Introduction
Long non-coding RNAs (lncRNAs) are >200 bp RNAs
that do not encode proteins, which have been demonstrated to fully
participate in the metabolic processes of cancer cells (1). Compared with RNA-encoding proteins,
lncRNAs possess a greater reservoir of transcriptional information
(2). EGFR-AS1 is a 2,821-kb lncRNA
located on human chromosome 7; its locus is in the opposite
direction of EGFR gene transcription according to the UCSC database
(http://genome.ucsc.edu/).
Since it was first studied in liver cancer (3), EGFR-AS1 has been comprehensively
studied in various solid cancer types including gastric, renal,
lung, colorectal, bladder and oral cancer (4–14).
Despite its coordination with EGFR to regulate cancer cell
metabolism, EGFR-AS1 has also been shown to possess independent
regulatory function (15). EGFR-AS1
can also be regulated via epigenetic modulation; it can be
activated by K27 acetylation, which suppresses the activation of
microRNA (miR)-2355-5p and promotes cervical cancer cell
proliferation via the Wnt pathway (16). EGFR-AS1 overexpression was also
reported to induce resistance to tyrosine kinase inhibitors in
squamous cell carcinoma (14).
Therefore, the function of EGFR-AS1 is similar to other oncogenes
and influences cancer cell prognosis. The present study was
conducted to comprehensively investigate the oncogenic role of
EGFR-AS1 in several types of solid tumour.
Materials and methods
Literature search
The articles screened in the present study were
identified following an electronic search of the PubMed (https://pubmed.ncbi.nlm.nih.gov/) database using
the keywords ‘cancer or carcinoma or neoplasm or tumour’ and ‘long
noncoding RNA EGFR-AS1 or noncoding RNA EGFR-AS1 or EGFR-AS1’ by
two investigators. These key words were applied in different
combinations. The bibliographies from any of the identified studies
were used to identify further studies. The screened articles were
limited to published studies or to studies where at least the
abstract was published in English. No contact was made with the
authors of the published studies to obtain unpublished data.
Selection of trials
A total of 22 articles were obtained from March,
2002 to March, 2023. Prior to finalizing the list of candidate
articles, several inclusion criteria were determined as follows: i)
The research contained at least one of the following clinical
characteristics: Patient sex, tumour size, distant metastasis,
lymph node metastasis, tumour-node-metastasis (TNM) stage,
pathological stage and overall survival (OS); ii) the expression of
EGFR-AS1 lncRNA was evaluated in tumour tissues only (not serum or
any other tissues) and the method used was reverse
transcription-quantitative PCR (RT-qPCR); iii) all analysed samples
were extracted from humans; and iv) all patients were divided into
groups based on high or low EGFR-AS1 expression.
Data extraction and quality
assessment
Two investigators independently extracted the data
from each original publication, including the first author's name,
the year of publication, the country of origin, the cancer type,
the total number of cases, the number of patients in the high and
low EGFR-AS1 expression groups, the detection method, the outcome
measures and the cut-off value for EGFR-AS1 expression levels.
Missing information was estimated according to the Cochrane
Handbook (https://china.cochrane.org/zh-hans/resources/cochrane-resources/cochrane-handbook).
Discrepancies between the two investigators (for example, whether a
certain study should be included) were resolved by discussion and
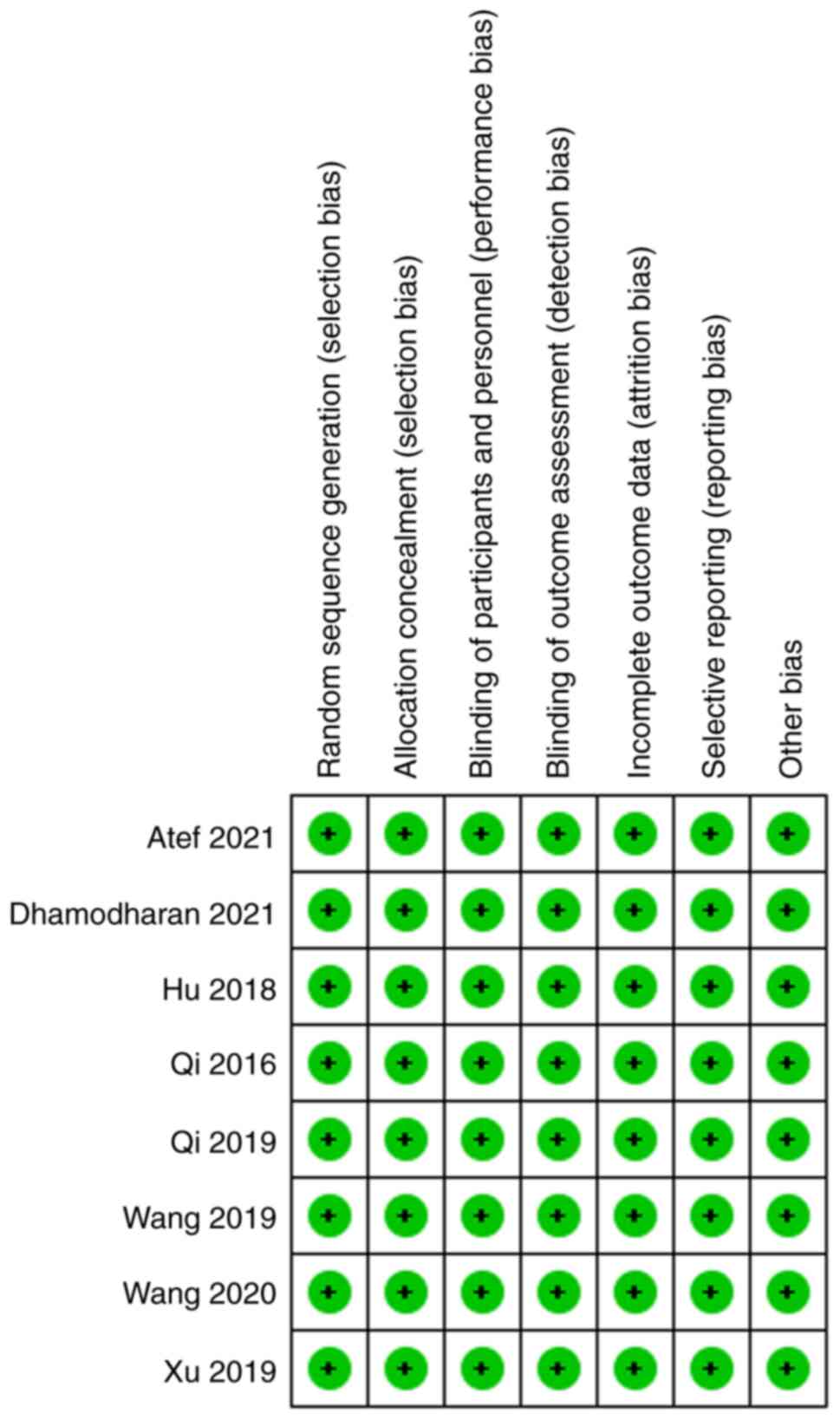
consensus. The quality of the included studies was assessed using
the Newcastle-Ottawa Quality Assessment Scale (NOQAS) (17).
Among the 8 included studies, only 5 provided
Kaplan-Meier curves and none provided hazard ratios (HRs) and
corresponding 95% confidence intervals (CIs), despite constructing
survival curves. Using widely proven and accepted scientific
methods, the data were extracted from the survival curves using
Engauge 4.1 (http://digitizer.sourceforge.net/) (18–20).
Subsequently, the extracted data were entered into the HR
calculation spreadsheet, which was created by Tierney et al
(21). The HRs, standard errors and
corresponding 95% CIs were then estimated from the survival
curves.
Statistical analysis
Risk ratios (RRs) with 95% CIs were estimated to
evaluate the correlation between high expression of EGFR-AS1 lncRNA
and the clinical outcomes of patients with cancer. The cancer
samples were classified into well-differentiated and
moderately/poorly differentiated according to their reported
differentiation grade status. According to the information provided
in each study, the tumour stageswere separated into two groups,
namely ≤T2 (early stage) and >T2 (advanced stage); the TNM stage
was divided into two groups, namely early stage (I–II) and advanced
stage (III–IV). Distant metastasis, lymph node metastasis and
vascular invasion were divided into ‘positive’ and ‘negative’
groups. All the extracted data were pooled using Review Manager 5.3
(The Cochrane Collaboration). According to the recommendations
provided by the Cochrane Handbook (https://training.cochrane.org/handbook/current/chapter-10#section-10-10-4-1),
a random-effects model was used to analyse the pooled results in
the present study.
Results
Characteristics of the included
trials
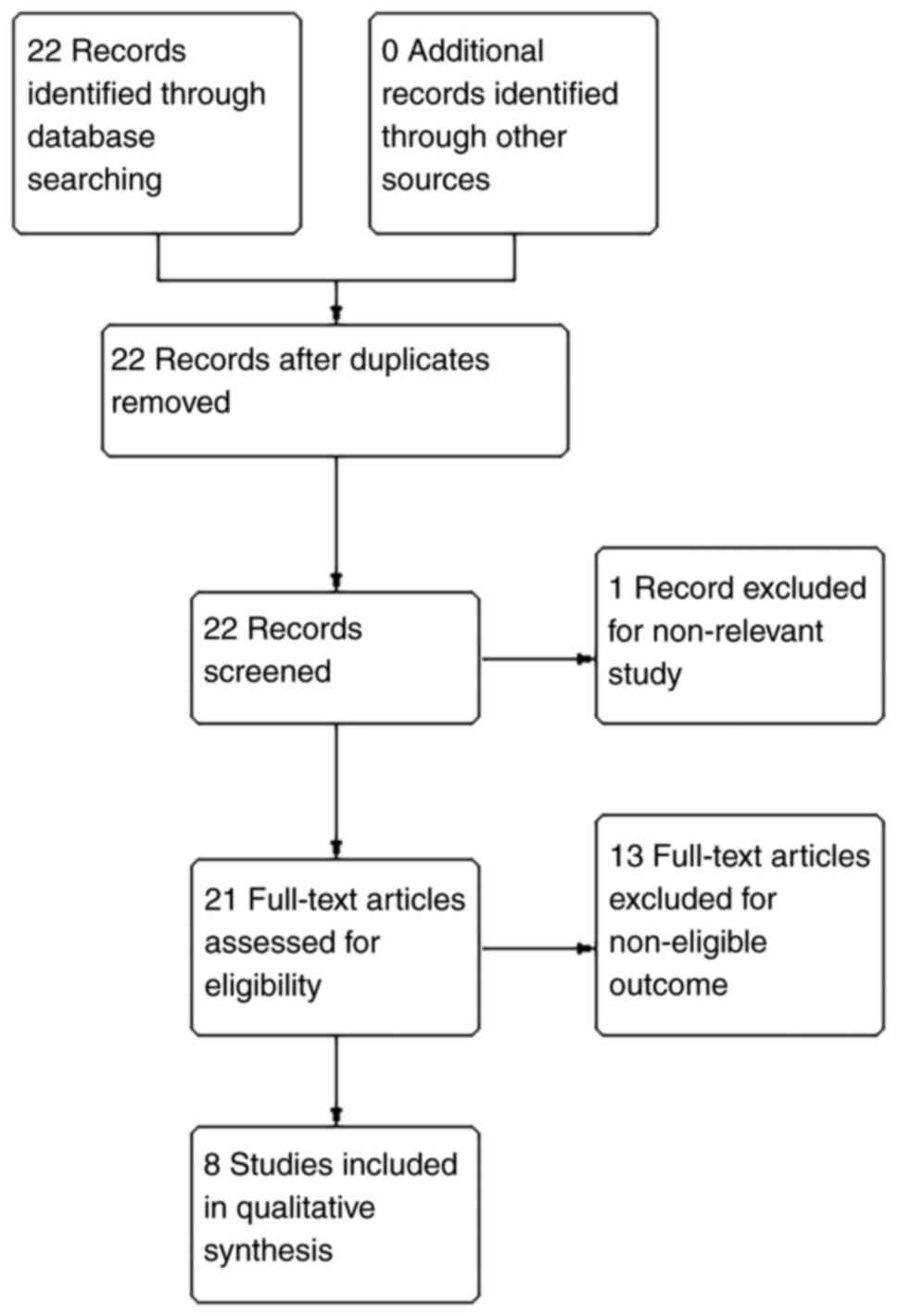
As shown in Fig. 1,
8 of the 22 identified studies completely fulfilled the inclusion
criteria of the present systematic review. A total of 14 trials
were excluded due to non-relevant study types or outcomes. The
included studies covered 7 types of cancer as follows: 1 for liver
cancer (3) 1 for gastric cancer
(13), 1 for renal cancer (10), 2 for non-small cell lung cancer
(NSCLC) (9,11), 1 for colorectal cancer (4), 1 for bladder cancer (6) and 1 for oral cancer (5).
As depicted in Table
I, the 8 included trials were from China, Egypt and India. The
trials included 773 patients in total, 420 with high ERGR-AS1
expression and 353 with low EGFR-AS1 expression. The EGFR-AS1
expression levels in patients were all evaluated by RT-qPCR
analysis. In total, 4 studies applied relative expression between
cancer and adjacent normal samples as the cut-off, 2 studies
applied median expression and 1 study applied 1.85 (the median mRNA
expression) as the cut-off for grouping.
 | Table I.Characteristics of the studies
included in the meta-analysis. |
Table I.
Characteristics of the studies
included in the meta-analysis.
|
|
|
|
| EGFR-AS1
expression |
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| First author,
year | Country | Cancer type | Total cases, n | High, n | Low, n | Detection method | Cut-off (high/low)
determination method | (Refs.) |
|---|
| Qi et al,
2016 | China | Liver cancer | 40 | 14 | 26 | RT-qPCR | Relative
Expression | (3) |
| Hu et al,
2018 | China | Gastric cancer | 58 | 32 | 26 | RT-qPCR | Relative
Expression | (13) |
| Wang et al,
2019 | China | Renal cancer | 204 | 102 | 102 | RT-qPCR | Median | (10) |
| Qi et al,
2019 | China | NSCLC | 87 | 36 | 51 | RT-qPCR | Relative
Expression | (9) |
| Xu et al,
2019 | China | NSCLC | 78 | 48 | 30 | RT-qPCR | Relative
Expression | (11) |
| Atef et al,
2021 | Egypt | Colorectal
cancer | 130 | 100 | 30 | RT-qPCR | 1.85 | (4) |
| Wang et al,
2020 | China | Bladder cancer | 128 | 64 | 64 | RT-qPCR | Median | (6) |
| Dhamodharan et
al, 2021 | India | Oral cancer | 48 | 24 | 24 | RT-qPCR | Median | (5) |
As shown in Table
II, the NOQAS scores of the included studies were not <7
points, indicating that the quality of the included studies was
relatively high, laying a solid foundation for the conclusions
drawn in the present study.
 | Table II.NOQAS scores for the studies included
in the meta-analysis. |
Table II.
NOQAS scores for the studies included
in the meta-analysis.
| First author,
year | Selection (out of
4) | Comparability (out
of 2) | Exposure (out of
4) | NOQAS score | (Refs.) |
|---|
| Qi et al,
2016 | 3 | 2 | 2 | 7 | (3) |
| Hu et al,
2018 | 3 | 2 | 2 | 7 | (13) |
| Wang et al,
2019 | 3 | 2 | 2 | 7 | (10) |
| Qi et al,
2019 | 3 | 2 | 2 | 7 | (9) |
| Xu et al,
2019 | 3 | 2 | 2 | 7 | (11) |
| Atef et al,
2021 | 3 | 2 | 2 | 7 | (4) |
| Wang et al,
2020 | 3 | 2 | 2 | 7 | (6) |
| Dhamodharan et
al, 2021 | 3 | 2 | 2 | 7 | (5) |
Association between EGFR-AS1
expression level and the general characteristics of patients with
cancer
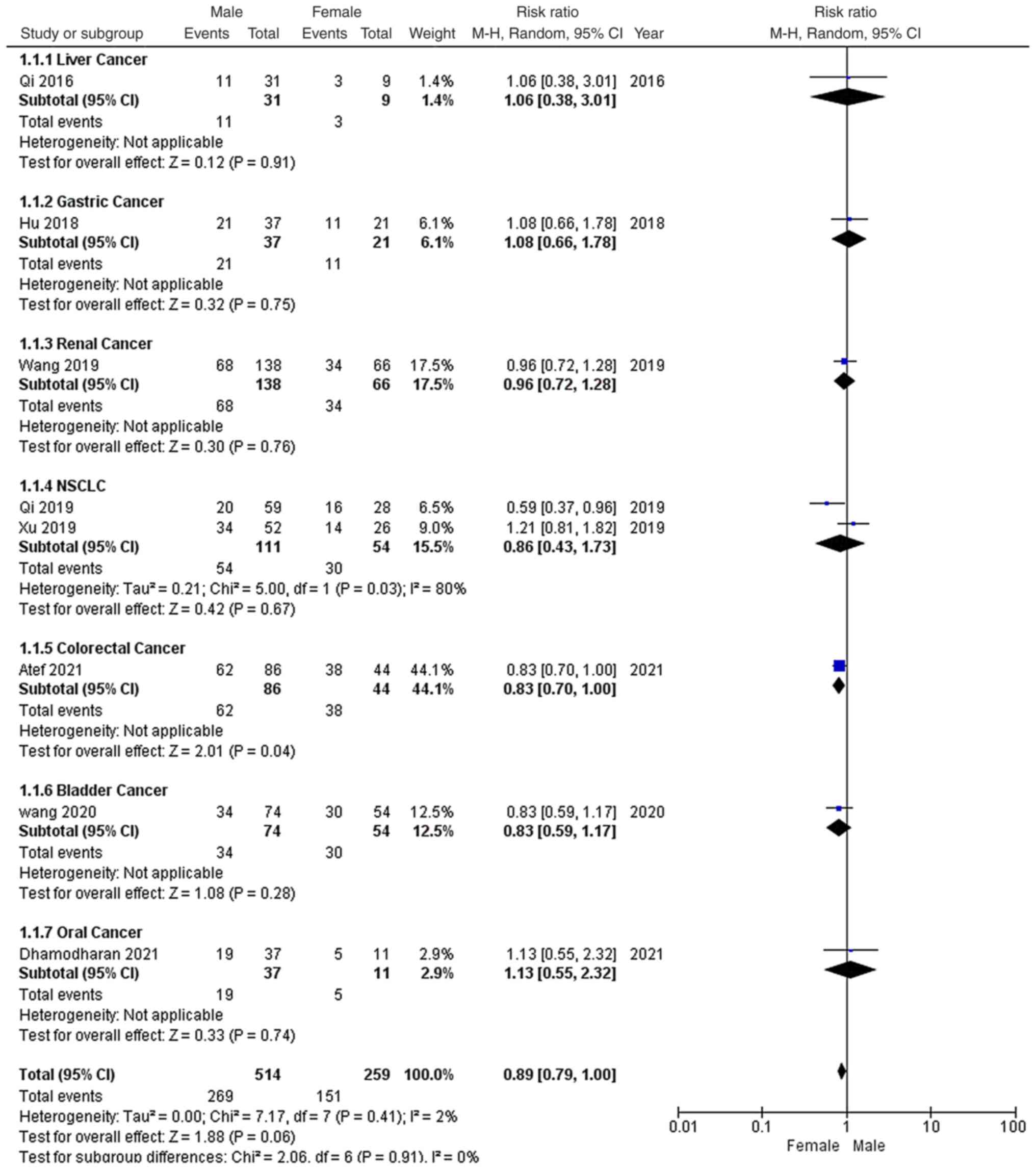
As shown in Fig. 2,
all the studies reported the sex distribution of the included
patients. Among the 773 patients, there were 514 males and 259
females. Furthermore, 269 male and 151 female patients had high
EGFR-AS1 expression. Due to low heterogeneity (P=0.41;
I2=2%) and the recommendations provided by the Cochrane
Handbook, a random-effects model was applied to evaluate the pooled
RR and 95% CI of EGFR-AS1 expression among the sexes. The pooled RR
of high EGFR-AS1 expression among males versus females was 0.89
with a 95% CI of 0.79–1.00, which indicated that high EGFR-AS1
expression was not associated with sex among these patients with
cancer.
As shown in Fig. 3,
among the 8 studies, only 3 studies reported the smoking status of
patients, which were 2 NSCLC studies and 1 oral cancer study. A
total of 60 smoking and 48 non-smoking patients with high EGFR-AS1
expression were identified out of the 213 patients. Due to high
heterogeneity (P=0.02; I2=75%), a random-effects model
was applied to evaluate the pooled RR and 95% CI of smoking status
and EGFR-AS1 expression. The pooled RR of high EGFR-AS1 expression
among smokers versus non-smokers was 0.78 with a 95% CI of
0.46–1.31, which indicated that high EGFR-AS1 expression was not
associated with the smoking status of these patients with
cancer.
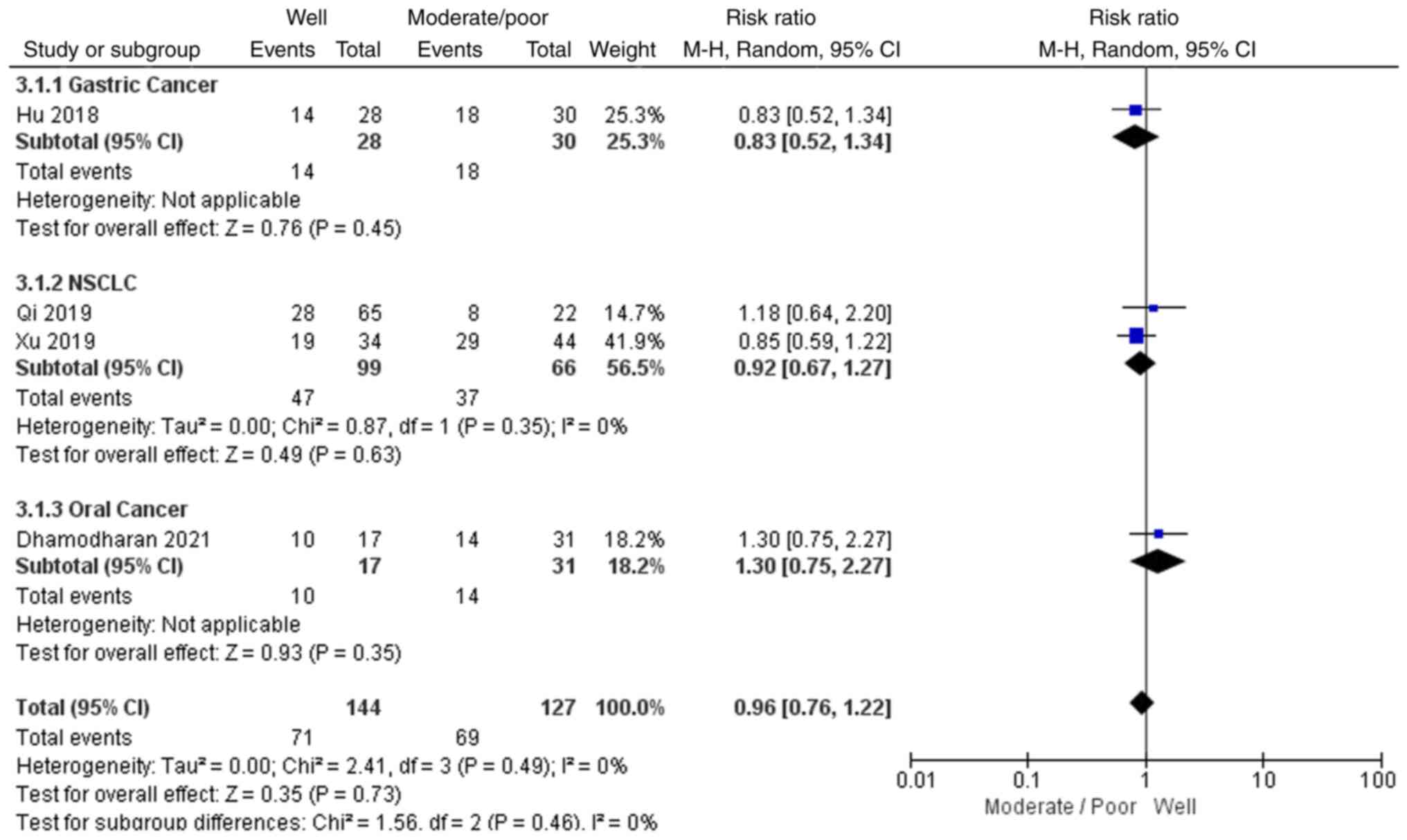
As shown in Fig. 4,
4 out of the 8 studies reported the differentiation grades of the
tumour samples, including 1 gastric cancer study, 2 NSCLC studies
and 1 oral cancer study. In total, 71 patients with
well-differentiated tumours and 69 with moderately or poorly
differentiated tumours had high EGFR-AS1 expression. Due to low
heterogeneity (P=0.49; I2=0) and the recommendations
provided by the Cochrane Handbook, a random-effects model was
applied to evaluate the pooled RR and 95% CI of tumour
differentiation and EGFR-AS1 expression. The pooled RR of high
EGFR-AS1 expression following a comparison of the
well-differentiated with the moderately/poorly differentiated
groups was 0.96 with a 95% CI of 0.76–1.22, which indicated that
high EGFR-AS1 expression was not associated with the
differentiation grade for these patients with cancer.
Association between EGFR-AS1
expression level and the clinical stages of patients with
cancer
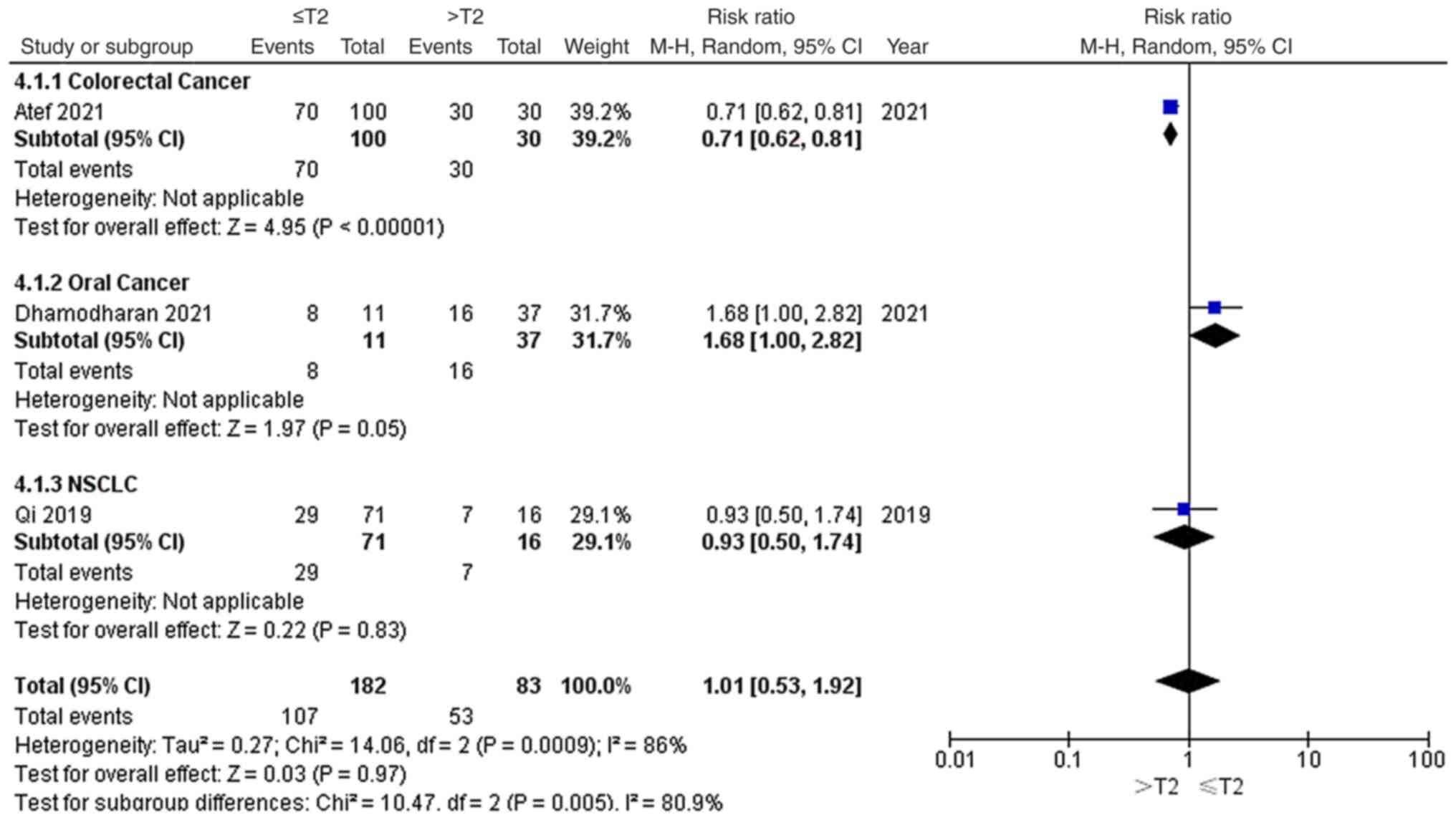
As shown in Fig. 5,
3 out of 8 studies reported the tumour stages of the included
patients, including 1 colorectal cancer study, 1 oral cancer study
and 1 NSCLC study. Among the 265 patients, 102 patients with ≤T2
stage and 53 patients with >T2 stage had high EGFR-AS1
expression. Due to significant heterogeneity (P=0.0009;
I2=86%), a random-effects model was applied to evaluate
the pooled RR and 95% CI of tumour grade and EGFR-AS1 expression.
The pooled RR of high EGFR-AS1 expression among the different
tumour stages was 1.01 with a 95% CI of 0.53–1.92, which indicated
that high EGFR-AS1 expression was not associated with tumour stage
for these patients with cancer.
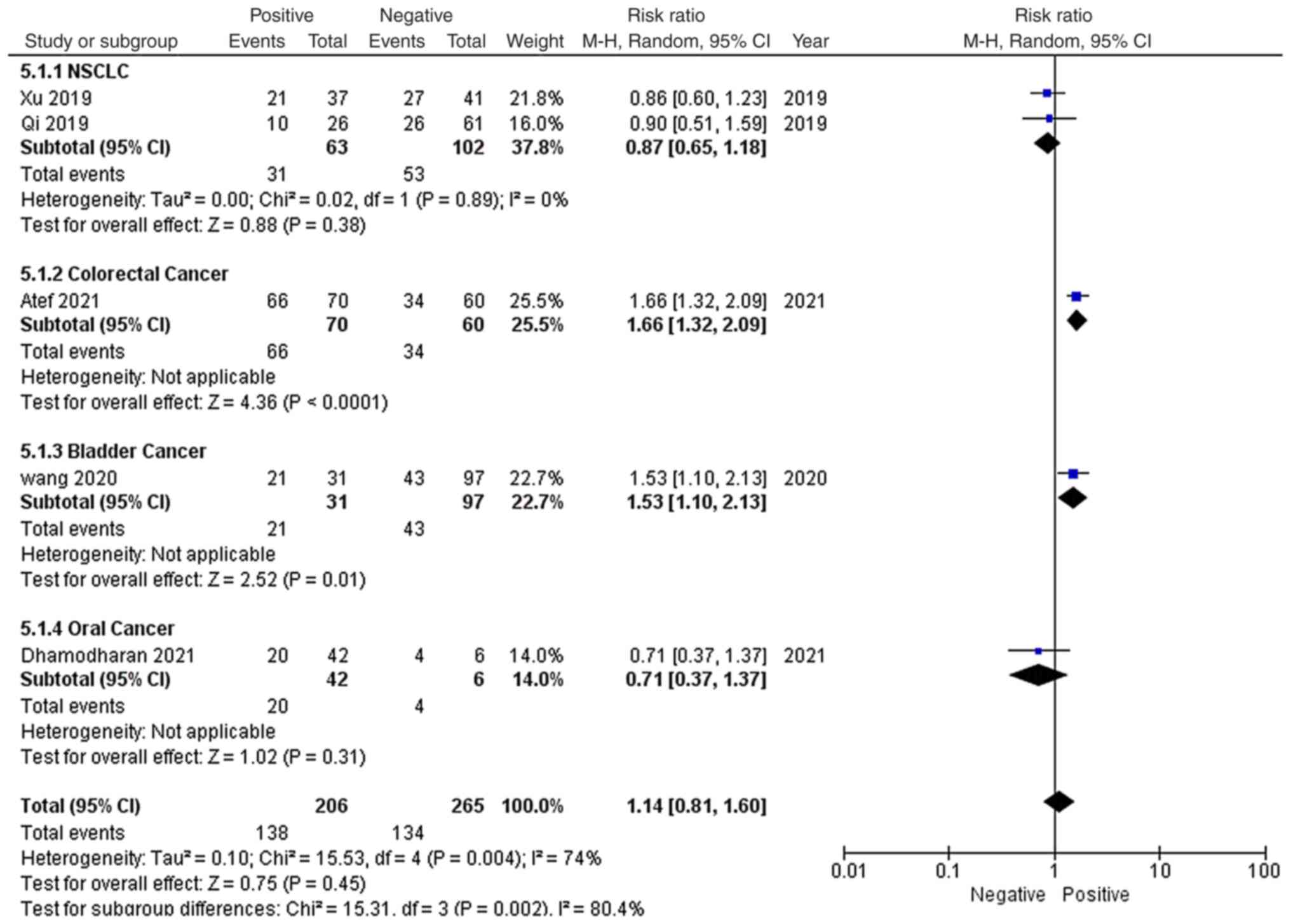
As shown in Fig. 6,
among the 8 studies, 5 reported the lymph node metastasis status of
patients, including 2 NSCLC studies, 1 colorectal cancer study, 1
bladder cancer study and 1 oral cancer study. Among the 471
patients, 138 patients with lymph node metastasis and 134 patients
without lymph node metastasis had high EGFR-AS1 expression. Due to
significant heterogeneity (P=0.004; I2=74%), a
random-effects model was applied to evaluate the pooled RR and 95%
CI of lymph node metastasis and high EGFR-AS1 expression. The
pooled RR of high EGFR-AS1 expression among the different lymph
node metastasis statuses was 1.14 with a 95% CI of 0.81–1.60, which
indicated that high EGFR-AS1 expression was not associated with
lymph node metastasis for these patients with cancer.
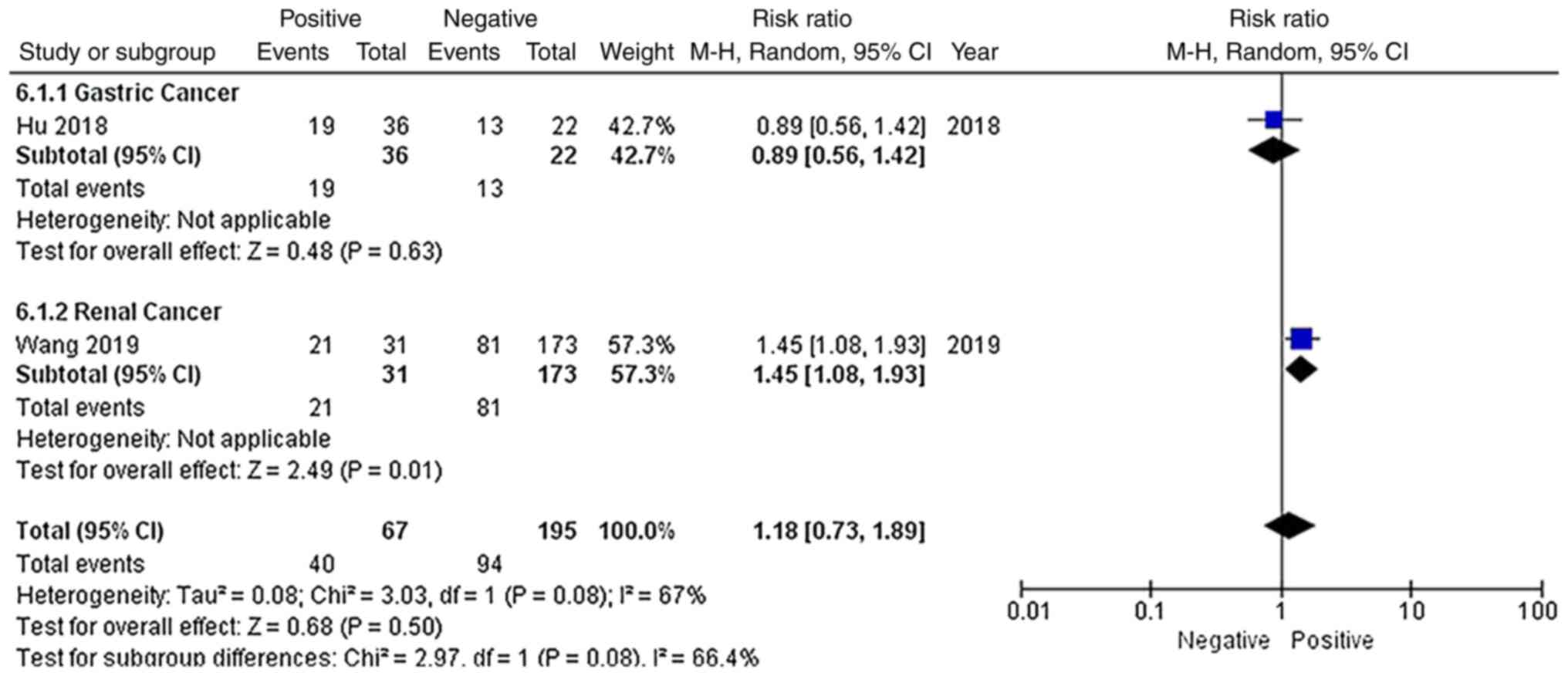
As shown in Fig. 7,
only 2 studies reported the distant metastatic status of patients,
including 1 gastric cancer study and 1 renal cancer study. Among
the 262 patients, 40 patients with distant metastasis and 94
patients without distant metastasis had high EGFR-AS1 expression.
Due to significant heterogeneity (P=0.08; I2=67%), a
random-effects model was applied to evaluate the pooled RR and 95%
CI of distant metastasis and EGFR-AS1 expression. The pooled RR of
high EGFR-AS1 expression among the different distant metastatic
statuses was 1.18 with a 95% CI of 0.73–1.89, which indicated that
high EGFR-AS1 expression was not associated with distant metastasis
for these patients with cancer.
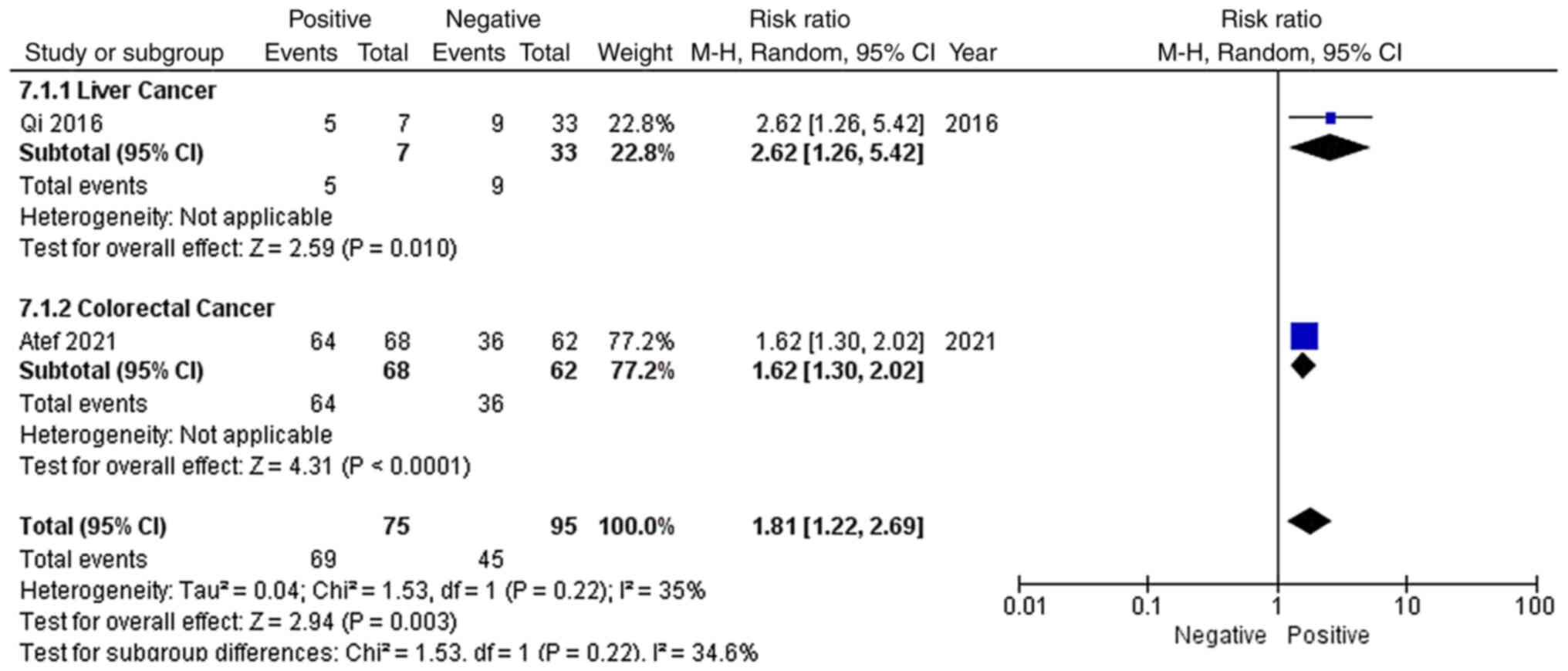
As shown in Fig. 8,
only 2 studies reported the vascular invasion status of the
included patients, including 1 liver cancer study and 1 colorectal
cancer study. Among the 170 patients, 69 patients with vascular
invasion and 45 patients without vascular invasion had high EGFR-AS
1 expression. Due to low heterogeneity (P=0.22; I2=35%)
and the recommendations provided by the Cochrane Handbook, a
random-effects model was applied to evaluate the pooled RR and 95%
CI of vascular invasion and EGFR-AS 1 expression. The pooled RR of
high EGFR-AS1 expression among the different vascular statuses was
1.81 with a 95% CI of 1.22–2.69 (P=0.003), which indicated that
high EGFR-AS1 expression was associated with vascular invasion.
As shown in Fig. 9,
4 out of 8 studies reported the TNM stages of the patients,
including 1 gastric cancer study, 1 renal cancer study and 2 NSCLC
studies. Among the 427 patients, 127 patients with stages ≤II and
91 patients with stages >II had high EGFR-AS1 expression. Due to
low heterogeneity (P=0.23; I2=30%) and the
recommendations provided by the Cochrane Handbook, a random-effects
model was applied to evaluate the pooled RR and 95% CI of TNM stage
and EGFR-AS1 expression. The pooled RR of high EGFR-AS1 expression
among different TNM statuses was 0.78 with a 95% CI of 0.60–1.01,
which indicated that high EGFR-AS1 expression was not associated
with TNM stage in these patients.
Association between the EGFR-AS1
expression level and the prognosis of patients with cancer
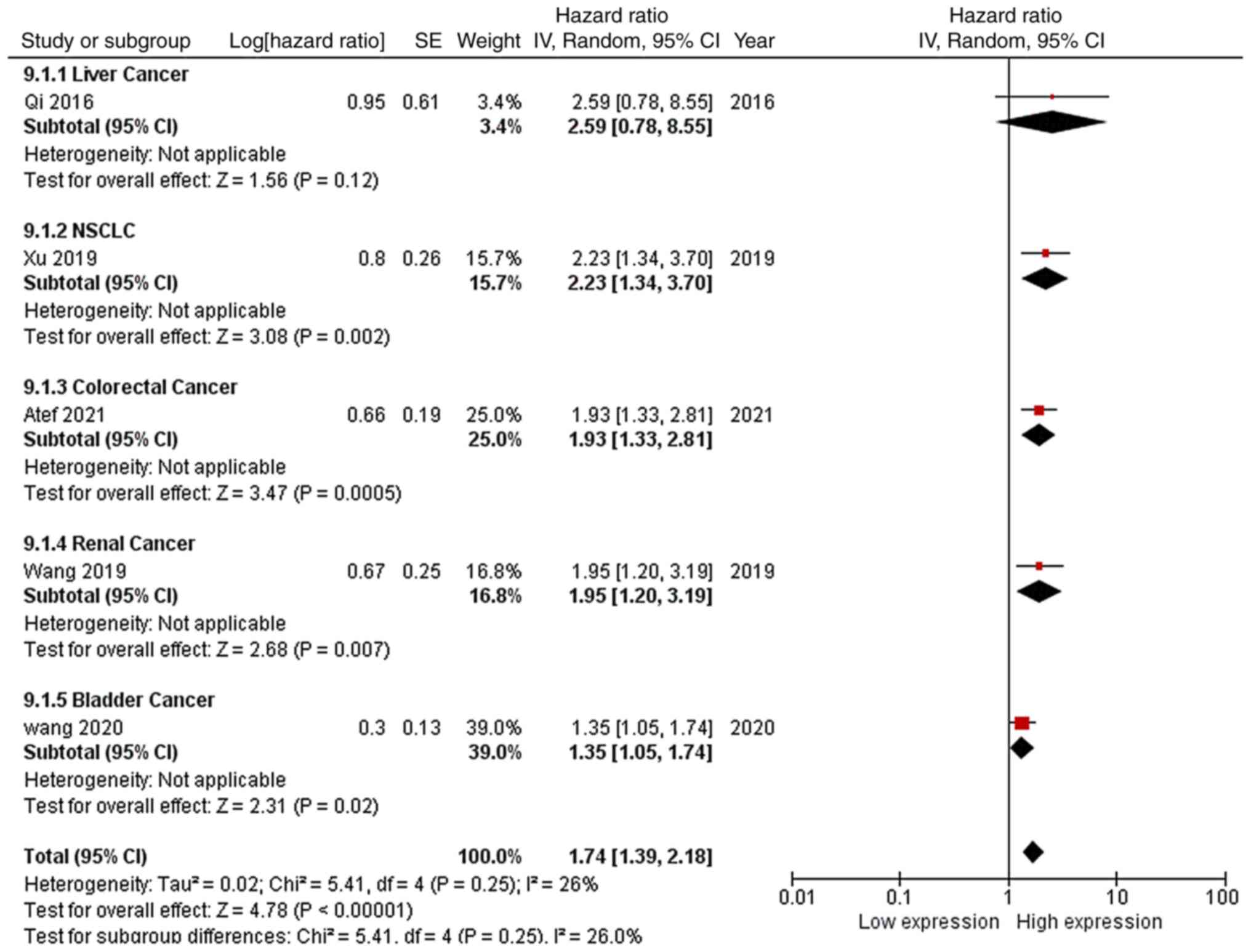
As shown in Fig.
10, 5 out of 8 studies reported the OS curves of the patients,
including 1 liver cancer study, 1 NSCLC study, 1 colorectal cancer
study, 1 renal cancer study and 1 bladder cancer study. Following
the acquisition of data from the respective OS curves, the
corresponding HRs and CIs of high EGFR-AS1 expression were
calculated. Due to low heterogeneity (P=0.25; I2=26%)
and the recommendations provided by the Cochrane Handbook, a
random-effects model was applied to evaluate the pooled HR and 95%
CI of OS and high EGFR-AS1 expression. The pooled HR of high
EGFR-AS1 expression for the OS of patients with cancer was 1.74
with a 95% CI of 1.39–2.18 (P<0.00001), which indicated that
high EGFR-AS1 expression was associated with a shorter OS time for
these patients.
Publication bias analysis
As shown in Figures
11 and 12, no apparent
publication bias was observed in the included studies for the
present meta-analysis. The funnel plot shown in Fig. 13 was symmetrical, indicating that
there was no notable publication bias.
Discussion
The results of the present study confirmed the
oncogenic role of EGFR-AS1 in several types of cancer, and high
EGFR-AS1 expression was demonstrated to impair the prognosis of
these patients with cancer. Although the research study reported by
Qi et al (9) demonstrated
that EGFR-AS1 levels were increased in patients with NSCLC who
smoked, the relationship between EGFR-AS1 expression and smoking
was not significant in this analysis. According to the pooled
results of the present study, high expression of EGFR-AS1 was
related to vascular invasion and poor prognosis in patients with
certain types of cancer. The meta-analysis outcomes were consistent
with the published results, indicating that EGFR-AS1 promoted
tumour cell growth and invasion in vivo and in vitro
(6,8). However, the role of EGFR-AS1 in the
TNM stage was equal to that in the tumour, node or metastasis
stages as no significant association was discovered between high
EGFR-AS1 expression and tumour size, lymph node metastasis or
distant metastasis. This result may be due to the lack of
sufficient studies or patients with cancer.
lncRNAs are widely spread throughout life, taking
part in various biological processes concerning glucose metabolism
and cancer (22). The biochemical
reaction processes are highly complicated and nearly any lncRNA
must combine with other molecules to employ its function. Due to
its genomic proximity to EGFR, EGFR-AS1 has the intrinsic ability
to regulate the expression of EGFR (6,10,13–14).
However, EGFR-AS1 also participates in other signalling pathways.
EGFR-AS1 has also been shown to regulate the expression of hypoxia
inducible factor 2A to increase NSCLC cancer stemness (9). Similar to lncRNAs, miRs are also a
type of non-coding RNA; moreover, miRs are typically related to
lncRNAs when regulating gene expression among cancer cells. It was
reported that EGFR-AS1 inhibited miR-381, leading to the
upregulation of Rho associated coiled-coil containing protein
kinase (ROCK)2 expression, to promote the invasiveness and
migration of bladder cancer cells (7). In oesophageal squamous cell carcinoma
cells, EGFR-AS1 increased the expression of ROCK1 by restraining
the activation of miR-145 to accelerate the progression of cancer
(8). In glioma cells, EGFR-AS1
restricted the activation of miR-133b to upregulate the expression
of RACK1, which promoted the proliferation and invasive ability of
cancer cells (12). In NSCLC,
EGFR-AS1 has been shown to impair autophagic lysosomal degradation
by downregulating the expression of miR-524-5p, leading to
significant cancer progression (23). EGFR-AS1 is also considered to
promote chemical resistance in NSCLC by directly binding to
miR-223, to increase the expression of insulin growth factor
receptor 1 (11).
To date, the oncogenic roles of EGFR-AS1 have been
fully identified in various patients with cancer. The aim of the
present meta-analysis was to merge the present studies to shed
light on the effects of EGFR-AS1 on patients with cancer. However,
certain limitations of the present study should be mentioned.
First, the studies regarding EGFR-AS1 consisted of several types of
solid tumours; however, insufficient studies have been reported for
individual tumour types to support the conclusions. Second, due to
the lack of original HR data presented for the published OS curves,
estimates were calculated using software, which may cause apparent
errors that would impair the reality of the final results. Lastly,
the cut-offs for high and low expression levels of EGFR-AS1 in each
study were not fully consistent. Only 4 included studies applied
relative expression as the cut-off, and the other 4 studies applied
median values or fixed values as the cut-off. There is therefore a
possibility that the patient was classified into the high
expression group due to the tumour EGFR-AS1 expression being above
the cut-off, even if the adjacent normal tissue exhibited higher
expression than that of the tumour tissue. This contradiction
resulted in the inability to use the median or fixed values as the
cut-off in the present study. Regardless of the aforementioned
limitations, to the best of our knowledge, the present analysis was
the first systematic review of the clinical roles of EGFR-AS1
expression in various human cancer types. However, numerous
large-scale, multicentre and high-quality prospective studies
should be conducted to comprehensively assess and sustain the
conclusions of the present study.
Acknowledgements
Not applicable.
Funding
This study was supported by ‘the Fundamental Research Funds for
the Central Universities’ (grant no. 2042021kf0103).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
HQ and GZ designed this research. LZ and JZ
processed the data. BW and HZ analyzed some of the data and wrote
the manuscript. LZ, JZ, HZ, GZ, BW, HQ confirm the authenticity of
the raw data. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nemeth K, Bayraktar R, Ferracin M and
Calin GA: Non-coding RNAs in disease: From mechanisms to
therapeutics. Nat Rev Genet. Nov 15–2023.doi:
10.1038/s41576-023-00662-1 (Epub ahead of print). PubMed/NCBI
|
|
2
|
Jin Y and Fan Z: New insights into the
interaction between m6A modification and lncRNA in cancer drug
resistance. Cell Prolif. e135782023.doi: 10.1111/cpr.13578 (Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qi HL, Li CS, Qian CW, Xiao YS, Yuan YF,
Liu QY and Liu ZS: The long noncoding RNA, EGFR-AS1, a target of
GHR, increases the expression of EGFR in hepatocellular carcinoma.
Tumour Biol. 37:1079–1089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Atef MM, Amer AI, Hafez YM, Elsebaey MA,
Saber SA and Abd ES: Long non-coding RNA EGFR-AS1 in colorectal
cancer: Potential role in tumorigenesis and survival via miRNA-133b
sponge and EGFR/STAT3 axis regulation. Br J Biomed Sci. 78:122–129.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dhamodharan S, Rose MM, Chakkarappan SR,
Umadharshini KV, Arulmurugan R, Subbiah S, Inoue I and Munirajan
AK: Genetic variant rs10251977 (G>A) in EGFR-AS1 modulates the
expression of EGFR isoforms A and D. Sci Rep. 11:88082021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang A, Jiang A, Gan X, Wang Z, Huang J,
Dong K, Liu B, Wang L and Chen M: EGFR-AS1 promotes bladder cancer
progression by upregulating EGFR. Biomed Res Int. 2020:66659742020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan S, Luan X, Chen H, Shi X and Zhang X:
Long non-coding RNA EGFR-AS1 sponges micorRNA-381 to upregulate
ROCK2 in bladder cancer. Oncol Lett. 19:1899–1905. 2020.PubMed/NCBI
|
|
8
|
Feng Z, Li X, Qiu M, Luo R, Lin J and Liu
B: LncRNA EGFR-AS1 upregulates ROCK1 by sponging miR-145 to promote
esophageal squamous cell carcinoma cell invasion and migration.
Cancer Biother Radiopharm. 35:66–71. 2020.PubMed/NCBI
|
|
9
|
Qi H, Wang S, Wu J, Yang S, Gray S, Ng C,
Du J, Underwood MJ, Li MY and Chen GG: EGFR-AS1/HIF2A regulates the
expression of FOXP3 to impact the cancer stemness of
smoking-related non-small cell lung cancer. Ther Adv Med Oncol.
11:4324970282019. View Article : Google Scholar
|
|
10
|
Wang A, Bao Y, Wu Z, Zhao T, Wang D, Shi
J, Liu B, Sun S, Yang F, Wang L and Qu L: Long noncoding RNA
EGFR-AS1 promotes cell growth and metastasis via affecting HuR
mediated mRNA stability of EGFR in renal cancer. Cell Death Dis.
10:1542019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu YH, Tu JR, Zhao TT, Xie SG and Tang SB:
Overexpression of lncRNA EGFR-AS1 is associated with a poor
prognosis and promotes chemotherapy resistance in non-small cell
lung cancer. Int J Oncol. 54:295–305. 2019.PubMed/NCBI
|
|
12
|
Dong ZQ, Guo ZY and Xie J: The lncRNA
EGFR-AS1 is linked to migration, invasion and apoptosis in glioma
cells by targeting miR-133b/RACK1. Biomed Pharmacother.
118:1092922019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu J, Qian Y, Peng L, Ma L, Qiu T, Liu Y,
Li X and Chen X: Long Noncoding RNA EGFR-AS1 Promotes Cell
Proliferation by Increasing EGFR mRNA stability in gastric cancer.
Cell Physiol Biochem. 49:322–334. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan D, Chong FT, Leong HS, Toh SY, Lau DP,
Kwang XL, Zhang X, Sundaram GM, Tan GS, Chang MM, et al: Long
noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor
addiction and modulates treatment response in squamous cell
carcinoma. Nat Med. 23:1167–1175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu D, Ouyang X, Zhang Y, Yu X, Su K and
Li L: A promising new cancer marker: Long noncoding RNA EGFR-AS1.
Front Oncol. 13:11304722023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J and Wang H: H3K27ac-activated
EGFR-AS1 promotes cell growth in cervical cancer through
ACTN4-mediated WNT pathway. Biol Direct. 17:32022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahn S, Pinheiro PS, Mcclure LA, Hernandez
DR, Caban-Martinez AJ and Lee DJ: An examination of psychometric
properties of study quality assessment scales in meta-analysis:
Rasch measurement model applied to the firefighter cancer
literature. PLoS One. 18:e2844692023. View Article : Google Scholar
|
|
18
|
Wang W, Yuan F and Xu J: The prognostic
role of long noncoding RNA CRNDE in cancer patients: A systematic
review and meta-analysis. Neoplasma. 66:73–82. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Oliveira IR, Santos-Jesus R, Po AL and
Poolsup N: Extracting numerical data from published reports of
pharmacokinetics investigations: Method description and validation.
Fundam Clin Pharmacol. 17:471–472. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qi HL, Zhuang BJ, Li CS and Liu QY:
Peri-operative use of sorafenib in liver transplantation: A
time-to-event meta-analysis. World J Gastroenterol. 21:1636–1640.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tierney JF, Stewart LA, Ghersi D, Burdett
S and Sydes MR: Practical methods for incorporating summary
time-to-event data into meta-analysis. Trials. 8:162007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hou XR, Zhang ZD, Cao XL and Wang XP: Long
noncoding RNAs, glucose metabolism and cancer (Review). Oncol Lett.
26:3402023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue Y, Zhang J, Hou J and Wang X: EGFR-AS1
promotes nonsmall cell lung cancer (NSCLC) progression via
downregulating the miR-524-5p/DRAM1 axis and inhibiting autophagic
lysosomal degradation. J Oncol. 2022:44025362022. View Article : Google Scholar : PubMed/NCBI
|