Introduction
Non-small-cell lung cancer (NSCLC) is the leading
cause of cancer-associated mortality in China and worldwide
(1–3). Although significant improvements have
been achieved due to the development of targeted therapies, the
5-year survival rate remains low (4), particularly in elderly patients with
refractory/relapsed (R/R) disease who are often unsuitable for most
conventional treatments, including surgery and chemotherapy
(5). Immunotherapy with programmed
death protein 1 (PD-1)/programmed death ligand 1 (PD-L1) blocking
antibodies has shown significant promise in treating patients with
R/R diseases (6,7). However, this benefit was only found in
a small subset of the patient population (8), thus highlighting the need for
alternative approaches to improve outcomes. The availability of
multi-form diagnostic platforms can be used to provide additional
information to improve decision-making regarding treatments, with
the aim of leading to improved remission.
Combination therapies, including those based on PD-1
blockade, have significantly improved treatment outcomes and
response rates in patients with cancer (9,10). It
has been reported that PD-1 blockade can be potentiated by
cytokine-induced killer (CIK) cell infusion (11–16),
and a combination of these two types of treatments has been trialed
in advanced NSCLC where it has demonstrated improved outcomes
(16–18). CIK cells are a group of
heterogeneous and major histocompatibility complex-unrestricted
cells with a mixed T/natural killer (NK) phenotype (19,20).
Compared with CAR-T cell therapy, which is only effective in
hematological malignancies, CIK therapy can be effective in the
treatment of both hematological and solid malignancies with low
toxicity (16–18,21–23).
In the present study, a novel, patient-tailored
approach was adopted to treat an elderly patient with relapsed
metastatic NSCLC (mNSCLC), who was ineligible for targeted therapy
(due to no suitable targeted drugs) and conventional chemotherapy
(due to their age and poor physical condition). Specifically,
multi-omics analysis for diagnosis, disease monitoring and guiding
a dynamic treatment regime, including the final utilization of
anti-PD-1/CIK cell combination therapy, was utilized for benefits
in both disease control and health improvement. The present case
describes an example of the integration of modern multi-omics
technologies for better therapeutic approaches and clinical
benefits in elderly patients with relapsed mNSCLC.
Case report
An 86-year-old female patient was admitted in May
2019 to the Elderly Respiratory Department, Henan Provincial
People's Hospital (Zhengzhou, China) due to coughing up bloody
sputum for ~2 weeks. They were then subjected to a set of
multi-omics analyses, including next-generation sequencing
(NGS)-based actionable gene panel sequencing, immunohistochemistry
(IHC) for PD-L1, circulating tumor cell (CTC) assay and flow
cytometric analysis of lymphocyte subsets (immune status) (Fig. 1). Conventional diagnostic methods
for cancer, including pathological analysis of tumor biopsy and
chest computed tomography (CT) were performed. In addition, routine
examinations, including blood tests, urine tests, tumor biomarker
detection, liver and kidney function tests, and an
electrocardiogram, were performed. Based on preliminary diagnostic
results, the patient was diagnosed with a relapsed NSCLC at stage
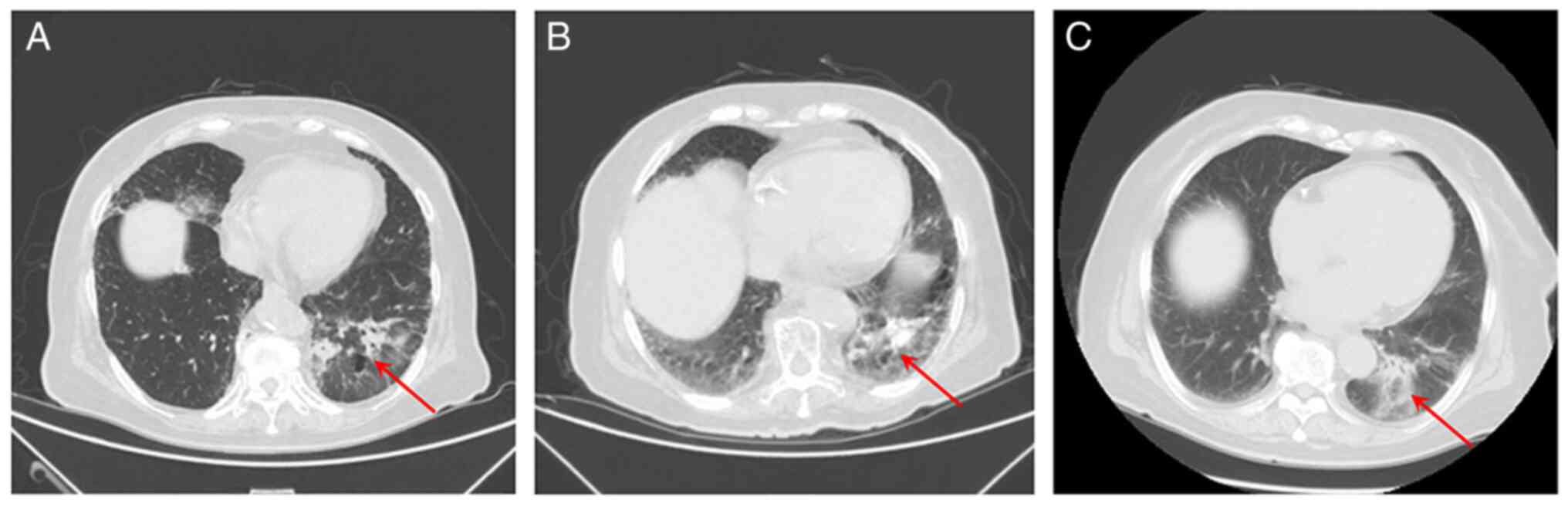
IV with multiple metastases. Using CT imaging, a tumor was observed
near the pulmonary hilum in the lower lobe of the left lung
(Fig. 2). In addition, multiple
nodules in both lungs spreading into the right anterior chest and
bilateral abdominal walls, thickened left adrenal gland, as well as
the destruction of multiple bones in C3 and L4 vertebrae and the
right sacral wing were observed. Increased plasma levels of tumor
markers cancer antigen (CA)199 (63.39 U/ml; reference value 0–30
U/ml) and CA153 (34.12 U/ml; reference value 0–24 U/ml) were
observed (Table I). Additionally,
15 CTCs (3 mesenchymal types and 12 hybrid types) were identified
in peripheral blood (Fig. 3A-C);
hematoxylin and eosin (H&E) staining showed both normal and
tumor cells (Fig. 3D); IHC revealed
a high content of cancer cells (80%) expressing PD-L1 (anti-PD-L1
antibody; monoclonal 22C3; Dako; Agilent Technologies, Inc.)
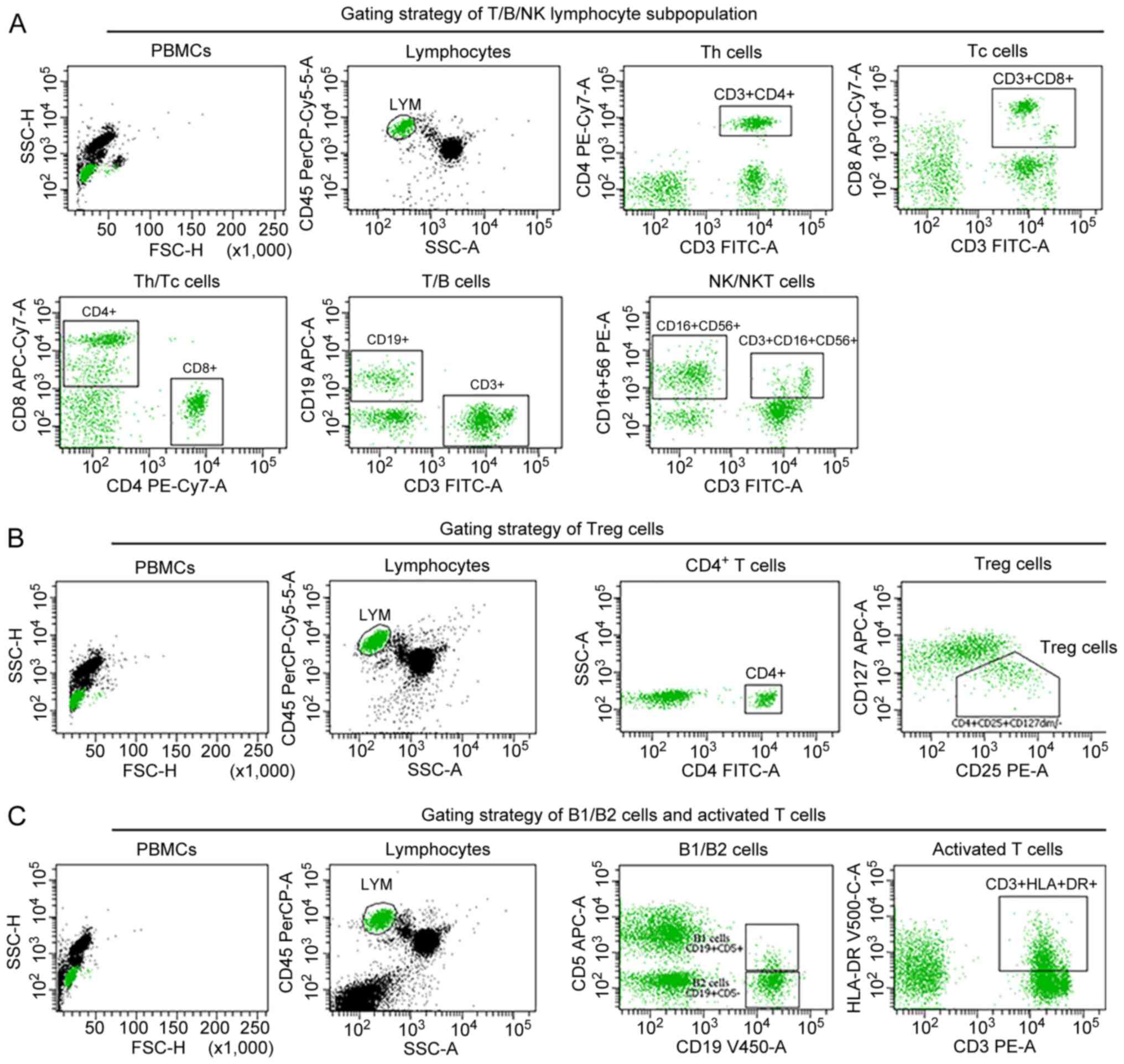
(Fig. 3E). Moreover, flow
cytometric analysis (Fig. 4)
suggested T-cell immunodeficiency with decreased counts for
lymphocytes (CD45+), total T cells (CD3+), T
helper cells (CD3+CD4+), a decreased
CD4+/CD8+ T-cell ratio, dysregulation of
regulatory T (Treg) cells
(CD45+CD4+CD25+CD127dim/−),
and an increased number of activated T cells and NK cells (Table II, month 0). While NGS-based
actionable gene sequencing (Burning Rock Biotech, Ltd.) detected no
targetable mutations, with the exception of a KRAS mutation
(c.35G>A, p.G12D). The patient also showed signs of poor health,
including multiple underlying diseases (cerebral infarction,
hypertension grade 3, hyperlipidemia, paroxysmal supraventricular
tachycardia and age-related dementia with brain atrophy) and
general conditions of ill health (pain, fatigue, malaise, a loss of
appetite and poor mental state).
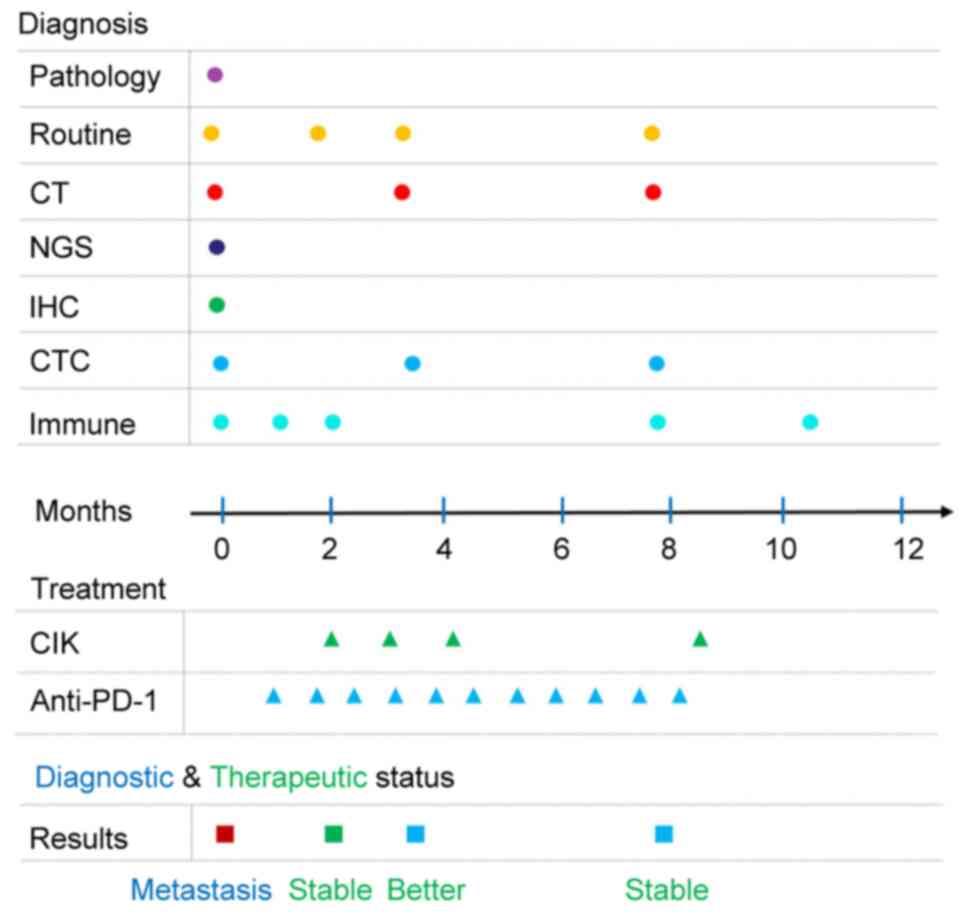 | Figure 1.Time chart of the diagnosis,
treatment and therapeutic outcomes of the patient during the entire
process of disease management. Routine refers to routine
inspection, including blood tests, urine tests, tumor biomarker
detection, liver and kidney function tests, and an
electrocardiogram. Immune refers to immune index detection via flow
cytometric analysis of lymphocyte subsets. CT, computed tomography;
IHC, immunohistochemistry; CTC, circulating tumor cell; CIK,
cytokine-induced killer; NGS, next generation sequencing; PD-1,
programmed death protein 1. |
 | Table I.Levels of blood cancer markers during
the treatment process. |
Table I.
Levels of blood cancer markers during
the treatment process.
|
| Months after
hospitalization |
|---|
|
|
|
|---|
| Blood cancer
marker | 0 | 2 | 3 | 8 |
|---|
| CEA (0–5
ng/ml) | 2.89 | 2.54 | 1.27 | 3.09 |
| AFP (0–7
ng/ml) | 3.01 | 1.82 | 2.75 | 3.56 |
| CA125 (0–25
U/ml) | 20.96 | 20.01 | 19.15 | 14.21 |
| CA199 (0–30
U/ml) | 63.39a | 40.34a | 24.43 | 23.81 |
| CA153 (0–24
U/ml) | 34.12a | 29.78a | 17.23 | 19.28 |
 | Table II.Immune status monitored by flow
cytometric analysis of lymphocyte subsets during the treatment
process. |
Table II.
Immune status monitored by flow
cytometric analysis of lymphocyte subsets during the treatment
process.
|
| Months after
hospitalization |
|---|
|
|
|
|---|
| Lymphocyte
subset | 0 | 1 | 2 | 8 | 10 |
|---|
| Lymphocyte
(CD45+) (20–50%) | 16.50%a | 16.40%a | 21.50%b | 35.60%b | 33.30%b |
| T cell
(CD3+) (55.62–84.84%) | 54.60%a | 54.80%a | 59.10%b | 39.90%a | 57.40%b |
| B cell
(CD19+) (6.58–24.52%) | 12.90%b | 5.90%a | 8.50%b | 9.30%b | 19.10%b |
| Th cell
(CD3+CD4+) (31.07–60.03%) | 24.40%a | 26.80%a | 34.40%b | 14.30%a | 35.90%b |
| Tc cell
(CD3+CD8+) (13.27–40.63%) | 25.10%b | 23.00%b | 21.60%b | 23.60%b | 19.90%b |
| NK cell
(CD3−/CD16+CD56+)
(5.15–27.08%) | 30.80%c | 36.30%c | 30.10%c | 48.30%c | 20.70%b |
| Treg cell
(CD45+CD4+CD25+CD127dim/−)
(5–10%) | 12.30%a | 17.20%a | 18.70%a | 26.20%a | 25.90%a |
| Activated T cell
(CD3+HLA-DR+) (0.37–13.98%) | 14.70%c | 8.20%b | 17.00%c | 5.00%b | 3.70%b |
| NKT cell
(CD3+/CD16+CD56+) (5.3–8.1%) | 6.90%b | 8.90%c | 5.40%b | 4.80%a | 3.30%a |
| CD4/CD8 (ratio
1.4–2.47) | 0.97a | 1.16a | 1.59b | 0.61a | 1.80b |
| B1
CD19+CD5+ (0–1.44%) | 0.90% b | 0.10% b | 0.50% b | - | 0.80% b |
| B2
CD19+CD5 (4.74–16.74%) | 13.30%b | 5.00%b | 7.70%b | - | 10.00%b |
Informed by integrative results from these
multi-omics analyses and the health condition of the patient, a
patient-tailored treatment plan using PD-1 blockade immunotherapy
(sintilimab; Innovent Biologics, Inc.; 200 mg each treatment every
3–4 weeks) was implemented, since PD-1 inhibitors have shown
efficacy in treating patients with NSCLC (24,25).
The response to treatment was also monitored periodically by
examinations including routine follow-up examinations for cancer,
CT, CTC analysis and flow cytometric analysis of immune status. The
curative effect, including complete response (CR), partial response
(PR), stable disease (SD) and progressive disease, was evaluated
according to the Response Evaluation Criteria in Solid Tumors
(version 1.1) guidelines (26). The
adverse reactions were judged according to the World Health
Organization grading standard (27)
of acute and subacute adverse reactions of anticancer drugs, such
as myelosuppression, digestive tract reaction, hypersensitivity,
fever, neuropathy and phlebitis. After two courses of sintilimab
treatment, SD was achieved; however, the symptoms, including
cancer-associated pain, fatigue, malaise, a loss of appetite and
poor mental state were not significantly improved. CIK cells have
been suggested to improve the quality of life and enhance the
effect of anti-PD-1 immunotherapy (28). Therefore, CIK cell infusions
(1–2×109 cells/day for 3 consecutive days in each
session, once per month and ≥3 days away from the treatment of PD-1
blockade) were added in combination with sintilimab to the
subsequent courses of treatment. After a total of four courses of
combined PD-1/CIK therapy, remission without severe adverse events
in the cancer and general condition of the patient was eventually
achieved, including alleviation of pain, decreased malaise, and
improvements in food intake and mental state. The diagnostic status
of the patient significantly improved, including a CTC count of 0,
normal serum levels of tumor biomarkers (Table I) and a significantly improved
status for T-cell immunodeficiency (Table II, month 10). The patient did not
require further treatment and continued to display an improved
condition for the next 12 months until the end of the follow-up.
These results demonstrated that the combination of CIK cell therapy
with PD-1 blockade therapy significantly improved therapeutic
efficacy, and multi-omics analysis for initial diagnosis and
follow-up disease monitoring can be used to inform the selection of
the appropriate regimen for an improved outcome. The treatment and
testing procedures for this elderly patient with mNSCLC are
summarized in Fig. 5.
The methods of the aforementioned multi-omics
analysis were performed as follows. First, NGS was performed as
described previously (29). This
method employed targeted capture of exonic and partially intronic
regions of eight genes that are recommended by the National
Comprehensive Cancer Network guidelines and are highly relevant to
NSCLC personalized treatment regimens (30). The eight-gene panel covered
oncogenic driver mutations of EGFR, ALK, BRAF, MET, RET, ROS1,
ERBB2 and KRAS (now upgraded to a nine-gene panel
Langke® CDx; NMPA certificate# 20223400343; Burning Rock
Biotech, Ltd.). Tissue DNA was extracted from formalin-fixed,
paraffin-embedded (FFPE) tumor tissue using a QIAamp DNA FFPE
tissue kit (Qiagen GmbH) according to the manufacturer's
instructions. DNA quantification was performed using a Qubit
fluorescence quantitative analyzer (Invitrogen; Thermo Fisher
Scientific, Inc.). Tissue DNA fragments between 200 and 400 bp were
purified (Agencourt AMPure XP Kit; cat. no. A63881; Beckman
Coulter, Inc.), hybridized with capture probe baits, selected with
magnetic beads and amplified. Target capture was performed using a
commercial panel consisting of 8 lung cancer-related genes (Burning
Rock Biotech, Ltd.). The loading concentration of the final library
was 1.6 pM for DNA sequencing. The quality and the size of the
fragments were assessed using a high sensitivity DNA kit and
Bioanalyzer 2100 (Agilent Technologies, Inc.). Indexed samples were
sequenced on the Nextseq 550 sequencing system (Illumina, Inc.)
using the NextSeq 500/550 kit (300 cycles; cat. no. 20024908;
Illumina, Inc.) with paired-end reads and average sequencing depth
of 1,000 × for tissue samples. Adapters were trimmed from the
sequence data with Trimmomatic v0.39 (http://www.usadellab.org/cms/index.php?page=trimmomatic)
and were assessed with FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
to ensure the ‘per base sequence quality’ of the reads was >30
(Phred quality score). Subsequently, sequence reads were mapped to
the human genome (hg19) using the BWA aligner 0.7.10 (http://maq.sourceforge.net). Picard (http://broadinstitute.github.io/picard/)
was utilized to mark and remove duplicates in the resultant
alignment file. Local alignment optimization and variant calling
were performed using Genome Analysis ToolKit 3.2 (http://www.broadinstitute.org/gsa/wiki/index.php/TheGenome_Analysis_Toolkit)
and VarScan (http://varscan.sourceforge.net). Variants were
filtered using the VarScan fpfilter pipeline. At least five
supporting reads were needed for insertion-deletions, while eight
supporting reads were needed for single nucleotide variants to be
called. According to the ExAC in gnomAD (https://gnomad.broadinstitute.org/), 1000 Genomes
(http://www.1000genomes.org), dbSNP
(https://www.ncbi.nlm.nih.gov/SNP/)
and ESP6500SI–V2 (https://esp.gs.washington.edu/drupal/) databases,
variants with a population frequency >0.1% were grouped as
single nucleotide polymorphisms and excluded from further analysis.
The remaining variants were annotated with ANNOVAR (https://annovar.openbioinformatics.org/)
and SnpEff v3.6 (http://sourceforge.net/projects/snpeff/files/snpEff_latest_core.zip).
DNA translocation analysis was performed using both Tophat 2
(http://ccb.jhu.edu/software/tophat/index.shtml) and
Factera (http://factera.stanford.edu), and
copy number variants were analyzed with CNVkit (https://cnvkit.readthedocs.io/en/stable/) (29).
Second, the characterization of CTCs was performed
using the CanPatrol® System and Tricolor RNA-ISH method
(certificate# 20142221528; Yishan Biotechnology Co., Ltd.) as
described previously (31).
Briefly, 5 ml peripheral blood was immediately collected and
transferred to EDTA-coated tubes. To remove red blood cells, a
lysis buffer (MilliporeSigma) was added. After 30 min,
centrifugation (600 × g for 5 min at room temperature) was
performed and the supernatant was removed. The remaining cells were
further separated using a CanPatrol® CTC enrichment
technique (Yishan Biotechnology Co., Ltd.). For the identification
of different CTC subtypes, cells were incubated at 42°C for 2.5 h
(ready to use according to the manufacturer's instructions) with
the following probes (Yishan Biotechnology Co., Ltd.): Alexa Fluor
594-conjugated epithelial biomarkers epithelial cellular adhesion
molecule (EpCAM; cat. no. Su-KC0824), cytokeratin (CK)8 (cat. no.
Su-KC0924), CK18 (cat. no. Su-KC1024) and CK19 (cat. no.
Su-KC1124); Alexa Fluor 488-conjugated mesenchymal biomarkers
Vimentin (cat. no. Su-KC1224) and Twist (cat. no. Su-KC1324); and
Alexa Fluor 647-conjugated leukocyte biomarker CD45 (cat. no.
Su-KC0724); and the nuclei were stained with DAPI (MilliporeSigma).
After staining with DAPI (MilliporeSigma) for 30 min at 4°C, the
cells were washed with 2% goat serum PBS (MilliporeSigma) solution
and images were captured at ×400 magnification using an Axio Imager
Z2 fluorescence microscope (Carl Zeiss AG).
Third, lymphocyte subset measurements using flow
cytometry were performed as described in detail previously
(32). Briefly, peripheral blood
mononuclear cells (PBMCs) were isolated by Ficoll 400 density
gradient liquid (Cytiva) centrifugation for 30 min at 600 × g and
room temperature, blocked for 15 min at 4°C with FcR (BD
Pharmingen; BD Biosciences) and incubated with BD Horizon™ Fixable
Viability Stain 575V (cat. no. 565694; BD Horizon; BD Biosciences)
for 15 min at room temperature. Cells were then washed twice with
fluorescence-activated cell sorter (FACS) buffer (BD Pharmingen; BD
Biosciences). For surface marker staining, BD Multitest™ 6-Color
TBNK (cat. no. 662967; BD Pharmingen; BD Biosciences), including
anti-human CD45-PerCP-Cy5.5, CD3-FITC, CD4-PE-Cy7, CD8-APC-Cy7,
CD19-APC and CD16+CD56-PE antibodies for the T/B/NK lymphocyte
subset; CD45-PerCP-Cy5.5 (cat. no. 340952; BD Pharmingen; BD
Biosciences), CD4-FITC (cat. no. 340133; BD Pharmingen; BD
Biosciences), CD25-PE (cat. no. 341010; BD Pharmingen; BD
Biosciences) and CD127-APC (cat. no. Z6410052; Beijing Tongsheng
Shidai Biotech Co., Ltd.) antibodies for Treg cells; CD45-PerCP
(cat. no. 340665; BD Pharmingen; BD Biosciences), CD3-PE (cat. no.
340662; BD Pharmingen; BD Biosciences), CD5-APC (cat. no. 340658;
BD Pharmingen; BD Biosciences), CD19-V450 (cat. no. 644492; BD
Horizon; BD Biosciences) and HLA-DR-V500 (cat. no. 561224; BD
Horizon; BD Biosciences) antibodies for B1/B2 cells and activated T
cells were added to cells at the recommended doses (ready to use),
and incubated for 20 min at room temperature. The cells were then
washed twice with FACS buffer. Cell analysis was performed using a
FACS machine (BD FACSCantoII; BD Biosciences) and cell populations
were analyzed using BD FACSDiva software v8.0.1 (BD Biosciences;
Fig. 4).
Finally, IHC was performed as described previously
(33). Briefly, the NSCLC tissue
(collected May 2019) was fixed in 10% neutral fixative at room
temperature for 3–4 h. An appropriately sized tissue section was
placed into an embedding box for dehydration. The dehydrated tissue
was then embedded in paraffin and sectioned with a microtome into
3-µm slices, which were subjected to immunohistochemical staining
using the PD-L1 IHC 22C3 pharmDx kit (cat. no. SK006; Dako; Agilent
Technologies, Inc.) on the Dako Autostainer Link 48 platform (Dako;
Agilent Technologies, Inc.). The paraffin-embedded sections were
dewaxed and hydrated, and antigen retrieval was performed for 1 h
at room temperature using the EnVision™ FLEX Target Retrieval
Solution (Dako; Agilent Technologies, Inc.). After incubation with
FLEX peroxidase non-specific binding blocking reagent (Dako;
Agilent Technologies, Inc.) for 5 min at room temperature, the
tissue was incubated with an anti-PD-L1 primary antibody (1:50;
cat. no. M365329; Dako; Agilent Technologies, Inc.) for 60 min at
room temperature, followed by incubation with the EnVision FLEX HRP
visualization reagent (cat. no. SM802; ready to use; Dako; Agilent
Technologies, Inc.) at room temperature for 30 min and color
development using DAB chromogenic solution at room temperature for
10 min. The tissue was then counterstained with hematoxylin at room
temperature for 2 min and a coverslip was added. All of the
aforementioned steps were followed by washes in EnVision FLEX Wash
Buffer 20X (1:20; Dako; Agilent Technologies, Inc.) for 5 min at
room temperature.
H&E staining was also conducted on
deparaffinized and rehydrated sections. The tissue was incubated
with hematoxylin aqueous solution (Baso Diagnostic, Inc.) for 5 min
at room temperature, differentiated in hydrochloric acid-ethanol
differentiation solution, rinsed and then incubated with eosin
staining solution (Baso Diagnostic, Inc.) for 3 min at room
temperature. After dehydration with alcohol, clearing with xylene
and sealing with a neutral resin, sections that underwent IHC and
H&E staining were examined under a light microscope (×200
magnification).
Discussion
With advances in medicine, novel therapies,
including targeted therapy, immunotherapy, CAR-T cell therapy and
vaccines, have been developed for various types of cancer in the
last few decades (34). This
progress requires an improvement in the development of diagnostic
methodologies that can be used to inform therapy (35). Notably, it is difficult to treat
elderly patients with R/R mNSCLC, especially when no targeted
therapies are available due to a lack of targeted mutations
(5). In the present case report, it
was demonstrated that an elderly patient benefited from the
integrative utilization of multi-omics analysis and dynamic
treatment regimens. Based on the diagnostic results, PD-1 blockade
alone was initially used to treat the patient, since it is a
well-established treatment option for patients with NSCLC,
particularly those with high levels of PD-L1 expression. At first,
autologous CIK cell infusion was not performed, as it is expensive
and requires time to manufacture. However, as informed by
periodical monitoring of the response to treatment, a combination
therapy of PD-1 blockade and infusion of autologous CIK cells was
finally adopted, and the addition of CIK cells resulted in a
significant improvement not only in the management of the patient's
cancer but also in their general health. Notably, anti-PD-1/CIK
cell combination therapy also induced significant clinical benefits
in three other published studies, including 3 patients with CR, 3
patients with PR and 5 patients with SD, out of a total of 15
patients with NSCLC (16–18).
The precise mechanisms underlying PD-1/CIK
combination therapy remain to be defined. Accumulating evidence has
demonstrated that blocking the PD-1/PD-L1 pathway is an effective
treatment option for multiple types of cancer, likely through the
restoration of tumor-specific immune activity, since tumor cells
may overexpress PD-L1 to co-opt the PD-1 pathway and to enable
evasion of the immune response (36). It has been shown that the tumor
regression achieved by anti-PD-1/PD-L1 drugs requires pre-existing
T cells within the tumor microenvironment, which have been
negatively regulated to confer tumor-mediated adaptive immune
resistance (37–39). Furthermore, evidence has suggested
that CIK cells, as a group of heterogeneous immune-active effector
cells, with the dual properties of both cytotoxic T lymphocytes and
NK cells (19), can promote the
efficacy of checkpoint inhibition involving PD-1 or other immune
checkpoints (12). Conversely,
in vitro studies have revealed that PD-1 blockade can also
directly enhance the cytotoxicity of CIK cells (13–15).
Therefore, it is likely that PD-1 blockade and CIK cell application
can potentiate each other to synergistically enhance the
therapeutic effect.
In the present case report, an example of the use of
multi-omics analysis in assisting the selection of an optimal
treatment plan is described. In this case, through integrating the
results of multi-omics analysis and the continued monitoring of the
response to treatment, dynamic therapeutic strategies were
employed, including the use of PD-1 blockade and CIK cell infusion
combination therapy, leading to significantly improved outcomes.
Therefore, the present study is informative as it is the first, to
the best of our knowledge, to describe an effective treatment
strategy for elderly patients who are usually unsuitable for most
treatment options. In addition, the present study demonstrated the
importance and benefit of a multi-omics approach for successful
cancer management. However, the present study has some limitations;
in particular, only one patient was reported on in the present case
report. Therefore, further studies should be performed to validate
this finding and to uncover the underlying mechanisms.
In conclusion, the present case report demonstrated
that a multi-omics analysis approach can inform patient-tailored
therapy to improve clinical outcomes in a hard-to-treat elderly
patient with mNSCLC physically unsuitable for surgery and most
types of chemotherapy, and genetically unsuitable for targeted
therapy.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The NGS data generated in the present study may be
found in the NCBI SRA database under the accession number
SRR27406315 or at the following URL: https://www.ncbi.nlm.nih.gov/sra/?term=SRR27406315.
All of the other data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YX contributed to statistical analysis, wrote the
original draft and assessed study quality. FQ, LH, JY and HZ
contributed to indicator detection, data extraction and literature
searching. YQ contributed to the establishment of a clinical
treatment plan, evaluation of efficacy and data extraction. ST and
YZ contributed to statistical analysis, and reviewed and edited the
manuscript. YX and YZ confirm the authenticity of all the raw data.
All authors contributed to the article, and have read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
The patient provided written informed consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
PD-1
|
programmed death protein 1
|
|
PD-L1
|
programmed death ligand 1
|
|
NGS
|
next-generation sequencing
|
|
CTC
|
circulating tumor cell
|
|
CIK
|
cytokine-induced killer
|
|
IHC
|
immunohistochemistry
|
|
SD
|
stable disease
|
|
CR
|
complete response
|
|
PR
|
partial response
|
|
NK
|
natural killer
|
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Colombet M, Soerjomataram I,
Parkin DM, Piñeros M, Znaor A and Bray F: Cancer statistics for the
year 2020: An overview. Int J Cancer. 149:778–789. 2021. View Article : Google Scholar
|
|
3
|
Sosa E, D'Souza G, Akhtar A, Sur M, Love
K, Duffels J, Raz DJ, Kim JY, Sun V and Erhunmwunsee L: Racial and
socioeconomic disparities in lung cancer screening in the United
States: A systematic review. CA Cancer J Clin. 71:299–314. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Khanna P, Blais N, Gaudreau PO and
Corrales-Rodriguez L: Immunotherapy comes of age in lung cancer.
Clin Lung Cancer. 18:13–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spagnuolo A and Gridelli C: The role of
immunotherapy in the first-line treatment of elderly advanced
non-small cell lung cancer. Cancers (Basel). 15:23192023.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xia L, Liu Y and Wang Y: PD-1/PD-L1
blockade therapy in advanced non-small-cell lung cancer: Current
status and future directions. Oncologist. 24 (Suppl 1):S31–S41.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Momotow J, Bühnen I, Trautmann-Grill K,
Kobbe G, Hahn D, Schroers R, Heinrich B, Gaska T, Forstbauer H,
Schmidt B, et al: Outcomes of anti-programmed death 1 treatment for
relapsed/refractory Hodgkin lymphoma: A German Hodgkin Study Group
multicentre real-world analysis. Br J Haematol. 198:401–404. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pan D, Hu AY, Antonia SJ and Li CY: A Gene
mutation signature predicting immunotherapy benefits in patients
with NSCLC. J Thorac Oncol. 16:419–427. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoffner B, Leighl NB and Davies M:
Toxicity management with combination chemotherapy and programmed
death 1/programmed death ligand 1 inhibitor therapy in advanced
lung cancer. Cancer Treat Rev. 85:1019792020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yi M, Zheng X, Niu M, Zhu S, Ge H and Wu
K: Combination strategies with PD-1/PD-L1 blockade: Current
advances and future directions. Mol Cancer. 21:282022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang K, Sun B, Luo N, Guo H, Hu J and
Peng J: Programmed death receptor 1 (PD1) knockout and human
telomerase reverse transcriptase (hTERT) transduction can enhance
persistence and antitumor efficacy of cytokine-induced killer cells
against hepatocellular carcinoma. Med Sci Monit. 24:4573–4582.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dehno MN, Li Y, Weiher H and Schmidt-Wolf
IGH: Increase in efficacy of checkpoint inhibition by
cytokine-induced-killer cells as a combination immunotherapy for
renal cancer. Int J Mol Sci. 21:30782020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Chen Y, Feng F, Chen C, Zeng H,
Wen S, Xu X, He J and Li J: Programmed cell death
protein-1/programmed death-ligand 1 blockade enhances the antitumor
efficacy of adoptive cell therapy against non-small cell lung
cancer. J Thorac Dis. 10:6711–6721. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Poh SL and Linn YC: Immune checkpoint
inhibitors enhance cytotoxicity of cytokine-induced killer cells
against human myeloid leukaemic blasts. Cancer Immunol Immunother.
65:525–536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu LW, Yang MY, Zhou M, Li JJ, Liu B and
Pan YY: Improvement of cytotoxicity of autologous CIKs from
patients with breast cancer to MCF-7 cells by suppressed PD-1
expression. Cancer Biomark. 20:609–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han Y, Mu D, Liu T, Zhang H, Zhang J, Li
S, Wang R, Du W, Hui Z, Zhang X and Ren X: Autologous
cytokine-induced killer (CIK) cells enhance the clinical response
to PD-1 blocking antibodies in patients with advanced non-small
cell lung cancer: A preliminary study. Thorac Cancer. 12:145–152.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Liu X, Till B, Sun M, Li X and Gao
Q: Combination of cytokine-induced killer cells and programmed cell
death-1 blockade works synergistically to enhance therapeutic
efficacy in metastatic renal cell carcinoma and non-small cell lung
cancer. Front Immunol. 9:15132018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao L, Han L, Zhang Y, Li T, Yang Y, Li
W, Shang Y, Lin H and Gao Q: Combination of PD-1 blockade and
RetroNectin®-activated cytokine-induced killer in
preheavily treated non-small-cell lung cancer: A retrospective
study. Immunotherapy. 10:1315–1323. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pievani A, Borleri G, Pende D, Moretta L,
Rambaldi A, Golay J and Introna M: Dual-functional capability of
CD3+CD56+ CIK cells, a T-cell subset that acquires NK function and
retains TCR-mediated specific cytotoxicity. Blood. 118:3301–3310.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mata-Molanes JJ, Sureda González M,
Valenzuela Jiménez B, Martínez Navarro EM and Brugarolas Masllorens
A: Cancer immunotherapy with cytokine-induced killer cells. Target
Oncol. 12:289–299. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Huang H, Han L, Li T, Wang Z and
Gao Q: Advanced renal-cell carcinoma pseudoprogression after
combined immunotherapy: Case report and literature review. Front
Oncol. 11:6404472021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao L, Li T, Song Y, Yang Y, Ma B, Zhang
Y, Shang Y, Xu B, Guo J, Qin P, et al: High complete response rate
in patients with metastatic renal cell carcinoma receiving
autologous cytokine-induced killer cell therapy plus
anti-programmed death-1 agent: A single-center study. Front
Immunol. 12:7792482022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharma A and Schmidt-Wolf IGH: 30 Years of
CIK cell therapy: Recapitulating the key breakthroughs and future
perspective. J Exp Clin Cancer Res. 40:3882021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao S, Li N, Gao S, Xue Q, Ying J, Wang S,
Tao X, Zhao J, Mao Y, Wang B, et al: Neoadjuvant PD-1 inhibitor
(Sintilimab) in NSCLC. J Thorac Oncol. 15:816–826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang F, Guo W, Zhou B, Wang S, Li N, Qiu
B, Lv F, Zhao L, Li J, Shao K, et al: Three-year follow-up of
neoadjuvant programmed cell death protein-1 inhibitor (Sintilimab)
in NSCLC. J Thorac Oncol. 17:909–920. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Helling M and Venulet J: Drug recording
and classification by the WHO research centre for international
monitoring of adverse reactions to drugs. Methods Inf Med.
13:169–178. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vaseq R, Sharma A, Li Y and Schmidt-Wolf
IGH: Revising the landscape of cytokine-induced killer cell therapy
in lung cancer: Focus on immune checkpoint inhibitors. Int J Mol
Sci. 24:56262023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng Y, Feng G, Lu X, Qian W, Ye J,
Manrique CA, Ma C and Lu Y; written on behalf of the AME Lung
Cancer Collaborative Group, : Exploratory analysis of introducing
next-generation sequencing-based method to treatment-naive lung
cancer patients. J Thorac Dis. 10:5904–5912. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsoulos N, Papadopoulou E, Metaxa-Mariatou
V, Tsaousis G, Efstathiadou C, Tounta G, Skapeti K, Bourkoula J,
Zarogoulidis P, Pentheroudakis GE, et al: Molecular profiling of
502 patient cohort with NSCLC using a 27 somatic gene panel. J Clin
Oncol. 35:e231932017. View Article : Google Scholar
|
|
31
|
Xing Y, Qin F, Zhai Y, Yang J, Yan Y, Li
D, Zhang H, Hu R, Xu X, Cao X, et al: Association of clinical
features of colorectal cancer with circulating tumor cells and
systemic inflammatory markers. Dis Markers. 2022:51055992022.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xing Y, Zhang X, Qin F, Yang J, Ai L, Wang
Q and Zhai Y: The clinical significance of circulating tumor cells
and T lymphocyte subtypes in pancreatic cancer patients.
Bioengineered. 13:2130–2138. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang X, Jiang L, Jin Y, Li P, Hou Y, Yun
J, Wu C, Sun W, Fan X, Kuang D, et al: PD-L1 expression in chinese
patients with advanced non-small cell lung cancer (NSCLC): A
multi-center retrospective observational study. J Cancer.
24:7390–7398. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Poondla N, Sheykhhasan M, Akbari M, Samadi
P, Kalhor N and Manoochehri H: The promise of CAR T-cell therapy
for the treatment of cancer stem cells: A short review. Curr Stem
Cell Res Ther. 17:400–406. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang Q, Zuo W, Wan C, Xiong S, Xu C, Yuan
C, Sun Q, Zhou L and Li X: Comprehensive genomic profiling of upper
tract urothelial carcinoma and urothelial carcinoma of the bladder
identifies distinct molecular characterizations with potential
implications for targeted therapy & immunotherapy. Front
Immunol. 13:10977302023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weber J: Immune checkpoint proteins: A new
therapeutic paradigm for cancer-preclinical background: CTLA-4 and
PD-1 blockade. Semin Oncol. 37:430–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Powles T, Eder JP, Fine GD, Braiteh FS,
Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, et
al: MPDL3280A (anti-PD-L1) treatment leads to clinical activity in
metastatic bladder cancer. Nature. 515:558–562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Herbst RS, Soria JC, Kowanetz M, Fine GD,
Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger
SN, et al: Predictive correlates of response to the anti-PD-L1
antibody MPDL3280A in cancer patients. Nature. 515:563–567. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tumeh PC, Harview CL, Yearley JH, Shintaku
IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu
V, et al: PD-1 blockade induces responses by inhibiting adaptive
immune resistance. Nature. 515:568–571. 2014. View Article : Google Scholar : PubMed/NCBI
|