Introduction
Pituitary adenylate cyclase-activating polypeptide
(PACAP) is a member of the vasoactive intestinal
polypeptide/glucagon/growth hormone-releasing factor/secretin
superfamily (1). This family of
proteins has numerous physiological roles, including functions in
the immune system, hormone secretion regulation, neurotrophic
effects and neural restoration (2–7).
PACAP can transfer extracellular signals into an intracellular
signal, through the activation of the PAC1 receptor (PAC1R)
(8). It has been previously shown
that PACAP can induce the growth of the rabbit trigeminal ganglion
and neurite outgrowth in vitro, and it has been verified
further by an in vitro study that the intracellular signal
can accelerate the recovery of corneal sensitivity (9). The development of PAC1R agonists as
novel therapeutic agents may therefore show potential for promoting
nerve regeneration. Research into the binding of ligands to PAC1R
is required for developing specific PAC1R agonists.
G protein-coupled receptors (GPCRs) are the drug
target for numerous diseases. At present, ~50% of known drug
targets are GPCRs (10). PAC1R is
a member of the class B GPCRs. Sun et al (11) used nuclear magnetic resonance
spectroscopy to analyze the crystal structure of PACAP (6–38) and
the PAC1R extracellular domains (11). However, the study failed to analyze
the structure of the whole PAC1R transmembrane domain. Furthermore,
the interactive mode between PACAP38 and PAC1R has not been studied
(12). A previous study used
site-directed mutagenesis to investigate the influence of three
amino acids of PACAP (6–38), Tyr10, Arg14 and Lys21, on the binding
of PACAP (6–38) to the extracellular domains of PAC1R (11). Beebe et al (13) reported the molecular structure of
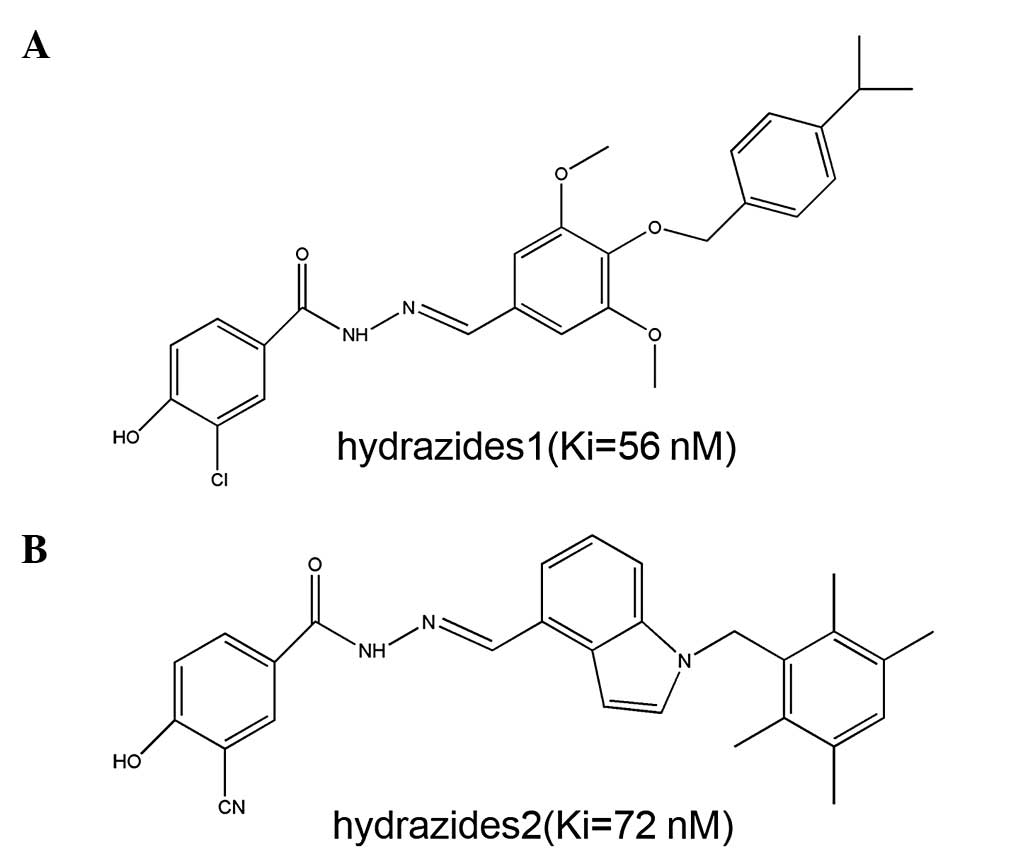
two small-molecule inhibitors, Hydrazides1 and Hydrazides2
(Fig. 1), and their interaction
with PAC1R. The binding strength IC50 value between the
small-molecule inhibitor and the receptor was subsequently
determined.
The present study aimed to predict and analyze the
physicochemical properties of the PAC1R protein, according to the
amino acid sequence of PAC1R. The transmembrane, intracellular and
extracellular spatial structures of the PAC1R protein were
constructed using Discovery Studio 2.5 (DS2.5; Accelrys Software
Inc., San Diego, CA, USA) software with a homology-modeling module.
The key binding sites and regions of PAC1R were preliminarily
studied by molecular docking methodology in order to provide
theoretical support for the study of the binding mode between
ligands and PAC1R, and for the further development of PAC1R
agonists.
Materials and methods
Materials
The primary sequence of PAC1R protein was obtained
from the Swiss-Prot database (available at http://www.uniprot.org/), and the homology template
was obtained through retrieving the protein database (http://www.rcsb.org/pdb/home/home.do).
Bioinformatics analysis of the amino acid
sequence
The physicochemical properties of PAC1R molecules
were analyzed with the ProtParam tool (http://web.expasy.org/protparam/). Hydrophobicity
analysis was performed for the primary sequence of PAC1R with the
Kyte and Doolittle algorithm within the ProtScale online software
(http://www.expasy.ch/tools/protscale.html). The
greater the positive value, the stronger the hydrophobicity, and
vice versa. Where the hydrophobic value was between −0.5 and +0.5,
the amino acid was considered to exhibit both hydrophilic and
hydrophobic characteristics (14)
Homology modeling and evaluation
The primary sequence of PAC1R was obtained from the
Swiss-Prot website (P41586-3) (15). The PAC1R extracellular domains (ID:
2JOD and 3N94) and the seven transmembrane regions (ID: 2KS9) were
identified through the Swiss-Model Alignment Mode tool on the
Swiss-Model website for homology search (http://swissmodel.expasy.org/). Homology modeling was
performed using the modeler module of the DS2.5 molecular
simulation software. The Blosum-62 matrix was used with a gap
penalty of 10 and a gap extension penalty of one. The modeler
module of DS2.5 was used to build the three-dimensional models of
PAC1R. The ‘number of models’ parameter was set to 100 and the
remaining parameters were set to the default values. The best-fit
models of the PAC1R were selected on the basis of the analysis
results of the internal scoring function of Modeler, the Profile-3D
program and the PROCHECK procedure. A suitable model was then
selected for further energy optimization and analysis (16).
Molecular docking
The PAC1R spatial structure obtained following the
optimization of homology modeling was used as the initial
conformation of the receptor protein. The appropriate module of the
DS2.5 molecular simulation software was applied for preprocessing
the Protein Data Bank file, including hydrogenation. The molecular
structures of the PAC1R inhibitors Hydrazides1 and Hydrazides2 were
drawn with DS2.5, and stored as a structure-data file following
energy optimization. The spatial structures of the protein and
small molecules were displayed using Pymol 1.5 software
(Schrödinger LLC, Portland, OR, USA).
Results
Physicochemical property analysis for the
PAC1R sequence Composition and physicochemical properties
The human PAC1R protein sequence was selected from
the Swiss-Prot database (ID: P41586-3, removing the signal peptide
from the front 20 amino acids prior to homology modeling). The
physicochemical properties of the PAC1R protein were predicted
using the ProtParam online tool: i) The molecular weight of the
PAC1R protein was 49073.8 Da; ii) the theoretical isoelectric point
was 6.07; iii) the atom composition was
C2,255H3,383N561O612S29;
iv) the molar digestion coefficient was 106,185 at 280 nm; v) the
instability index was 47.45; vi) the fat coefficient was 88.95 and
vii) the total average hydrophilicity was 0.183. The amino acid
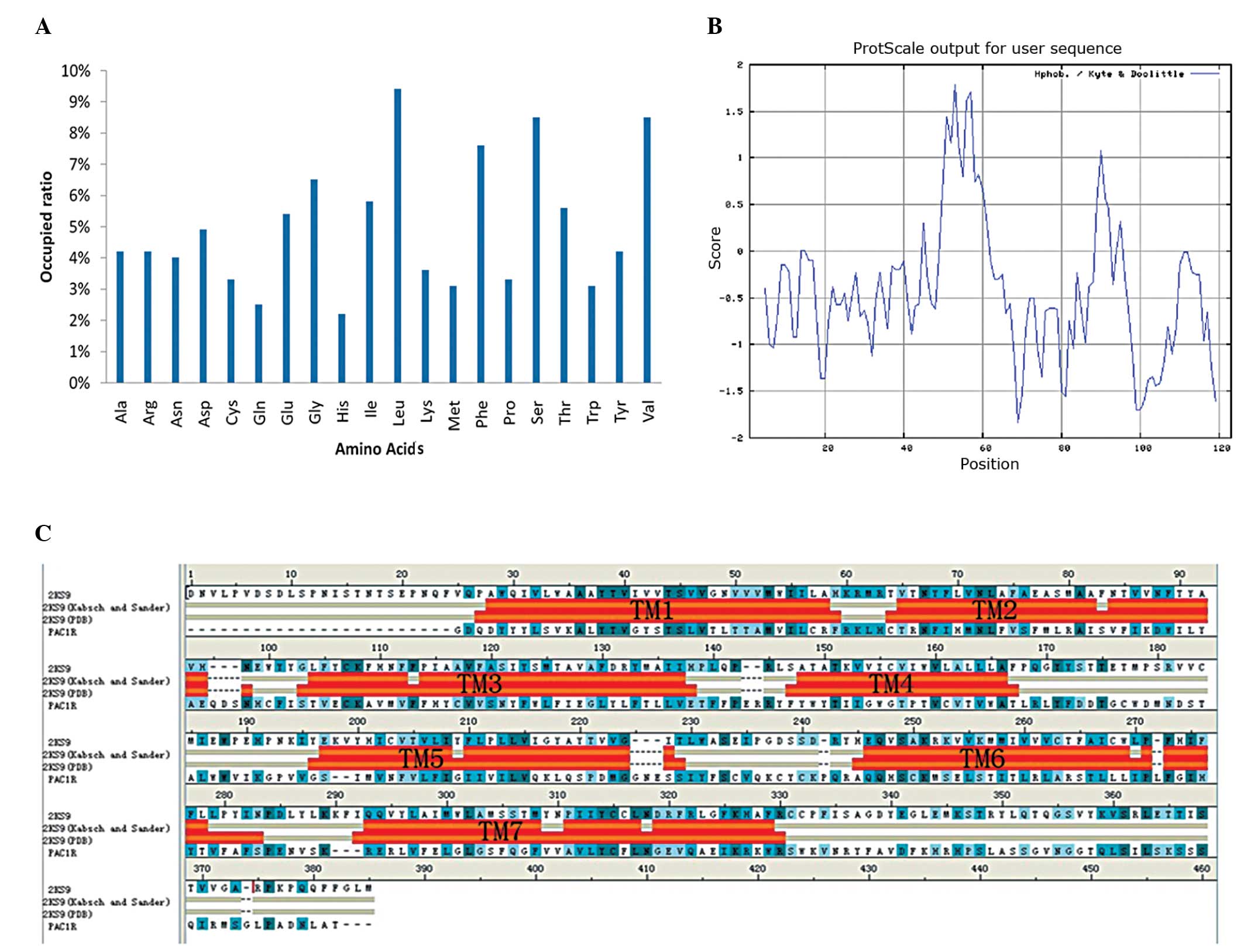
composition of PAC1R is shown in Fig.
2A, in which the amino acid residues with a positive charge
accounted for 35% and the amino acid residues with a negative
charge accounted for 40%. The content of Leu, Val, Ser and Phe was
high, whereas that of His and Gln was low.
Hydrophobicity analysis
The hydrophilicity and hydrophobicity of the PAC1R
protein sequence were analyzed using the ProtScale online tool
provided by Expasy (Swiss Institute of Bioinformatics), and the
distribution of hydrophilic and hydrophobic amino acids of the
protein was identified, as well a prediction of the secondary
structure, including the transmembrane helices and the distribution
of the amino acids on the surface of the protein. The maximum
hydrophobicity of the protein was 3.6 and the minimum
hydrophobicity was −2.4 (Fig. 2B).
It was observed that there were eight evident hydrophobic domains
in the protein, among which seven were the transmembrane domains of
the protein.
Homology modeling of the PAC1R
sequence
The homology modeling of PAC1R was performed using
DS Modeling 1.2. Similarity searches for the sequence were
conducted using Position-Specific Iterative Basic Local Alignment
Search Tool (PSI-BLAST) provided by the National Center for
Biotechnology Information and, following the selection of the
NCBI-BLAST contrast results, the sequence was labeled as 2JOD and
3N94. The A-chain homology of the extracellular domain and 2JOD and
3N94 was 99% (E-value=5×10−69) and 92%
(E-value=1×10−66) respectively; however, there was no
available (high homology) spatial structure template in the
transmembrane domain and intracellular region of PAC1R.
It has been reported in the literature that the
parathyroid hormone receptor I (PTHR1) from the family of B class
GPCRs has been built with the human β2-adrenergic G protein-coupled
receptor at a low matching ratio identity (8.5%; similarity, 25.5%)
in the transmembrane domain as the module through segmented
homology (17,18). However, since GPCRs have seven
highly conservative spiral transmembrane domains and each domain
has one to two conservative residues with conservative spatial
structure, the complete spatial structure of PTH1R transmembrane
domain was successfully constructed with the dual-template method.
The PAC1R transmembrane and intracellular regional sequences were
uploaded to the Swiss-Model website, and the template search
contrast was then conducted using the Swiss-Model Alignment Mode.
The results showed that the sequences had the highest sequence
homology with the A-chain of 2KS9 (same rate, 15%; similar to the
rate, 35.7%). Sequence alignment between the sequence and 2KS9 was
performed and both the primary and secondary structures were
analyzed using Align123 in DS Modeling 1.1, followed by a manual
modification based on the hydrophobicity analysis result for
sequence alignment. The final alignment was carefully evaluated and
evidenced to be a match for the conserved residue data for the
class B GPCR (19). The predicted
transmembrane helices of the transmembrane protein module were
analyzed and edited with DS2.5. The prediction identified that the
transmembrane domain of the PAC1R was overlapping with the
transmembrane domain of 2KS9 (Fig.
2C). During the homology modeling process for the construction
of homology models using DS2.5, the transmembrane domain factor was
added. The number of models was set as 100, the refine loops
parameter was set as true and 200 models were constructed, whilst
the remaining parameters were set to default.
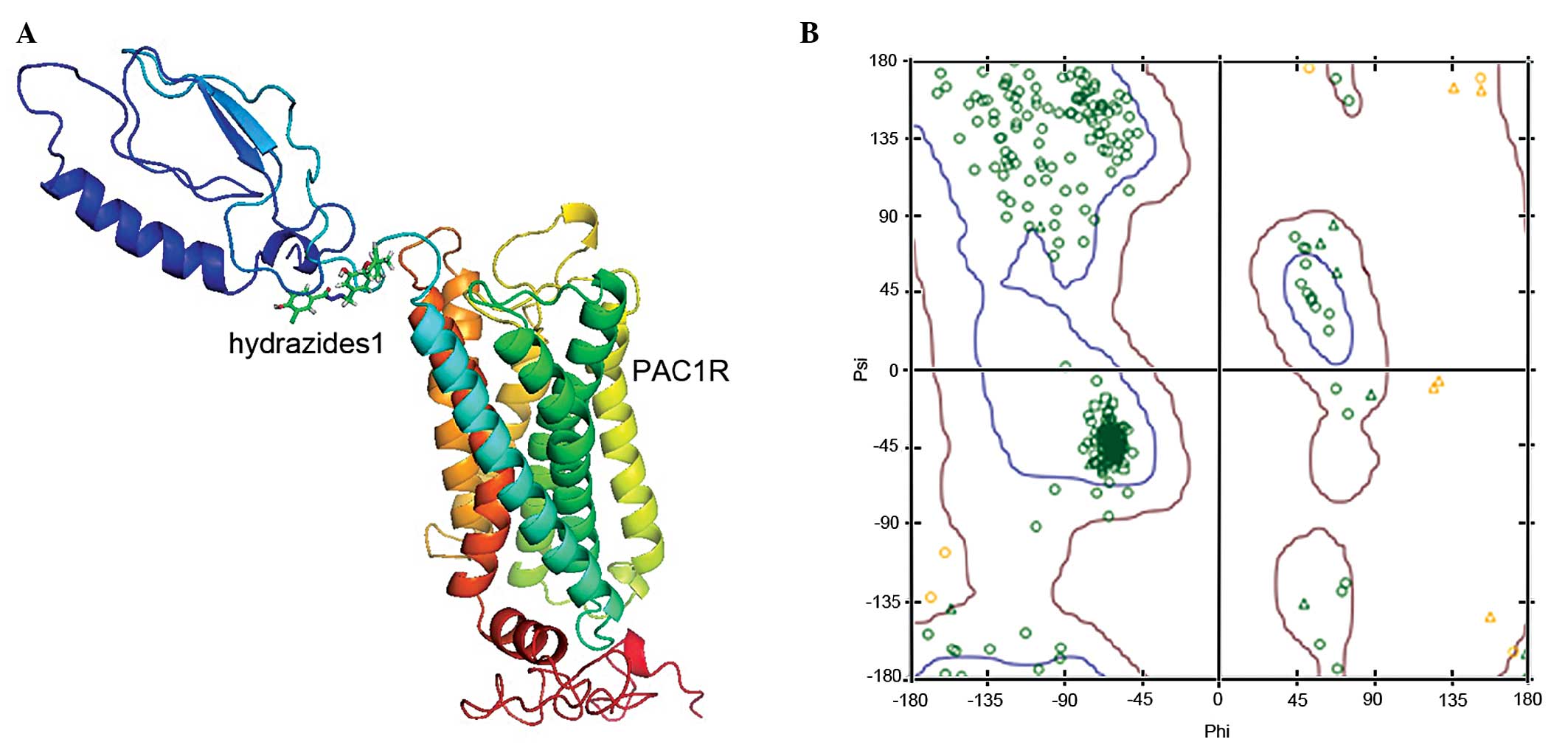
The probability density function energy of the PAC1R
model was checked, indicating a valid structure for the entire
model, and the Ramachandran plots for local backbone conformation
of each residue in the final models were produced by PROCHECK. The
three-dimensional structural compatibility principle of the protein
constructed was evaluated using the Profile-3D module of DS2.5
(20), and the average score was
144.02, between the maximum of 194.85 and minimum of 87.68. The
stereochemical features of the model were constructed, including
the evaluation of the stereochemical stability of the main and side
chains, which was conducted using the Ramachandran module of DS2.5
software. A total of ≥90% of the ϕ and ψ stereochemistry in the
main chain was distributed within the allowed region, and was in
accordance with the principles of stereochemistry, suggesting that
the model (Fig. 3A) was
theoretically reliable. The Ramachandran plot, generated by the
DS2.5 software, is shown in Fig.
3B; ~98.1% of the amino acids of the optimal model selected
were within the allowed region (red region), suggesting that the
model constructed may reflect the three-dimensional structure of
the protein.
Small-molecule inhibitor and PAC1R
binding
The optimal spatial structure of PAC1R protein,
obtained through homology modeling, was modified with the Clean
Protein tool in the DS2.5 software package. Under physiological
conditions (pH 7.0), the hydrogenation could guarantee the
protonation processing, which would be the initial conformation of
the acceptor molecule following further energy optimization. The
ligand molecules, Hydrazides1 and Hydrazides2, were compiled and
stored as sd files with DS molecular simulation software, and the
energy optimization for small molecules in the CHARMM force field
was conducted. The binding between the small molecules and the
receptor was implemented using the LibDock program of the DS
simulation software package, and the active site of binding was
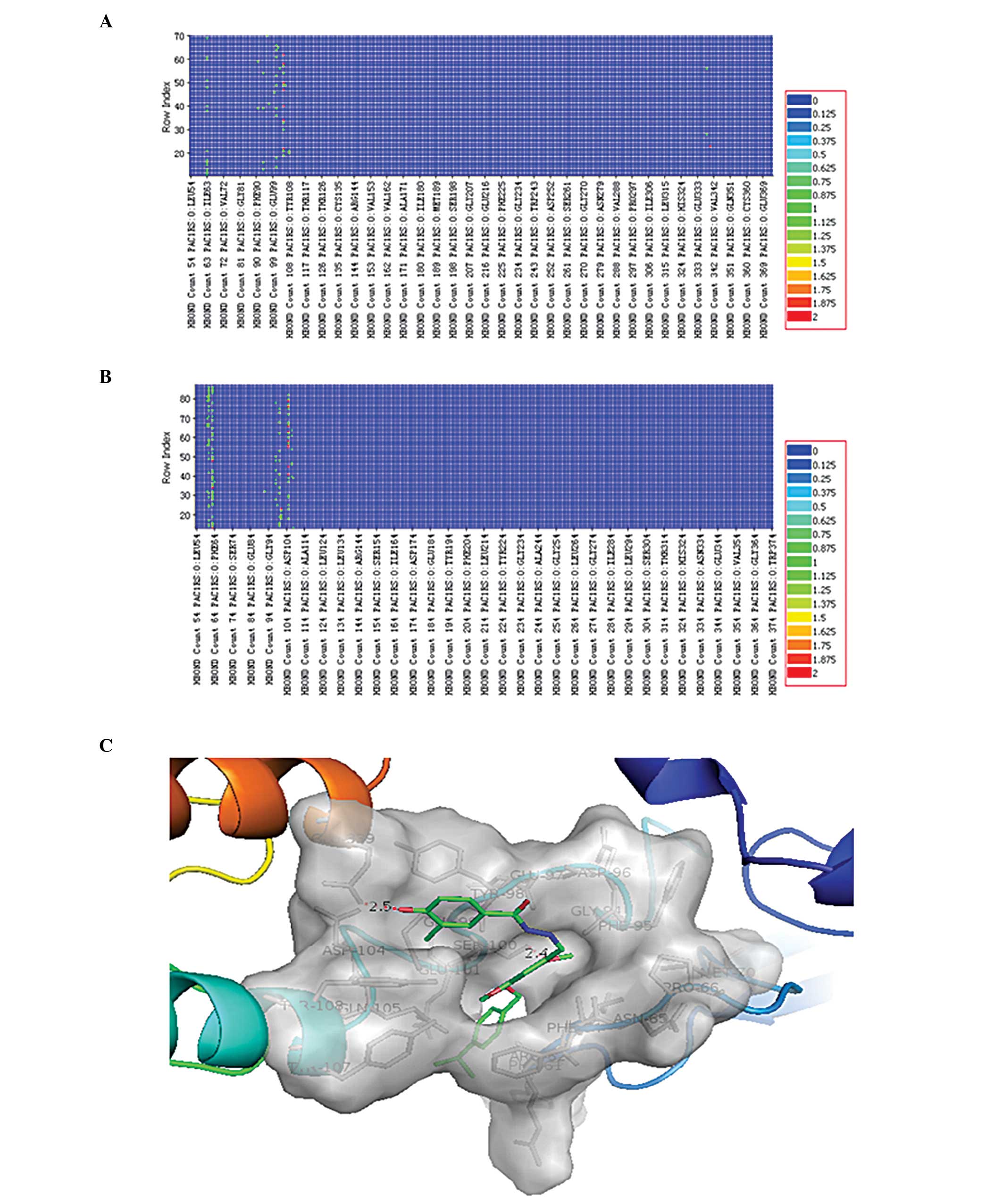
predicted by the DS software. According to the binding result,
there were 76 conformations in the binding of Hydrazides1 and the
receptor, while there were 89 conformations in the binding of
Hydrazides2 and the receptor. The hydrogen bond formed between the
two small molecules and the receptor docking conformation was
calculated with the ‘Analyze Ligand Poses’ process analysis. The
hydrogen bonding heat map is shown in Fig. 4A and B. It can be observed that the
Ile63, Ser100 and Gln105 residues of PAC1R formed a hydrogen bond
with most Hydrazides1 and Hydrazides2 docking conformations. The
conformation with the highest LibDock score for the docking of
Hydrazides1 with the receptor was selected for the display
(Fig. 4C). According to the
docking results, in the selected conformation, Hydrazides1 formed
hydrogen-bond interactions at the Ser100 and Glu339 of PAC1R, and
the chain lengths were 2.4 and 2.5 Å respectively.
Discussion
PACAP performs numerous biological functions,
including promotion of nerve regeneration, neuroprotection,
prevention of arteriosclerosis and regulation of energy metabolism
(21–24). Following the binding of PACAP to
its receptor, the extracellular signal is transferred into the
cell, generating a biological response. The complete spatial
structure of the seven-transmembrane receptors has not been
analyzed, which has restricted the research and development of
drugs targeting the PAC1R (25).
The first high-resolution template that was suitable
for homology modeling of GPCRs became available in the year 2000
with the crystallization of bovine rhodopsin (26,27).
The three-dimensional model of PTHR1 and the octopamine receptor 2
were constructed through two steps of homology modeling methods
(17–19). The transmembrane regions exhibited
low matching rates with 2RH1 as a template; however, the GPCR had
the seven highly conserved, spiral transmembrane regions, and each
transmembrane region had 1–2 conserved residues and the spatial
structure was additionally conserved (17,27,28).
Therefore, the three-dimensional model of the PAC1R was constructed
by employing two-mode plate methods.
According to previous research (29), one of the active modes between the
GPCR small-molecule inhibitors and the receptor took the
extracellular domain of the receptor as the target, such as the
binding model of small-molecule T-0632 with glucagon-like peptide 1
receptor (29). The Ki values of
the small molecules Hydrazides1 and Hydrazides2 with the
extracellular domain of the PAC1R protein, were 56 and 72 nM
respectively (as shown in Fig. 1),
with strong binding capability. A series of docking conformations
were generated with the molecule docking method, and the binding
conformation of the two molecules and the receptor was subsequently
analyzed. The heat map of the hydrogen bonds generated in the
receptor molecule is shown in Fig.
4. When analyzing the heat map of the hydrogen bonds, the key
amino acids involved in the binding between the small molecules
Hydrazides1 and Hydrazides2 and the receptor structure were
directly observed (as shown in Fig. 4A
and B). As observed from the position of the binding of
Hydrazides1 and Hydrazides2 to the receptor (Fig. 3A), it can be predicted that these
small-molecule inhibitors can prevent the normal interactions
between the ligand agonist in the extracellular domain and the
active region formed by the seven transmembrane regions of PAC1R
through the free activity in the extracellular domain, thus
achieving the inhibitory effect. In addition, according to the
literature, the key domain of the binding ligand PACAP38 of PAC1R
comprises residues 116–120, in the extracellular domain (11). It was suggested by the heat map of
the hydrogen bonds that the hydrogen bond was formed between the
two small-molecule inhibitors Hydrazides1 and Hydrazides2 and the
Ser100 of PAC1R (the first 20 amino acids were the signal peptide
and was removed, and the site Ser100 was Ser120). Each
small-molecule inhibitor produces a steric hindrance in the binding
of PACAP to PAC1R, which can inhibit the biological activity of
PACAP competitively. The manner in which PACAP can bind with the
PAC1R, the changes in the C-backbone of PAC1R, as well as how the
extracellular signal is transferred into the cell through
structural changes remains to be studied using methods such as
molecular dynamics, isothermal titration calorimetry or surface
plasmon resonance.
Acknowledgements
This study was supported by the Science and
Technology project of Guangzhou (grant no. 2011J4300107).
References
|
1
|
Henle F, Fischer C, Meyer DK and Leemhuis
J: Vasoactive intestinal peptide and PACAP38 control
N-methyl-D-aspartic acid-induced dendrite motility by modifying the
activities of Rho GTPases and phosphatidylinositol 3-kinases. J
Biol Chem. 281:24955–24969. 2006. View Article : Google Scholar
|
|
2
|
Kimura H, Kawatani M, Ito E and Ishikawa
K: PACAP facilitate the nerve regeneration factors in the facial
nerve injury. Regul Pept. 123:135–138. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
May V, Lutz E, MacKenzie C, Schutz KC,
Dozark K and Braas KM: Pituitary adenylate cyclase-activating
polypeptide (PACAP)/PAC1HOP1 receptor activation coordinates
multiple neurotrophic signaling pathways: Akt activation through
phosphatidylinositol 3-kinase gamma and vesicle endocytosis for
neuronal survival. J Biol Chem. 285:9749–9761. 2010. View Article : Google Scholar
|
|
4
|
Bourgault S, Vaudry D, Botia B, et al:
Novel stable PACAP analogs with potent activity towards the PAC1
receptor. Peptides. 29:919–932. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmidt DT, Rühlmann E, Waldeck B, et al:
The effect of the vasoactive intestinal polypeptide agonist Ro
25–1553 on induced tone in isolated human airways and pulmonary
artery. Naunyn Schmiedebergs Arch Pharmacol. 364:314–320. 2001.
|
|
6
|
Deutsch PJ and Sun Y: The 38-amino acid
form of pituitary adenylate cyclase-activating polypeptide
stimulates dual signaling cascades in PC12 cells and promotes
neurite outgrowth. J Biol Chem. 267:5108–5113. 1992.
|
|
7
|
Deutsch PJ, Schadlow VC and Barzilai N:
38-Amino acid form of pituitary adenylate cyclase activating
peptide induces process outgrowth in human neuroblastoma cells. J
Neurosci Res. 35:312–320. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rawlings SR: PACAP, PACAP receptors, and
intracellular signalling. Mol Cell Endocrinol. 101:C5–C9. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukiage C, Nakajima T, Takayama Y,
Minagawa Y, Shearer TR and Azuma M: PACAP induces neurite outgrowth
in cultured trigeminal ganglion cells and recovery of corneal
sensitivity after flap surgery in rabbits. Am J Ophthalmol.
143:255–262. 2007. View Article : Google Scholar
|
|
10
|
Klabunde T and Hessler G: Drug design
strategies for targeting G-protein-coupled receptors. Chembiochem.
3:928–944. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun C, Song D, Davis-Taber RA, et al:
Solution structure and mutational analysis of pituitary adenylate
cyclase-activating polypeptide binding to the extracellular domain
of PAC1-RS. Proc Natl Acad Sci USA. 104:7875–7880. 2007. View Article : Google Scholar
|
|
12
|
Kumar S, Pioszak A, Zhang C, Swaminathan K
and Xu HE: Crystal structure of the PAC1R extracellular domain
unifies a consensus fold for hormone recognition by class B
G-protein coupled receptors. PLoS One. 6:e196822011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beebe X, Darczak D, Davis-Taber RA, et al:
Discovery and SAR of hydrazide antagonists of the pituitary
adenylate cyclase-activating polypeptide (PACAP) receptor type 1
(PAC1-R). Bioorg Med Chem Lett. 18:2162–2166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wilkins MR, Gasteiger E, Bairoch A, et al:
Protein identification and analysis tools in the ExPASy server.
Methods Mol Biol. 112:531–552. 1999.PubMed/NCBI
|
|
15
|
Dautzenberg FM, Mevenkamp G, Wille S and
Hauger RL: N-terminal splice variants of the type I PACAP receptor:
isolation, characterization and ligand binding/selectivity
determinants. J Neuroendocrinol. 11:941–949. 1999. View Article : Google Scholar
|
|
16
|
Kufareva I, Rueda M, Katritch V, Stevens
RC and Abagyan R; GPCP Dock 2010 participants. Status of GPCR
modeling and docking as reflected by community-wide GPCR Dock 2010
assessment. Structure. 19:1108–1126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Yang HQ, Li J and Fang HS:
Homology modeling of three-dimensional structure of human CCR5.
Pharmaceutical Biotechnology. 19:432–436. 2012.(In Chinese).
|
|
18
|
Lin KJ, Zhu DJ, Leng YG and You QD:
Structural characterization of human parathyroid hormone 1
receptor. Acta Phys Chim Sin. 28:1783–1789. 2012.
|
|
19
|
Mirzadegan T, Benkö G, Filipek S and
Palczewski K: Sequence analyses of G-protein-coupled receptors:
similarities to rhodopsin. Biochemistry. 42:2759–2767. 2003.
View Article : Google Scholar
|
|
20
|
Bowie JU, Lüthy R and Eisenberg D: A
method to identify protein sequences that fold into a known
three-dimensional structure. Science. 253:164–170. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seaborn T, Masmoudi-Kouli O, Fournier A,
Vaudry H and Vaudry D: Protective effects of pituitary adenylate
cyclase-activating polypeptide (PACAP) against apoptosis. Curr
Pharm Des. 17:204–214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fila T, Trazzi S, Crochemore C, Bartesaghi
R and Ciani E: Lot1 is a key element of the pituitary adenylate
cyclase-activating polypeptide (PACAP)/cyclic AMP pathway that
negatively regulates neuronal precursor proliferation. J Biol Chem.
284:15325–15338. 2009. View Article : Google Scholar
|
|
23
|
Deguil J, Jailloux D, Page G, et al:
Neuroprotective effects of pituitary adenylate cyclase-activating
polypeptide (PACAP) in MPP+-induced alteration of
translational control in Neuro-2a neuroblastoma cells. J Neurosci
Res. 85:2017–2025. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Somogyvári-Vigh A and Reglodi D: Pituitary
adenylate cyclase activating polypeptide: a potential
neuroprotective peptide. Curr Pharm Des. 10:2861–2889.
2004.PubMed/NCBI
|
|
25
|
Hruby VJ: Designing peptide receptor
agonists and antagonists. Nat Rev Drug Discov. 1:847–858. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Palczewski K, Kumasaka T, Hori T, et al:
Crystal structure of rhodopsin: a G protein-coupled receptor.
Science. 289:739–745. 2000. View Article : Google Scholar
|
|
27
|
Elbegdorj O, Westkaemper RB and Zhang Y: A
homology modeling study toward the understanding of
three-dimensional structure and putative pharmacological profile of
the G-protein coupled receptor GPR55. J Mol Graph Model. 39:50–60.
2013. View Article : Google Scholar
|
|
28
|
Okada T, Sugihara M, Bondar AN, Elstner M,
Entel P and Buss V: The retinal conformation and its environment in
rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol.
342:571–583. 2004. View Article : Google Scholar
|
|
29
|
Tibaduiza EC, Chen C and Beinborn M: A
small molecule ligand of the glucagon-like peptide 1 receptor
targets its amino-terminal hormone binding domain. J Biol Chem.
276:37787–37793. 2001.PubMed/NCBI
|