no. RMA-0731) |
| PPARδ Mouse
Antibody | Abmart
Pharmaceutical Technology Co., Ltd. (cat. no. MN51264) |
| p-PPARδ (Thr256)
Rabbit Antibody | Abmart
Pharmaceutical Technology Co., Ltd. (cat. no. TA4331) |
| PPARγ Rabbit
Antibody | Abmart
Pharmaceutical Technology Co., Ltd. (cat. no. TD6073) |
| p-PPARγ (Ser273)
Rabbit Antibody | Abmart
Pharmaceutical Technology Co., Ltd. (cat. no. TA3675) |
Lentivirus production
The GV248 lentiviral vector containing a short
hairpin RNA (shRNA) targeting PGBD5 (sh-PGBD5) and the negative
control (NC) vector (sh-NC) containing a non-targeting shRNA
sequence were established by Shanghai GeneChem Co., Ltd. and were
ready to directly infect the cells. The shRNA sequence (and
negative control sequence) is provided in Table I and matches the sequence reported
in a previous study (29). The
element sequence of the vector is hU6-MCS-Ubiquitin-enhanced green
fluorescent protein (EGFP)-IRES-puromycin.
Lentivirus infection
A172, U251 and U87 cells (5×104/well)
were evenly spread into 6-well plates for culture. After ~10 h,
when the cells completely adhered to the well, the level of cell
fusion was controlled at ~30%. The following formula was used to
calculate the required viral titer: Virus volume=(MOI x cell
count)/virus titer, where MOI refers to multiplicity of infection,
and a MOI of 10 was used to infect the cells. Subsequently, the
original culture medium was replaced with culture medium containing
lentiviruses for further culture. The culture medium was refreshed
again after 12 h at 37°C. Because the EGFP gene sequence is present
in lentiviruses, its expression could be observed in all three cell
lines 72 h after infection under a fluorescence microscope equipped
with appropriate filter sets for GFP detection. In addition, the
lentivirus carries a puromycin resistance gene sequence; therefore,
to further eliminate the influence of wild-type cells on subsequent
experiments, 2 μg/ml puromycin (Dalian Meilun Biology
Technology Co., Ltd.) was used to screen the cells. After
undergoing more than three sub-culturing cycles, the cell
population predominantly exhibited antibiotic resistance. Western
blotting and RT-qPCR were used to evaluate the protein and mRNA
expression levels of PGBD5 in cells infected with sh-PGBD5 and
sh-NC.
Transwell assay
Precooled FBS-free medium was added to the lower
chamber of a 24-well Transwell chamber (Corning Life Sciences) to
evenly hydrate the basement membrane, and precooled FBS-free medium
was also used to dilute Matrigel (Corning Life Sciences) at a
dilution ratio of 7:1. Subsequently, 100 μl prepared
Matrigel was added to the upper chamber, and the chamber was
incubated at 37°C for 1 h to solidify the matrix. The Transwell
chamber was then removed from the incubator, the medium in the
upper chamber was aspirated carefully, and 600 μl complete
medium supplemented with 10% FBS was added to the lower chamber. A
certain cell suspension volume was prepared, and 200 μl of
the suspension, containing a cell density of 1×105
cells/ml, was added evenly to each well in the upper chamber. After
24 h at 37°C, the chamber was removed, and the culture medium,
cells and Matrigel in the upper chamber were carefully wiped with a
cotton swab. Cells that had migrated to the lower chamber were
fixed with 600 μl 4% paraformaldehyde (Beijing Solarbio
Science & Technology Co., Ltd.) for 30 min at room temperature.
After fixation, the chamber was stained with crystal violet
solution (Beijing Solarbio Science & Technology Co., Ltd.) for
30 min at room temperature. Finally, the chamber was washed three
times with PBS. After air-drying, five high-power fields were
randomly selected under a light microscope for observing and
counting the cells. Matrigel was added to the upper chamber for the
invasion assay, whereas it was not added to the upper chamber for
the migration assay.
Cell apoptosis analysis
A172, U251 and U87 cells were collected by digesting
and centrifuging. After the binding buffer was diluted with
deionized water at a 3:1 ratio, the cells were resuspended and the
concentration was adjusted to 4×106/ml. In a 5-ml flow
tube, 100 μl cell suspension was added along with 5
μl Annexin V/PE. After pipetting and mixing, the suspension
was incubated at room temperature in the dark for 5 min.
Subsequently, the suspension was stained with 10 μl 20
μg/ml 7AAD and 400 μl PBS in the dark for 15 min at
room temperature. An Annexin V-PE/7-AAD apoptosis detection kit
(Nanjing KeyGen Biotech Co., Ltd.) was used, followed by detection
using a Novocyte 2060r Flow Cytometer (ACEA Bisociences; Agilent
Technologies, Inc.) and data analysis using FlowJo 10.4 (FlowJo
LLC).
Cell cycle distribution analysis
A172, U251 and U87 cells were collected by digesting
and centrifuging. Next, 500 μl 1X staining buffer with RNase
A (25 μg/ml) was added to resuspend the cells and 5
μl PI dye was added to label the cells in the dark for 30
min at 37°C. For this assay, a Cell Cycle Detection Kit (Beijing 4A
Biotech Co., Ltd.) was used for cell cycle analysis, followed by
detection using a Novocyte 2060r Flow Cytometer and data analysis
using FlowJo 10.4.
Tumor xenograft assay
Nude BALB/c mice (age, 6 weeks; male) were purchased
from the Animal Experiment Center at Kunming Medical University.
The mice were raised in a controlled temperature (20±2°C) and
50-60% humidity under a 12-h light/dark cycle, and food and water
were provided ad libitum. The nude mice were randomly
divided into two groups (n=5/group; weight, 18-21 g): U87-sh-PGBD5
and U87-sh-NC groups. The U87-sh-PGBD5 and U87-sh-NC cells were
cultured separately, and when the cells were in the logarithmic
growth phase, they were digested and centrifuged at 200 x g for 5
min at 4°C, resuspended in sterile PBS and the cell suspension
concentration was adjusted to 2.0×106 cells/100
μl. Subsequently, 100 μl cell suspension was injected
subcutaneously into the right armpit of the nude mice. The tumors
were checked daily, and the measurements of the longest and
shortest diameters were recorded once a week using vernier
calipers. The tumor volume was calculated as follows: Tumor
volume=0.52 x L x W2; where L is the longest diameter of
the tumor and W is the shortest diameter of the tumor. After 5
weeks of monitoring, or upon reaching humane endpoints (defined as
mice losing 20% body weight or exhibiting moribund behavior), the
mice were sacrificed. Prior to cervical dislocation, the mice were
deeply anesthetized with 5% isoflurane to minimize any potential
pain or distress during the procedure. The tumors were then
collected, and the tumor volume curve was prepared.
Immunohistochemistry
Nude mouse subcutaneous tumors were immediately
fixed in 4% paraformaldehyde for 48 h at 4°C, dehydrated in graded
alcohol and xylene, embedded in paraffin and sliced into
3-μm serial sections. The samples were heated in an oven for
30 min at 60°C, and were then dewaxed in different concentrations
(100, 95 and 75%) of xylene (10 min) and graded alcohol (2 min).
After rinsing with tap water, the sections were immersed in EDTA
antigen retrieval solution (MXB Biotechnologies) for 3 min at 95°C
and allowed to cool naturally. Subsequently, 3%
H2O2 was added to the sections at room
temperature for 15 min, followed by washing three times with PBS.
The sections were first blocked for nonspecific binding using 5%
normal goat serum (cat. no. 566380; MilliporeSigma) in PBS for 20
min at room temperature, then incubated with Ki67 antibody at a
dilution of 1:100 (MXB Biotechnologies; cat. no. RMA-0731) at 26°C
for 75 min and were washed three times with PBS, after which, they
were incubated with horseradish peroxidase-conjugated goat
anti-mouse IgG antibody (Dako; Agilent Technologies, Inc.) for 30
min and then washed three times with PBS. Subsequently, the
sections were incubated with a freshly prepared DAB staining
solution (MXB Biotechnologies) for 15 min at room temperature. This
was followed by washing three times with PBS and incubation with
hematoxylin solution for 14 sec at room temperature. The sections
were then treated with hydrochloric acid and alcohol
differentiation solution, and rinsed with tap water. Finally, the
sections were dehydrated with graded ethanol, permeabilized with
xylene, mounted with neutral balm and a coverslip was placed over
the sections. Subsequently, images were captured under a light
microscope. For hematoxylin and eosin staining, sections were
stained with hematoxylin (0.1% w/v in water) for 4 min at room
temperature to highlight the nuclei, followed by a brief rinse in
running tap water to remove excess stain. Subsequently, eosin (1%
w/v in water) staining was applied for 2 min at room temperature to
visualize cytoplasmic and extracellular matrix components. After
staining, sections were dehydrated, cleared, and mounted with a
coverslip using a xylene-based mounting medium. Stained sections
were examined under a light microscope.
Transcriptomics study
The sh-NC and sh-PGBD5 A172 and U87 cell groups were
simultaneously cultured in T25 culture flasks for transcription
analysis. When the cell flasks reached 100% confluence, RNA was
extracted from the cells using RNAIso Plus (Takara Biotechnology,
Ltd.; cat. no. 9108). RNA sequencing was performed at MajorBiotech.
After quality verification with a BioAnalyzer 2100 (Agilent
Technologies, Inc.), RNA libraries at a final concentration of 300
pM were sequenced using an Illumina NovaSeq 6000 SP Reagent kit
v1.5 (300 cycles; cat. no. 20028400; Illumina, Inc.) in 150 bp
paired-end reads. Reference gene source: Homo sapiens;
reference genome version: GRCh38; reference genome source:
http://asia.ensembl.org/Homo_sapiens/Info/Index. The
clean reads of these samples were compared with the designated
reference genome. Based on the quantitative expression results,
differential gene analyses were performed between groups to
identify the differentially expressed genes between each pair.
DESeq2 software (Bioconductor, Inc.; https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
was used for differential gene analysis, and the screening
threshold was: log2FC ≥1/log2FC ≤-1, Padjust
<0.05. The P-values were adjusted for multiple testing using the
Benjamini-Hochberg method. Principal component analysis (PCA) was
employed to reduce the dimensionality of the transcriptomic data,
enabling the identification of the principal components within the
dataset. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analyses were performed. GO analysis was
performed using DAVID (https://david.ncifcrf.gov) to understand the functions
of the identified genes. KEGG pathway analysis was performed using
KAAS (https://www.genome.jp/kaas-bin/kaas_main) to identify
which pathways the genes were involved in.
Statistical analysis
The data in the present study are presented as mean
± standard deviation for continuous variables, and were analyzed
using SPSS 22.0 statistical software (IBM Corp.). The experiments
were repeated a minimum of three times. Comparisons between two
groups were performed using a paired or unpaired Student's t-test.
Multi-group comparisons were performed using one-way ANOVA with the
Tukey's multiple comparison test. P<0.05 was considered to
indicate a statistically significant difference.
Results
PGBD5 expression in glioma cells and
tissues
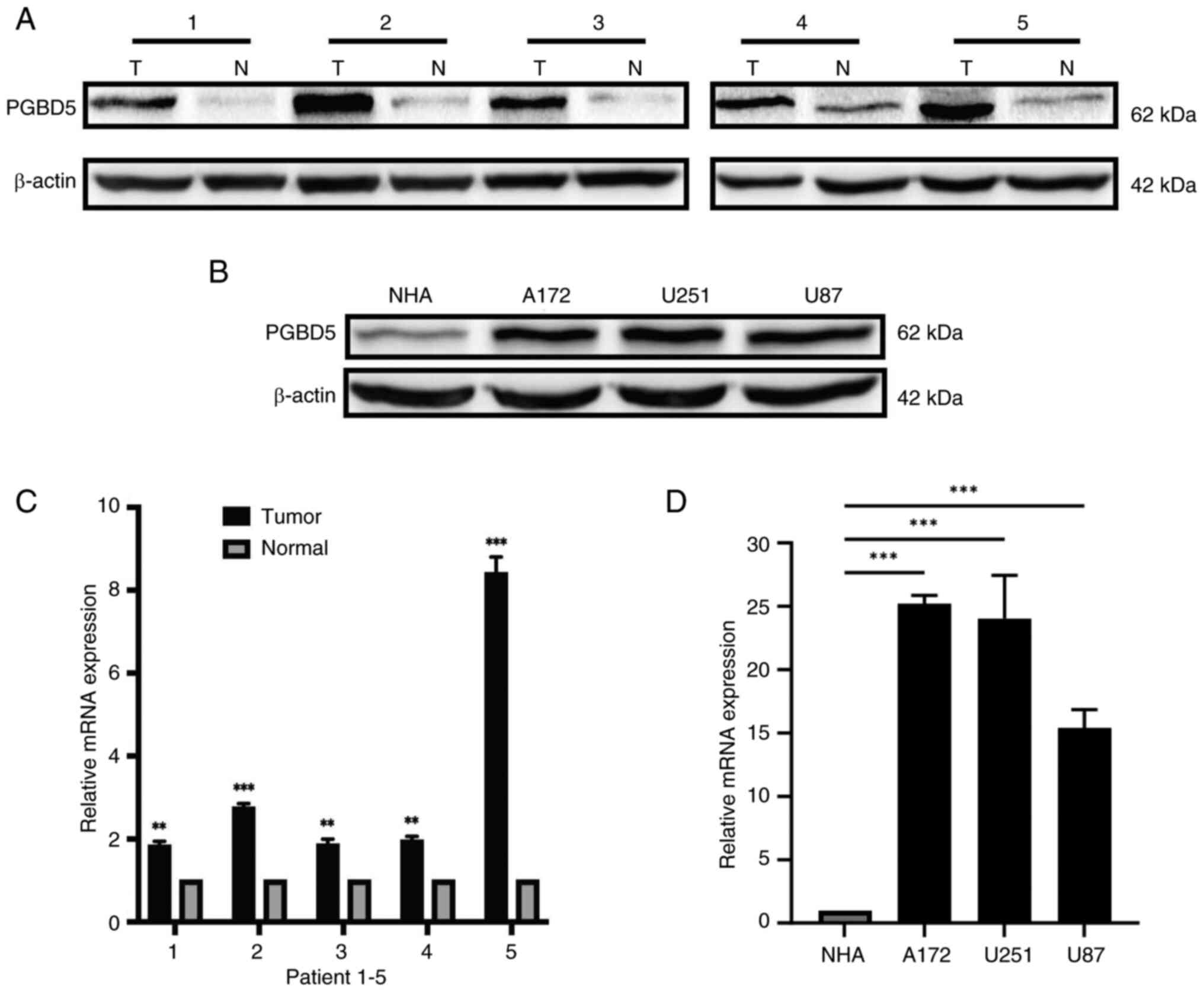
The rela-tive protein and mRNA expression levels of
PGBD5 were significantly elevated in glioma tissues compared with
those in para-cancerous tissues (P<0.01; Figs. 1A and C, and S1A). The relative protein and mRNA
expression levels of PGBD5 were also significantly higher in glioma
cell lines (A172, U251 and U87) compared with those in NHAs
(P<0.001; Figs. 1B and D, and
S1B). The results suggested that
the protein and mRNA expression levels of PGBD5 were increased in
both glioma tissues and cell lines.
Lentivirus-mediated PGBD5 silencing in
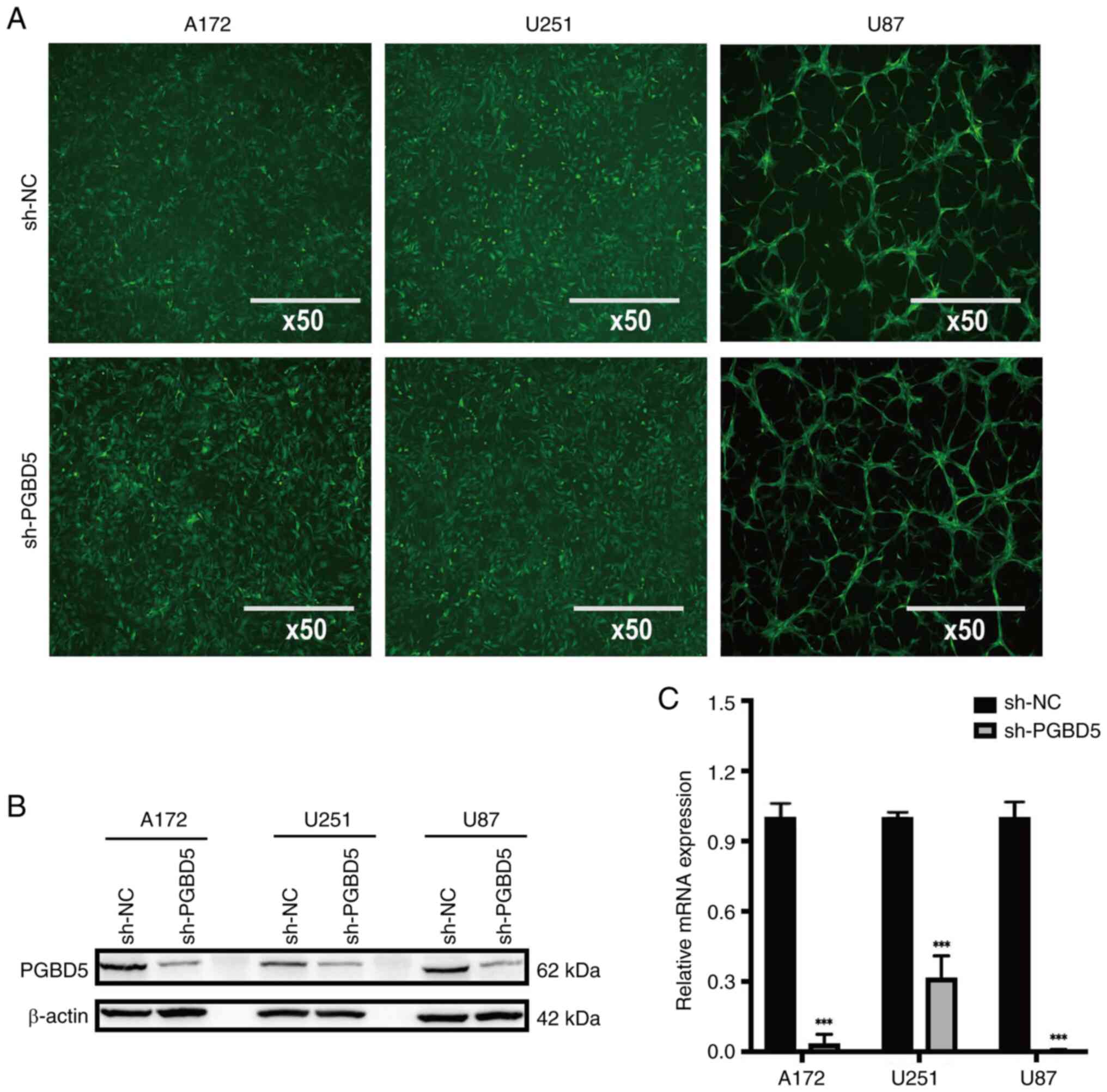
glioma cells
The detection of GFP staining indicated successful
lentiviral infection (Fig. 2A).
In addition, the protein (Figs.
2B and S1C) and mRNA
(Fig. 2C) expression levels of
PGBD5 were markedly decreased in A172, U251 and U87 glioma cells in
the sh-PGBD5 group, compared with those in the sh-NC group
(P<0.001). These results suggested that lentivirus-mediated
PGBD5 silencing was successful in glioma cells.
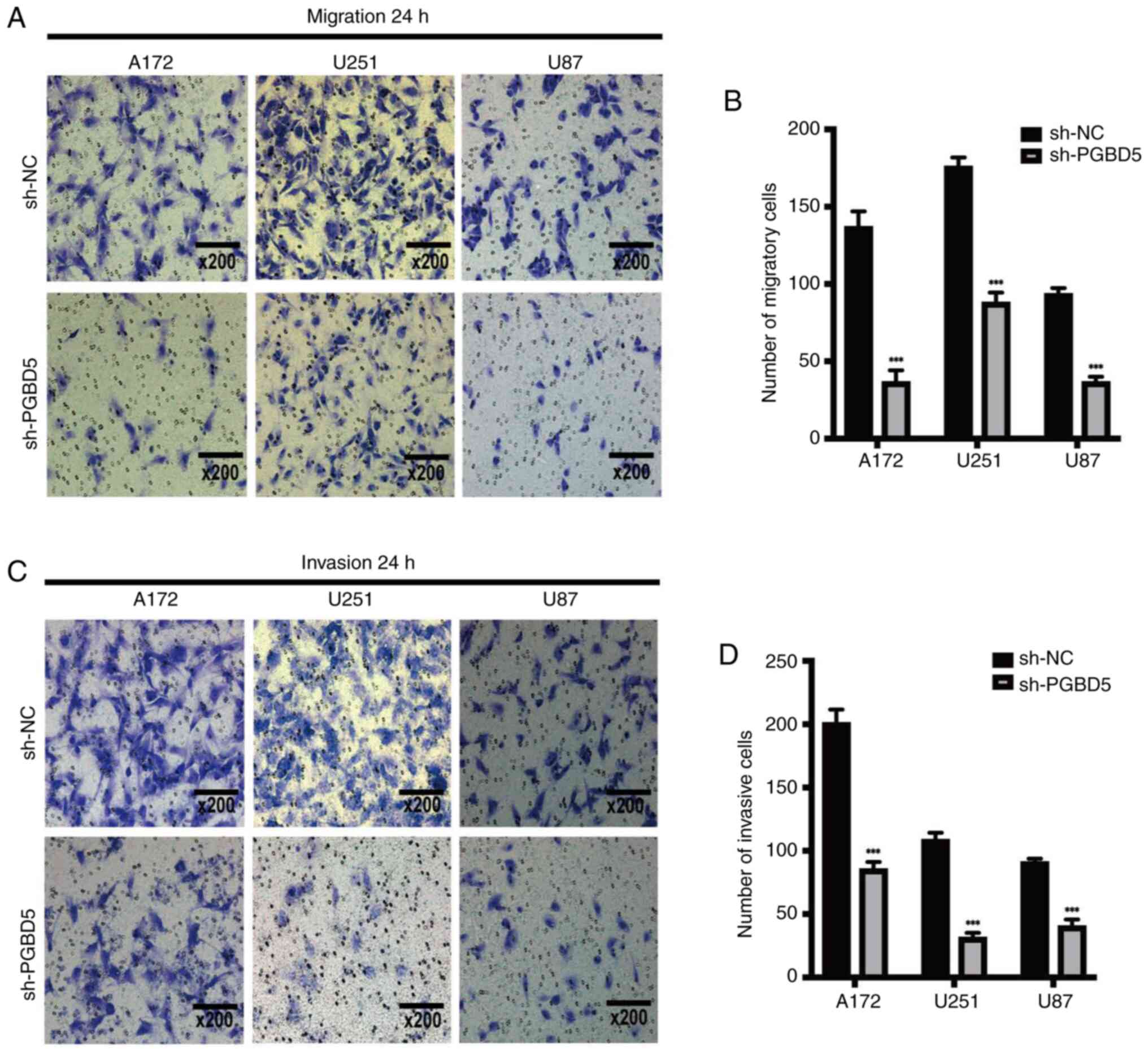
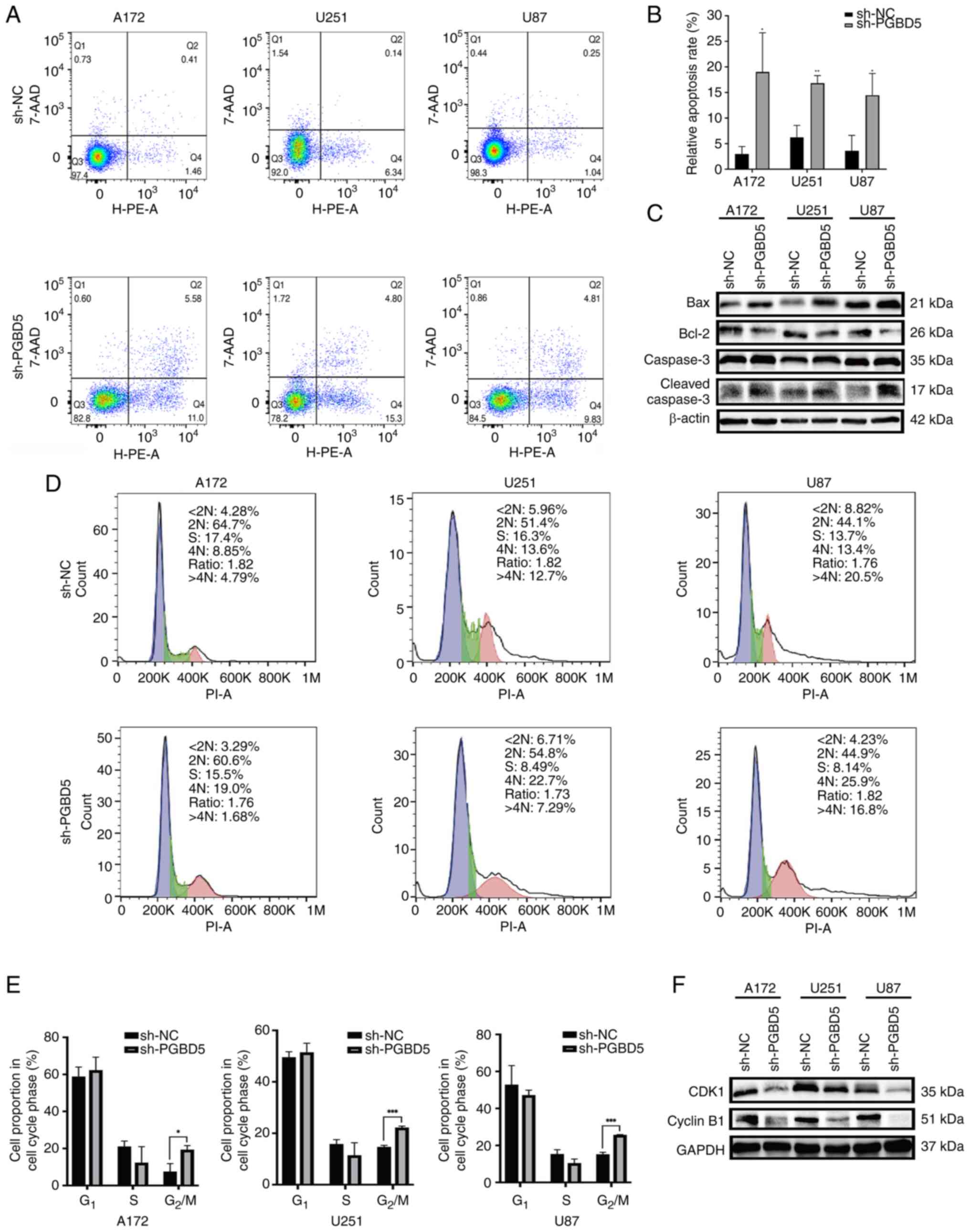
Effects of PGBD5 knockdown on glioma
cells
The knockdown of PGBD5 inhibited glioma cell
migration (Fig. 3A and B) and
invasion (Fig. 3C and D) compared
with those in the sh-NC group (P<0.001). In addition, PGBD5
knockdown significantly induced the apoptosis of glioma cells
compared with that in the sh-NC group (P<0.05; Fig. 4A and B). The knockdown of PGBD5
could also cause glioma cell cycle arrest in the G2/M
phase (P<0.05; Fig. 4D and E).
PGBD5 knockdown not only promoted the expression levels of
apoptosis-related proteins, such as Bax and cleaved caspase-3, but
also inhibited the expression levels of anti-apoptotic proteins and
carcinogenic cell cycle regulators, such as Bcl-2, CDK1 and cyclin
B1 (Figs. 4C and F, and S1D-I).
PGBD5 knockdown inhibits glioma growth in
vivo
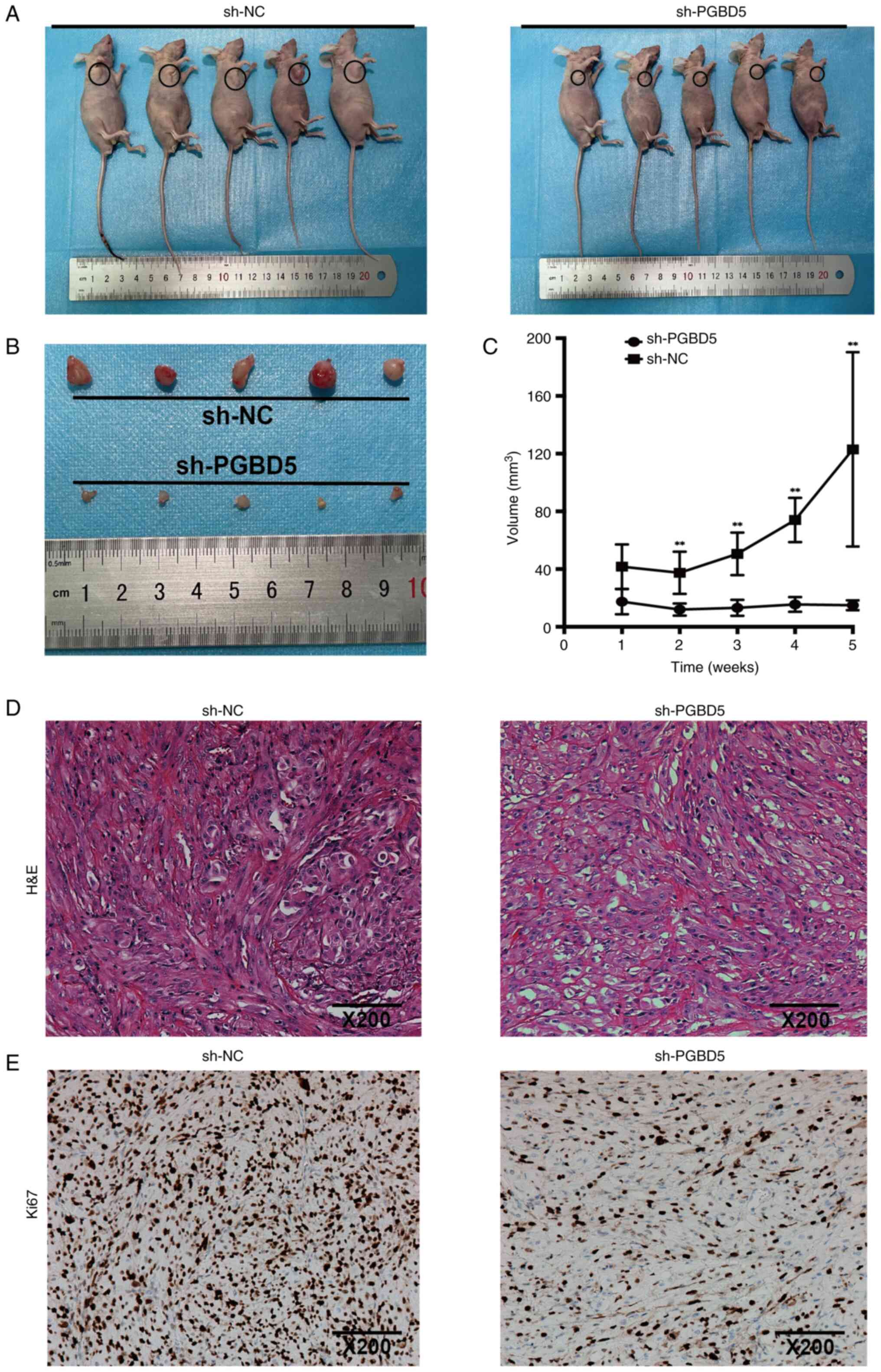
The growth rate and volume of the subcutaneous
tumors in nude mice from the sh-PGBD5 group were significantly
reduced compared with those in the sh-NC group (P<0.01; Fig. 5A-C). Hematoxylin and eosin
staining was performed to analyze the subcutaneous tumors of nude
mice, which revealed characteristic histological features
associated with tumor growth, such as increased cellularity,
irregular nuclear morphology and aberrant tissue architecture
(Fig. 5D). In addition,
immunohistochemical analysis showed that, in the sh-PGBD5 group,
the expression levels of Ki67 were inhibited compared with those in
the sh-NC group in vivo (Figs.
5E and S1N), which may have
markedly restricted the growth of subcutaneous tumors.
PGBD5 promotes glioma progression via the
PPAR signaling pathway
PCA was conducted on four groups of samples, the
separation of the four groups underscores the unique transcriptomic
signatures, highlighting the potential for underlying biological
differences caused by PGBD5 knockdown (Fig. 6A). Venn analysis revealed that
after PGBD5 knockdown, 1,230 genes were differentially expressed in
the A172 cell group, and 1,308 genes were differentially expressed
in the U87 cell group, of which 242 genes overlapped (Fig. 6B). Furthermore, transcriptome
analysis revealed that compared with that in the sh-NC group,
Knocking down PGBD5 significantly affected 'cellular metabolic
process' (GO:0044237) and the 'PPAR signaling pathway'
(KEGG:03320), indicating a crucial role of PGBD5 in regulating
metabolic processes and signaling mechanisms related to PPAR
(Fig. 6C-F). In contrast with
that in the sh-NC group, the phosphorylated and total expression
levels of PPARδ and PPARγ were increased in the sh-PGBD5 group
(Figs. 6G and S1J-M), both indicating enhanced
activity of PPARδ and PPARγ. Furthermore, the phosphorylated and
total expression levels of PPARδ were increased in response to
PGBD5 knockdown in A172 and U87 cells, but not in U251 cells
(Fig. 6G). In vivo
experiments involving subcutaneous tumors in nude mice revealed
that PPARγ and PPARδ levels in the sh-PGBD5 group were markedly
higher than those in the sh-NC group (P<0.05; Figs. 6H, and S1O-P). Fig. S1 shows statistical analysis of
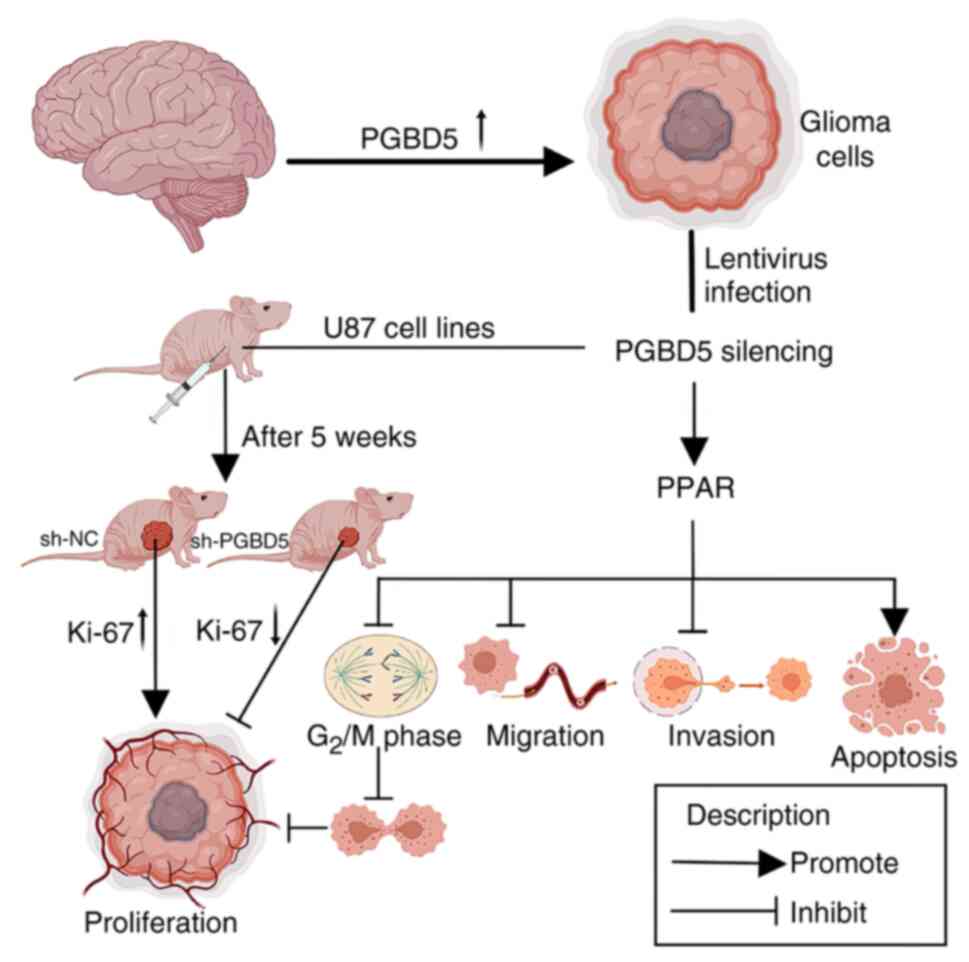
all western blots. A graphical overview of the research findings
can be found in Fig. 7.
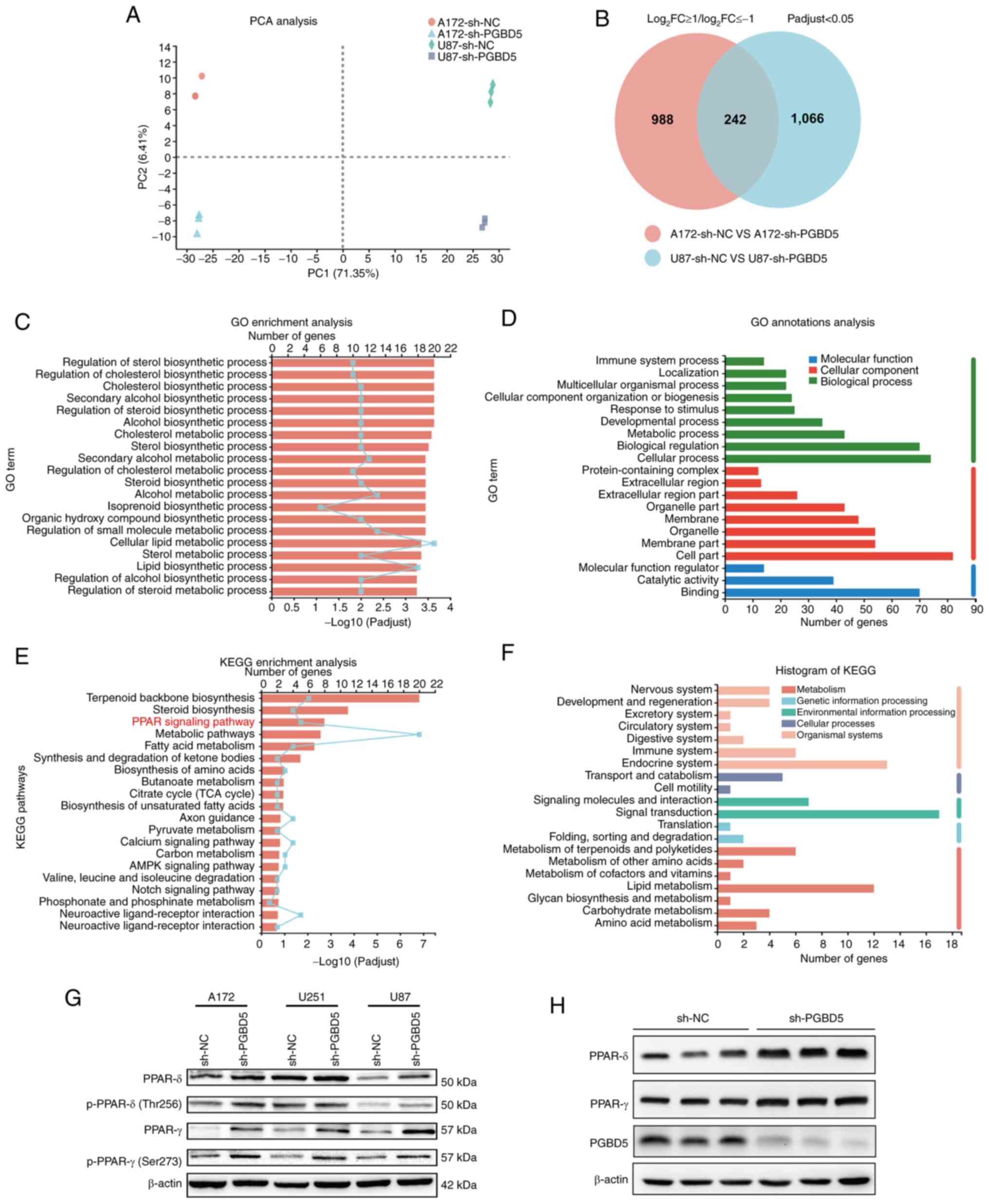 | Figure 6PPAR is involved in the effect of
PGBD5 on the occurrence and development of glioma. (A) PCA of data
from four groups of samples. (B) Venn diagram showed 1,230 altered
genes in the A172 cells and 1,308 in the U87 cells, among which 242
gene alterations were overlapping. (C) GO enrichment analyses and
(D) GO annotation analyses of differentially expressed genes
revealed significant associations with cellular metabolic process.
(E) KEGG enrichment analyses and (F) KEGG enrichment analyses of
differentially expressed genes revealed significant associations
with the PPAR signaling pathway. (G) Western blotting was performed
to detect PPAR-related protein expression. β-actin was used as the
protein loading control. The experiments were repeated three times.
(H) PGBD5, PPARγ and PPARδ levels in in vivo subcutaneous
tumors after PGBD5 knockdown. GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; NC, negative control; p-,
phosphorylated; PCA, principal component analysis; PGBD5, piggyBac
transportable element derived; PPAR, peroxisome
proliferator-activated receptor; sh, short hairpin. |
Discussion
In the present study, the biological function and
molecular mechanism of action of PGBD5 in glioma were assessed
using RT-qPCR, western blotting, Transwell assay, flow cytometry
and histopathology. The results revealed that PGBD5 was upregulated
in glioma tissues and cell lines, and in vitro experiments
revealed that, after PGBD5 knockdown, some of the biological
functions of glioma cells were affected, which could inhibit the
growth of glioma in vivo. Furthermore, transcriptomic
analysis demonstrated that PGBD5 knockdown could activate the PPAR
signaling pathway and upregulate the expression of PPAR-related
proteins, thus exerting an anticancer effect.
Glioma is a common tumor originating from glial
cells, which exhibits a high degree of malignancy. The malignant
behavior of glioma may be related to the expression of multiple
genes and various signaling pathways, such as TP53, EGFR and IDH1
(43). At present, surgery with
radiotherapy and chemotherapy remains the major treatment strategy
for glioma. Although numerous potential therapeutic targets have
been identified, additional clinical investigation is required to
determine effective treatment strategies (44). PGBD5 is a member of the piggyBac
transposon family, and the cell transformation it induces has been
linked to the occurrence of chromosome deletion, inversion and
translocation (28). In our
preliminary unpublished experiments, it was observed that PGBD5
overexpression in tumor-bearing mice and glioma cells did not
significantly alter tumor progression or severity, whereas knocking
down PGBD5 substantially reduced tumor size. The lack of impact
from PGBD5 overexpression may be due to high endogenous levels of
PGBD5 in the models, thus resulting in a saturation effect, or it
may indicate that the function of this gene is context-dependent
and influenced by other factors. Given these findings and the
complex nature of glioma biology, the present study focused on the
effects of PGBD5 knockdown.
PPAR is a transcription factor from the nuclear
hormone receptor superfamily that is activated by fatty acids and
regulates energy metabolism. PPAR is also expressed in immune cells
and serves an important role in their differentiation (45). Because of its strong
pharmacological activity, PPAR has been identified as a therapeutic
target in multiple diseases, such as metabolic disorders,
cardiovascular diseases and inflammatory diseases (46). An increasing body of evidence has
shown that PPAR exerts a key regulatory effect on various aspects
of immunity, inflammation, vascular function, oxidative stress,
cell proliferation, differentiation, development, apoptosis and
cancer (47,48).
In several studies, PPAR, as a regulator, has been
confirmed to serve important roles in the proliferation and
differentiation of cancer cells (41,49). Because PPAR occasionally acts as a
carcinogen and a suppressor in different tumor types, there may be
conflicting results from different studies (50,51). Furthermore, PPARδ not only
maintains metabolic activity in peripheral organs and tissues
(52), but also exhibits high
expression in the brain (53).
Other studies have shown that the absence of PPARδ expression in
mice can cause defects in brain development (54). PPAR expression is also associated
with some age-related diseases, and PPARγ has a role as a biomarker
in patients with cerebral ischemia (55). It may also serve an important role
in the inhibition of tumors of neuroectodermal origin, since PPARδ
and its ligand erucic acid have been shown to exert antitumor,
neuroprotective and myelina-ting effects in neuroblastoma,
glioblastoma and Parkinson's disease (56). Erucic acid at therapeutic
concentration can also reduce the proliferation of C6 glioma cells
in vitro and induce the death of C6 glioma cells (57). A previous study also showed that
PPAR induces a synergistic effect on the differentiation of C6
glioma cells into oligodendrocytes (58). Moreover, PPARδ may promote the
expression of both PPARα and PPARγ to increase the expression of
oligodendrocyte-specific markers and enzymes necessary for myelin
synthesis in C6 glioma cells (58). Two major classes of agonists of
PPARγ, including thiazolidinedione and non-thiazolidinedione, can
block the migration and invasion of glioma cells and other
highly-invasive solid tumors; however, the specific mechanism
remains unclear (59). Other
studies have also revealed that the PPAR-mediated signaling pathway
and gene expression defects may affect the normal expression and
induction of catalase in malignant glioma (60,61); however, further investigation
showed that PPAR agonists can significantly improve the expression
level and activity of catalase in normal astrocytes, but exert no
significant effect on glioma cells. Nevertheless, this suggests
that PPAR agonists may serve as a potential drug for glioma
treatment (60). Another study on
clinical glioma tissues demonstrated that PPARγ exhibits positive
expression rates of 94.1% in LGG and 39.6% in HGG, and its
expression is closely related to microvessel density; thus, PPARγ
may be involved in the regulation of angiogenesis in glioma
(62).
The mechanisms through which PGBD5 knockdown
influences the PPAR pathway may involve various cellular processes.
It is plausible that PGBD5 affects the stability and
transcriptional activity of PPARs or their co-regulators through
direct or indirect interactions. Alterations in PGBD5 could lead to
changes in chromosomal structure or gene expression profiles,
thereby modulating the PPAR pathway. Additionally, as PGBD5
influences chromosomal stability, its knockdown may lead to a more
stable genomic environment, reducing the mutations or alterations
that could lead to dysregulated PPAR activity, a common occurrence
in cancer cells.
The present study has numerous limitations. First,
the sample size of glioma tissues was relatively low, and a study
with a larger sample size is needed. The most notable limitation
was the lack of rescue experiments for PGBD5 gene expression or
PPAR signaling, which will be focused upon in future research. The
potential protein-protein interaction between PGBD5 and PPAR also
needs further investigation. Additionally, the specific mechanism
through which the PPAR pathway promotes glioma cell migration and
invasion was not fully elucidated within the current study. In
future studies, we aim to focus on assessing the signaling cascades
downstream of PPAR activation. This would involve a detailed
analysis of the PPAR-responsive elements within the glioma genome,
and how agonists modulate the transcription of genes involved in
cell motility, adhesion and extracellular matrix remodeling, which
are critical for migration and invasion.
Although PGBD5 is a highly conserved gene, it has
been shown that it has long lost its transposase activity (63); thus, its complete knockout by
genome editing technologies such as CRISPR-Cas9 will exert a
limited effect on the organism. Establishing a rat model of
intracranial in situ tumor, and observing the changes and
primary blood indicators following PGBD5 gene knockout are the next
steps. This will help eliminate the limitations caused by
immunosuppression in nude mice to a certain extent. Given the
molecular diversity and complexity of glioma, understanding
individual molecular targets such as PGBD5 can lead to more
personalized and effective treatments. Patients whose glioma
exhibits significant upregulation of PGBD5 may benefit from
therapies specifi-cally designed to target this gene, leading to
more tailored and potentially more effective treatment strategies.
In our future work, we aim to develop drugs or gene therapies that
can effectively target PGBD5. This may involve screening for small
molecules that inhibit PGBD5, developing RNA interference
technologies or other gene-editing techniques, such as CRISPR, to
knockdown or edit the gene in tumor cells.
In conclusion, the present study demonstrated that
the expression of PGBD5 was upregulated in glioma tissues and
cells. Knockdown of PGBD5 expression exerted an anticancer effect
mediated by the promotion of PPAR expression in vitro, which
not only accelerated the apoptosis of glioma cells, but also
inhibited their migration, invasion and induced cell cycle arrest.
PGBD5 silencing could also significantly restrict tumor growth
in vivo. The present study first evaluated the biological
role of PGBD5 in glioma and preliminarily explored its related
molecular mechanisms. To some extent, the findings of the current
study provided a theoretical reference for exploring novel
therapeutic targets in human glioma.
Supplementary Data
Availability of data and materials
The data generated in the present study may be found
in the BioProject database under accession number PRJNA953025 or at
the following URL: https://www.ncbi.nlm.nih.gov/biopro-ject/PRJNA953025/
Authors' contributions
PL, JY and SZ designed the study. PL, JY and LJ
carried out the experiments. JD, SY and CL participated in the data
analysis. PL, JY and SZ wrote the manuscript. JD, SY and CL revised
the manuscript critically for important intellectual content. PL
and SZ confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The use of NHAs, paired tumor tissues and
para-cancerous tissues were approved by the Ethics Committee of
Kunming University of Science and Technology School of Medicine
(approval no. KMUST-MEC-092). All enrolled patients provided
written informed consent. The animal experiments were conducted in
accordance with the national legislation and associated guidelines,
and the procedures were approved by the Ethics Committee of Kunming
University of Science and Technology School of Medicine [approval
no. PZWH (DIAN) K2021-0021]. The study was conducted in accordance
with the guidelines of The Declaration of Helsinki.
Patient consent for publication
Written informed consent has been obtained from the
patients to publish this paper.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This research was supported by the Xingdian Talent Support Plan
Project (grant no. XDYC-QNRC-2022-0320) and the Joint Projects of
Kunming University of Science and Technology and Medical Science
(grant no. KUST-KH2023028Y).
References
|
1
|
Louis DN, Perry A, Wesseling P, Brat DJ,
Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM,
Reifenberger G, et al: The 2021 WHO classification of tumors of the
central nervous system: A summary. Neuro Oncol. 23:1231–1251. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weller M and Le Rhun E: How did lomustine
become standard of care in recurrent glioblastoma? Cancer Treat
Rev. 87:1020292020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duffau H and Taillandier L: New concepts
in the management of diffuse low-grade glioma: Proposal of a
multistage and individualized therapeutic approach. Neuro Oncol.
17:332–342. 2015.
|
|
4
|
Campian J and Gutmann DH: CNS tumors in
neurofibromatosis. J Clin Oncol. 35:2378–2385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin L, Cai J and Jiang C: Recent advances
in targeted therapy for glioma. Curr Med Chem. 24:1365–1381. 2017.
View Article : Google Scholar
|
|
7
|
Gottesman MM, Robey RW and Ambudkar SV:
New mechanisms of multidrug resistance: An introduction to the
cancer drug resistance special collection. Cancer Drug Resist.
6:590–595. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu A, Jiang B, Song C, Zhong Q, Mo Y,
Yang R, Chen C, Peng C, Peng F and Tang H: Isoliquiritigenin
inhibits circ0030018 to suppress glioma tumorigenesis via the
miR-1236/HER2 signaling pathway. MedComm (2020).
4:e2822023.PubMed/NCBI
|
|
9
|
Tang H, Liu Q, Liu X, Ye F, Xie X, Xie X
and Wu M: Plasma miR-185 as a predictive biomarker for prognosis of
malignant glioma. J Cancer Res Ther. 11:630–634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Q, Fu W, Fu Y, Ye W, Yan H, Yu Z, Li R,
Cai Y, Chen Y, Wang L, et al: BNIP3 as a potential biomarker for
the identification of prognosis and diagnosis in solid tumours. Mol
Cancer. 22:1432023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang B, Zhao J, Wang Y, Xu H, Gao BO,
Zhang G, Han B, Song G, Zhang J and Meng W: CHRM3 is a novel
prognostic factor of poor prognosis and promotes glioblastoma
progression via activation of oncogenic invasive growth factors.
Oncol Res. 31:917–927. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cordaux R and Batzer MA: The impact of
retrotransposons on human genome evolution. Nat Rev Genet.
10:691–703. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smit AF: Interspersed repeats and other
mementos of transposable elements in mammalian genomes. Curr Opin
Genet Dev. 9:657–663. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Muñoz-López M and García-Pérez JL: DNA
transposons: Nature and applications in genomics. Curr Genomics.
11:115–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schrader L and Schmitz J: The impact of
transposable elements in adaptive evolution. Mol Ecol.
28:1537–1549. 2019. View Article : Google Scholar
|
|
16
|
Percharde M, Sultana T and Ramalho-Santos
M: What doesn't kill you makes you stronger: Transposons as dual
players in chromatin regulation and genomic variation. Bioessays.
42:e19002322020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morales ME, Servant G, Ade C and Roy-Engel
AM: Altering genomic integrity: Heavy metal exposure promotes
transposable element-mediated damage. Biol Trace Elem Res.
166:24–33. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deniz Ö, Frost JM and Branco MR:
Regulation of transposable elements by DNA modifications. Nat Rev
Genet. 20:417–431. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saleh A, Macia A and Muotri AR:
Transposable elements, inflammation, and neurological disease.
Front Neurol. 10:8942019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Burns KH: Our conflict with transposable
elements and its implications for human disease. Annu Rev Pathol.
15:51–70. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Payer LM and Burns KH: Transposable
elements in human genetic disease. Nat Rev Genet. 20:760–772. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yusa K: piggyBac transposon. Microbiol
Spectr. 3:MDNA3-0028-20142015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Helou L, Beauclair L, Dardente H, Piégu B,
Tsakou-Ngouafo L, Lecomte T, Kentsis A, Pontarotti P and Bigot Y:
The piggyBac-derived protein 5 (PGBD5) transposes both the closely
and the distantly related piggyBac-like elements Tcr-pble and Ifp2.
J Mol Biol. 433:1668392021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Majumdar S, Singh A and Rio DC: The human
THAP9 gene encodes an active P-element DNA transposase. Science.
339:446–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Henssen AG, Henaff E, Jiang E, Eisenberg
AR, Carson JR, Villasante CM, Ray M, Still E, Burns M, Gandara J,
et al: Genomic DNA transposition induced by human PGBD5. Elife.
4:e105652015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pavelitz T, Gray LT, Padilla SL, Bailey AD
and Weiner AM: PGBD5: A neural-specific intron-containing piggyBac
transposase domesticated over 500 million years ago and conserved
from cephalochordates to humans. Mob DNA. 4:232013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sarkar A, Sim C, Hong YS, Hogan JR, Fraser
MJ, Robertson HM and Collins FH: Molecular evolutionary analysis of
the wide-spread piggyBac transposon family and related
'domesticated' sequences. Mol Genet Genomics. 270:173–180. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Henssen AG, Koche R, Zhuang J, Jiang E,
Reed C, Eisenberg A, Still E, MacArthur IC, Rodríguez-Fos E,
Gonzalez S, et al: PGBD5 promotes site-specific oncogenic mutations
in human tumors. Nat Genet. 49:1005–1014. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie W, Zeng Y, Hu L, Hao J, Chen Y, Yun X,
Lin Q and Li H: Based on different immune responses under the
glucose metabolizing type of papillary thyroid cancer and the
response to anti-PD-1 therapy. Front Immunol. 13:9916562022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu Z, Du M and Lu L: A novel 16-genes
signature scoring system as prognostic model to evaluate survival
risk in patients with glioblastoma. Biomedicines. 10:3172022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Phua WWT, Wong MXY, Liao Z and Tan NS: An
aPPARent functional consequence in skeletal muscle physiology via
peroxisome proliferator-activated receptors. Int J Mol Sci.
19:14252018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Montaigne D, Butruille L and Staels B:
PPAR control of metabolism and cardiovascular functions. Nat Rev
Cardiol. 18:809–823. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang YX: PPARs: Diverse regulators in
energy metabolism and metabolic diseases. Cell Res. 20:124–137.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mirza AZ, Althagafi II and Shamshad H:
Role of PPAR receptor in different diseases and their ligands:
Physiological importance and clinical implications. Eur J Med Chem.
166:502–513. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan Y, Wang M, Yang K, Chi T, Liao Z and
Wei P: PPAR-alpha modulators as current and potential cancer
treatments. Front Oncol. 11:5999952021. View Article : Google Scholar
|
|
36
|
Dong YW, Wang XP and Wu K: Suppression of
pancreatic carcinoma growth by activating peroxisome
proliferator-activated receptor gamma involves angiogenesis
inhibition. World J Gastroenterol. 15:441–448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang N, Li G, Li X, Xu L and Chen M:
Circ5379-6, a circular form of tumor suppressor PPARα, participates
in the inhibition of hepatocellular carcinoma tumorigenesis and
metastasis. Am J Transl Res. 10:3493–3503. 2018.
|
|
38
|
Andrejeva D, Kugler JM, Nguyen HT,
Malmendal A, Holm ML, Toft BG, Loya AC and Cohen SM: Metabolic
control of PPAR activity by aldehyde dehydrogenase regulates
invasive cell behavior and predicts survival in hepatocellular and
renal clear cell carcinoma. BMC Cancer. 18:11802018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen SZ, Ling Y, Yu LX, Song YT, Chen XF,
Cao QQ, Yu H, Chen C, Tang JJ, Fan ZC, et al: 4-Phenylbutyric acid
promotes hepatocellular carcinoma via initiating cancer stem cells
through activation of PPAR-α. Clin Transl Med. 11:e3792021.
View Article : Google Scholar
|
|
40
|
Wagner N and Wagner KD: PPAR beta/delta
and the hallmarks of cancer. Cells. 9:11332020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chi T, Wang M, Wang X, Yang K, Xie F, Liao
Z and Wei P: PPAR-γ modulators as current and potential cancer
treatments. Front Oncol. 11:7377762021. View Article : Google Scholar
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
43
|
Li X and Meng Y: Analyses of
metastasis-associated genes in IDH wild-type glioma. BMC Cancer.
20:11142020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang K, Wu Z, Zhang H, Zhang N, Wu W, Wang
Z, Dai Z, Zhang X, Zhang L, Peng Y, et al: Glioma targeted therapy:
Insight into future of molecular approaches. Mol Cancer. 21:392022.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Christofides A, Konstantinidou E, Jani C
and Boussiotis VA: The role of peroxisome proliferator-activated
receptors (PPAR) in immune responses. Metabolism. 114:1543382021.
View Article : Google Scholar
|
|
46
|
Xi Y, Zhang Y, Zhu S, Luo Y, Xu P and
Huang Z: PPAR-mediated toxicology and applied pharmacology. Cells.
9:3522020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jimenez R, Toral M, Gómez-Guzmán M, Romero
M, Sanchez M, Mahmoud AM and Duarte J: The role of Nrf2 signaling
in PPARβ/δ-mediated vascular protection against
hyperglycemia-induced oxidative stress. Oxid Med Cell Longev.
2018:58527062018. View Article : Google Scholar
|
|
48
|
Botta M, Audano M, Sahebkar A, Sirtori CR,
Mitro N and Ruscica M: PPAR agonists and metabolic syndrome: An
established role? Int J Mol Sci. 19:11972018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cheng HS, Tan WR, Low ZS, Marvalim C, Lee
JYH and Tan NS: Exploration and development of PPAR modulators in
health and disease: An update of clinical evidence. Int J Mol Sci.
20:50552019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Heudobler D, Rechenmacher M, Lüke F,
Vogelhuber M, Pukrop T, Herr W, Ghibelli L, Gerner C and Reichle A:
Peroxisome proliferator-activated receptors (PPAR)γ agonists as
master modulators of tumor tissue. Int J Mol Sci. 19:35402018.
View Article : Google Scholar
|
|
51
|
Wang W, Wang R, Zhang Z, Li D and Yut Y:
Enhanced PPAR-gamma expression may correlate with the development
of Barrett's esophagus and esophageal adenocarcinoma. Oncol Res.
19:141–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Feige JN, Gelman L, Michalik L, Desvergne
B and Wahli W: From molecular action to physiological outputs:
Peroxisome proliferator-activated receptors are nuclear receptors
at the crossroads of key cellular functions. Prog Lipid Res.
45:120–159. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Braissant O, Foufelle F, Scotto C, Dauça M
and Wahli W: Differential expression of peroxisome
proliferator-activated receptors (PPARs): Tissue distribution of
PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology.
137:354–366. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Peters JM, Lee SS, Li W, Ward JM,
Gavrilova O, Everett C, Reitman ML, Hudson LD and Gonzalez FJ:
Growth, adipose, brain, and skin alterations resulting from
targeted disruption of the mouse peroxisome proliferator-activated
receptor beta(delta). Mol Cell Biol. 20:5119–5128. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bhullar KS and Rupasinghe HP: Polyphenols:
Multipotent therapeutic agents in neurodegenerative diseases. Oxid
Med Cell Longev. 2013:8917482013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Altinoz MA, Elmaci I, Hacimuftuoglu A and
Ozpinar A, Hacker E and Ozpinar A: PPARδ and its ligand erucic acid
may act anti-tumoral, neuroprotective, and myelin protective in
neuroblastoma, glioblastoma, and Parkinson's disease. Mol Aspects
Med. 78:1008712021. View Article : Google Scholar
|
|
57
|
Altinoz MA, Bilir A and Elmaci I: Erucic
acid, a component of Lorenzo's oil and PPAR-δ ligand modifies C6
glioma growth and toxicity of doxorubicin. Experimental data and a
comprehensive literature analysis. Chem Biol Interact. 294:107–117.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Leisewitz AV, Urrutia CR, Martinez GR,
Loyola G and Bronfman M: A PPARs cross-talk concertedly commits C6
glioma cells to oligodendrocytes and induces enzymes involved in
myelin synthesis. J Cell Physiol. 217:367–376. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Seufert S, Coras R, Tränkle C, Zlotos DP,
Blümcke I, Tatenhorst L, Heneka MT and Hahnen E: PPAR gamma
activators: Off-target against glioma cell migration and brain
invasion. PPAR Res. 2008:5139432008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Khoo NKH, Hebbar S, Zhao W, Moore SA,
Domann FE and Robbins ME: Differential activation of catalase
expression and activity by PPAR agonists: Implications for
astrocyte protection in anti-glioma therapy. Redox Biol. 1:70–79.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang X, Wang G, Shi Y, Sun L, Gorczynski
R, Li YJ, Xu Z and Spaner DE: PPAR-delta promotes survival of
breast cancer cells in harsh metabolic conditions. Oncogenesis.
5:e2322016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang J, Yang W, Zhao D, Han Y, Liu B,
Zhao H, Wang H, Zhang Q and Xu G: Correlation between TSP-1, TGF-β
and PPAR-γ expression levels and glioma microvascular density.
Oncol Lett. 7:95–100. 2014. View Article : Google Scholar
|
|
63
|
Kolacsek O, Wachtl G, Fóthi Á, Schamberger
A, Sándor S, Pergel E, Varga N, Raskó T, Izsvák Z, Apáti Á and
Orbán TI: Functional indications for transposase
domestications-characterization of the human piggyBac transposase
derived (PGBD) activities. Gene. 834:1466092022. View Article : Google Scholar
|