Introduction
The origin and mechanism of development of ovarian
tumors remains unclear. In recent years, a method to classify
ovarian cancer into two types, type I and II tumors, has been
proposed. Type I tumors comprise low-grade serous adenocarcinoma,
low-grade endometrioid adenocarcinoma, clear cell adenocarcinoma
and mucinous carcinomas and Brenner tumors. They are generally
indolent, present in stage I at diagnosis (tumor confined to the
ovary) and characterized by specific mutations, including mutations
of KRAS, BRAF and ERBB2, which target specific cell signaling
pathways. Type II tumors comprise high-grade serous adenocarcinoma,
high-grade endometrioid adenocarcinoma and malignant mixed
mesodermal tumors (carcinosarcomas) and undifferentiated
carcinomas. They are aggressive, present in an advanced stage at
diagnosis and have a very high frequency of TP53 mutations, but
rarely harbor the mutations detected in type I tumors (1).
In contrast to patients with high-grade disease,
low-grade serous ovarian cancer occurs at a younger age, typically
exhibits a low mitotic index and is largely resistant to
chemotherapy (2–5). Mutational analyses may provide further
insight into the development sequence of low-grade serous
carcinomas.
Ovarian serous borderline tumors are relatively rare
in Japan and are somewhat more difficult to diagnose. The
differentiation of serous borderline tumors from serous
adenocarcinoma rests on whether destructive stromal invasion is
present or not. According to reports, BRAF gene mutation is
detected in ~30–50% of serous borderline tumors, but in very few
serous adenocarcinomas. Furthermore, the reported rate of BRAF or
KRAS mutation in serous borderline tumors is 61% (6), while neither BRAF nor KRAS mutation is
found often in invasive high-grade serous adenocarcinomas (6–8). It
has recently been shown by whole exome sequencing that low-grade
serous ovarian tumors contain very few mutations other than those
involving BRAF and KRAS, indicating that MAPK activation is of
central importance in the pathogenesis of these neoplasms (9).
The advent of specific inhibitors of mutant BRAF
with clinical activity against malignant melanoma (10) and preclinical data indicating that
inhibition of the MAPK pathway in BRAF-mutated ovarian tumors may
yield clinical benefit, suggest that reliable and sensitive
detection of the V600E mutation may be of clinical relevance in
serous ovarian tumors (11,12). Therefore, an accurate and practical
assay is urgently needed to detect this molecular subset of ovarian
cancer. Currently, the methods available for detecting the mutation
are: polymerase chain reaction (PCR)-based assays such as Sanger
sequencing (Sas), pyrosequencing, and qPCR. PCR is a single
detection test, detecting gene mutations; however, it generally
requires multiple tissue sections, dissection for tumor cell
enrichment and a multiplex system. Competitive allele-specific
TaqMan PCR technology (CAST) for BRAF mutation detection is highly
specific and has a sensitivity of <1% of mutation rate (13). However, it may require a high
quality laboratory infrastructure with well-trained staff and it is
also expensive. Recently, it has been demonstrated that
mutation-specific antibodies can reliably detect the exchange of
even single amino acids in routinely processed, formalin-fixed and
paraffin-embedded (FFPE) tumor tissues (14,15).
To increase the sensitivity, we modified the BRAF V600E protein
expression detection system by immunohistochemistry (IHC) using
specific monoclonal antibody, VE1 and Dako EnVision™ FLEX, using a
linker in a Japanese cohort (16).
A good concordance of the results has been demonstrated between IHC
and direct sequencing (16,17). However, to date, there have been no
reported studies that included all three methods, or compared the
pros and cons of each. In this study, we matched the results of
Sas, IHC and the mutation rate of the BRAF gene detected by
CAST-PCR in ovarian serous borderline tumor cases.
Materials and methods
Patient samples
The study group included 11 patients with ovarian
serous borderline tumors who had undergone surgery and were
diagnosed as having serous borderline tumor by pathological
diagnosis at the Department of Obstetrics and Gynecology, Nagoya
City University Hospital, between February 1995 and August 2013.
Additionally, 20 randomly selected ovarian cancers, including 6
serous adenocarcinomas, 3 endometrioid adenocarcinomas, 3 clear
cell adenocarcinomas, 1 mucinous adenocarcinoma, 2 mucinous
borderline tumors, 1 granulosa cell tumor, 1 carcinosarcoma, 1
mixed germ-cell tumor, 1 undifferentiated carcinoma and 1
metastatic colorectal cancer were analyzed by the CAST-PCR BRAF
assay. This study was conducted with the approval of the Research
Ethics Committee of Nagoya City University Graduate School of
Medical Sciences.
VE-1 IHC was previously established in a cohort of
26 lung cancer cases (16), and
that study confirmed the presence of BRAF V600E in 5 cases. We used
these 5 cases with BRAF V600E and 30 cases of wild-type BRAF, as
determined by Sas, as the control samples for CAST-PCR.
Direct sequencing for detecting BRAF and
KRAS gene mutations
Genomic DNA was extracted from FFPE tumor tissue
using the Qiagen PCR purification kit (Qiagen, Tokyo, Japan)
according to the manufacturer’s instructions. DNA concentration was
determined with the NanoDrop ND-1000 Spectrophotometer (NanoDrop
Technologies Inc., Rockland, DE, USA). The primer sequences for the
BRAF gene at exon 15 were: forward primer, 5′-TCATA
ATGCTTGCTCTGATAGGA-3′ and reverse primer, 5′-GGCC
AAAAATTTAATCAGTGGA-3′. The cycling conditions were: initial
denaturation at 95°C for 3 min, followed by 40 cycles at 95°C for
45 sec, 58°C for 45 sec and 72°C for 45 sec. The PCRs were
performed using the rTaq kit (Takara Bio Inc, Shiga, Japan) in a 50
μl volume of the reaction mixture. The primer sequences for the
KRAS gene were: forward primer, 5′-TCATTATTTTTATTATAAGGCCTGCTGAA-3′
and reverse primer, 5′-CAAAGACTGGTCCTGCACCAGTA-3′. The cycling
conditions were: initial denaturation at 95°C for 3 min, followed
by 40 cycles at 94°C for 45 sec, 60°C for 45 sec, 72°C for 30 sec.
The PCR reactions were performed using an LA-Taq kit (Takara Bio
Inc.) in a 50 μl volume of the reaction mixture.
The products were purified using the Qiagen PCR
purification kit (Qiagen, Valencia, CA, USA). The amplified cDNAs
were separated on 1% agarose gels and the bands were visualized by
ethidium bromide and photographed under ultraviolet
transillumination. Sequencing was then carried out with the ABI
prism 3100 analyzer (Applied Biosystems Japan Ltd., Tokyo, Japan)
and analyzed by BLAST and manual review of chromatograms.
BRAF V600E protein IHC
The 11 tumor specimens were immunostained by
automated methods (Dako Japan Co.) for BRAF V600E expression, using
the mouse monoclonal BRAF V600E clone, VE1 (Spring Bioscience,
Pleasanton, CA, USA). Unstained 4-μm sections of FFPE tumor tissue
that were slided from the same FFPE block that were used for
extracting genomic DNA were submitted for the analysis. The Dako
EnVision™ FLEX detection system included pretreatment with Dako PT
Link (pretreatment module) and Target Retrieval Solution, High pH
(K8004, Dako Co., Tokyo, Japan) at 97°C for 20 min, followed by
incubation with ×200 diluted mouse anti-BRAF V600E (clone VE1) with
Antibody Diluent (K8006, Dako Co.) overnight at 4°C. Antibody
incubation was followed by standard signal amplification, including
with rabbit LINKER (K8019) at room temperature for 15 min,
HRP-conjugated EnVision™ FLEX at room temperature for 20 min, DAB
reaction for 10 min and counterstaining with hematoxylin for 3 min.
An IHC score was assigned to each case according to the following
criteria based on the staining pattern: 3+, intense, granular
cytoplasmic staining; 2+, moderate, smooth cytoplasmic staining;
1+, faint cytoplasmic staining; and 0, no staining. The staining
area was categorized as 0–10, 25, 50, 75 and 90%. Tumors showing
positive staining involving a minimum of 50% of the tumor cells
were considered to be positive for BRAF V600E expression. These
criteria were determined by one of the co-authors who was blinded
to the clinical data.
CAST-PCR
CAST-PCR was carried out using the Mutation
Detector™ (Life Technologies) according to the manufacturer’s
instructions. CAST is a real-time quantitative Clamp-based PCR
technology (qPCR) (18). qPCR
allows measurement of a quantitative variable, namely, the
quantification cycle (Cq) that quantifies the presence of the
molecular variant. Each TaqMan mutation detection assay contained a
separate set of wild-type and mutant probe mix. qPCRs were run in a
final volume of the reaction mixture of 10 μl in 96-well plates
containing 5 μl of 2× TaqMan genotyping master mix (Life
Technologies), 1 μl of 10× assay mix for allele 1 (or 2), 2.5 μl of
deionized water and 10 ng of the DNA template. Runs were performed
on an Applied Biosystems 7500 real-time PCR System. The cycling
conditions for the CAST-PCR were: initial denaturation at 95°C for
10 min, followed by 5 cycles at 92°C for 15 sec, 58°C for 1 min,
then 40 cycles at 92°C for 15 sec, 60° for 1 min. Specific assays
for wild-type and mutant BRAF were commercially obtained from Life
Technologies. The data were analyzed with the SDS 2.0 software
program (13).
Results
BRAF and KRAS gene alterations in ovarian
serous borderline tumors
We carried out sequencing of exon 15 of the BRAF
gene in histopathology specimens of the 11 ovarian serous
borderline tumors. Of the 11 tumors, we found 2 cases of BRAF V600E
by direct sequencing of DNA samples, which involved a change of
nucleotide 1799 from thymine to adenine, resulting in a change of
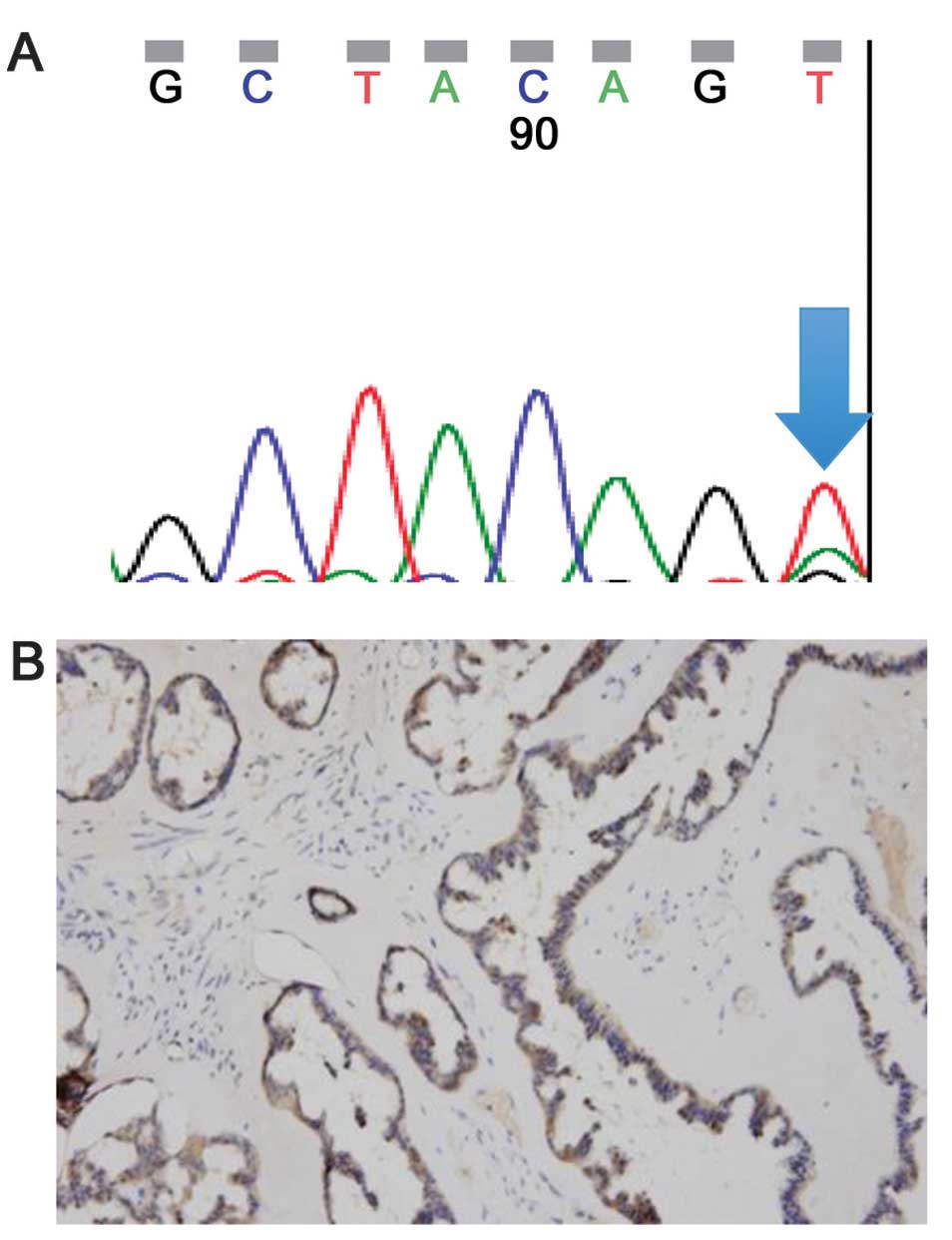
the amino acid valine to glutamic acid (case 4, Fig. 1A). No BRAF mutations were detected
in the remaining 9 patients. We also found 2 cases of KRAS mutation
among the 11 tumors, consisting of one case of KRAS glycine to
valine substitution at codon 12 (GGT>GTT, G12V) (case 6,
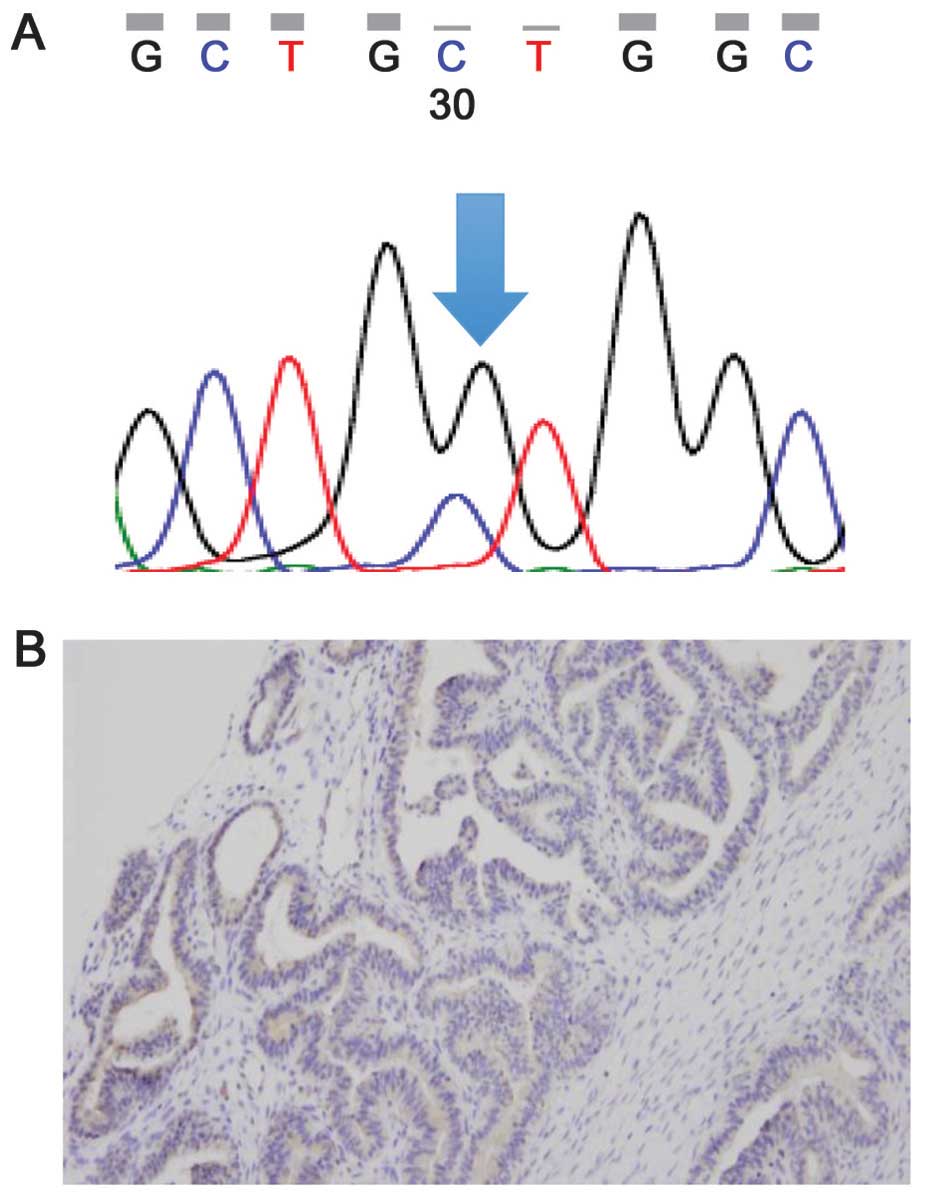
Fig. 2) and the other of KRAS
glycine to alanine substitution at codon 12 (GGT>GCT, G12A)
(case 7, Fig. 3A).
Both of the tumors that showed KRAS mutation were
BRAF wild-type. Thus, the BRAF and KRAS mutations were mutually
exclusive.
IHC for BRAF V600E
Of the 11 tumors, 3 were found by IHC to be positive
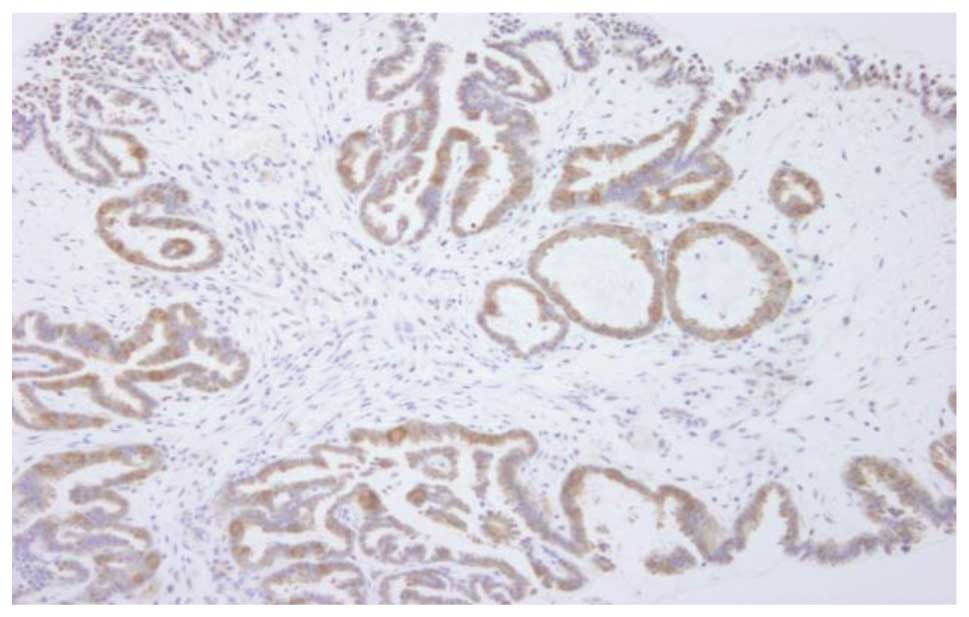
for BRAF mutation; one of the cases showed 3+ staining intensity
with an involved area of 90% (case 11, Fig. 4) and the remaining two showed 2+
staining intensity with an involved area of 75% (case 4, Fig. 1A). The staining pattern was diffuse
and cytoplasmic. One of the 3 positive cases showed a negative
result for BRAF mutation by direct sequencing (case 11).
In addition, one case showed a 1+ staining intensity
with an involved area of 25–50% (case 7, Fig. 3B) and was negative for BRAF mutation
by direct sequencing (stained <50%).
CAST-PCR for BRAF
CAST-PCR using DNA samples from 35 lung cancer cases
revealed the BRAF V600E mutation in 5 cases. The mutation ratios
were 8, 8.9, 10, 14.9 and 45.2%, respectively, and all cases were
positive by IHC. All 30 cases assessed as wild-type BRAF by Sas
were found to be negative by CAST-PCR (data not shown). We used
these samples for positive and negative controls.
Of the 11 ovarian serous borderline cases, CAST-PCR
carried out using DNA samples revealed BRAF V600E in 3 cases. The
mutation ratios were 17.7% (case 11), 16.3% (case 4) and 12.7%
(case 9) and all cases were positive by IHC. One case was
false-negative by direct sequencing (case 11). All the results are
summarized in Table I. The 20
randomly selected ovarian cancers were all wild-type BRAF.
 | Table IResults of IHC, direct sequencing and
CAST-PCR. |
Table I
Results of IHC, direct sequencing and
CAST-PCR.
| Case (no.) | Age (years) | BRAF | KRAS direct
sequence |
|---|
|
|---|
| IHC | Direct sequence | CAST-PCR (mutation
rate) |
|---|
| 1 | 55 | N | N | N | N |
| 2 | 44 | N | N | N | N |
| 3 | 40 | N | N | N | N |
| 4 | 48 | P++ | P | P (16.3%) | N |
| 5 | 51 | N | N | N | N |
| 6 | 26 | N | N | N | P |
| 7 | 55 | P | N | N | P |
| 8 | 27 | N | N | N | N |
| 9 | 25 | P++ | P | P (12.7%) | N |
| 10 | 65 | N | N | N | N |
| 11 | 27 | P+++ | N | P (17.7%) | N |
Discussion
From the results of the detection of the BRAF
mutation using 3 methods, one case was false-negative in direct
sequencing (case 11). In this study, we used modified methods for
the detection of BRAF V600E, namely, EnVision™ FLEX IHC, a
sensitive method using a linker and the results showed excellent
concordance of the results with CAST-PCR. As shown by our study as
well as previous studies, IHC serves as a highly specific and
sensitive test for mutated BRAF and may be used to confirm the
diagnosis. The results of BRAF mutation rate in CAST-PCR were not
those in the tumor tissues but those of the FFPE graft which we
used for extracting DNA. Under controlled conditions, the same FFPE
block was used for all 3 methods. When examined with both normal
and tumor tissue, IHC and CAST-PCR were the most effective at
detecting BRAF mutations. We also confirmed that IHC was the best
of the three methods in terms of accuracy, convenience and economic
advantage.
Currently, the presence of point mutations in
clinically relevant genes is assessed by various molecular-biologic
techniques, including allele-specific PCR, single-strand
conformational polymorphism, as well as conventional Sas and
pyrosequencing, all of which require a high quality laboratory
infrastructure with well-trained staff and are time-consuming.
Currently, Sas is probably the most frequently used technique owing
to its reliability and high specificity. However, potential sources
of error of molecular-diagnostic techniques include poor DNA
quality and an inadequate number of tumor cells in the sample, a
relevant problem in the case of low-grade serous ovarian tumors,
which was also encountered in our study. The sensitivity rate of
Sas generally does not allow for the detection of mutations
existing at a frequency of <15–20% of tumor cells (19,20).
Mutant-specific PCR (MS-PCR) may detect mutations in a greater
proportion of cases than routine Sas (21). According to one report, BRAF V600E
was detected by Sas in only 32.1% of melanomas, while 75.9% of the
melanomas had V600E mutations as assessed by MS-PCR (21). Notably, intra- and inter-tumor
heterogeneity of BRAF V600E mutations in melanomas has also been
reported by the group (21).
Amplification refractory mutation system (ARMS)-PCR detected BRAF
V600E in only 55% (30/55) cases of papillary thyroid carcinoma,
whereas Sas detected the mutation in 27 of 30 cases (22). We selected CAST-PCR, a technology
based on small-fragment amplification and qPCR suitable for low
quantities of DNA templates (16).
CAST-PCR allows efficient amplification from FFPE samples, the
probes are highly specific and all assays have a sensitivity of
<1% of mutation (16). However,
formalin fixation may reduce the DNA quality and potentially affect
the performance of the test. The existence of cross reactivity
between probes and the consequences of false-positive results need
to be evaluated (16).
Recently, mutation-specific antibodies were raised
against specific peptide sequences generated by missense mutations
(23,24). In particular, BRAF V600E is an ideal
target, as it occurs in a broad range of neoplasms and is the most
common genetic mutation in human cancers. IHC is a tissue-based,
cost-effective technique, which is easy to perform and routinely
available in most pathology laboratories (25). Capper et al (14,15)
developed a monoclonal mouse antibody (clone VE1) that recognizes
the BRAF V600E protein. Several studies have shown that this
antibody is reliable for the detection of BRAF V600E in malignant
neoplasms by IHC in routinely processed FFPE tissues (14–16,26).
VE-1 IHC was previously established in a cohort of 26 Japanese lung
cancer cases (16). If the
immunohistochemical staining technique is established properly, the
sensitivity and specificity may be expected to be equal to those of
established molecular-biologic techniques and use of this method
may be advantageous for cases with a low percentage of neoplastic
cells. A previous report indicated that 22 of 31 serous borderline
tumors showed VE-1 IHC positivity, whereas 20 of the 31 tumors were
confirmed to show the mutation by allele-specific PCR (17). Two VE1-positive cases with low
epithelial cell content required repeat microdissection to confirm
the presence of the mutation (17).
VE-1 IHC positivity was detected in 39 out of 265 colorectal cancer
cases (14.7%), while only 24 of the 39 were confirmed by Sas and
only 13 of 15 were confirmed by ultra-deep sequencing (26).
The V600E mutation rate in ovarian serous borderline
tumor specimens obtained from the Japanese patients was lower than
that in previous Caucasian reports [44.6% (27) and 30% (28)], which could be attributed to racial
differences in the prevalence of the BRAF mutation. However, V600E
substitution accounts for all the BRAF mutations in serous
borderline tumors (27). Serous
borderline tumors and low-grade serous ovarian cancers are
typically chemotherapy-resistant and the reported response rates to
cytotoxic chemotherapy are 4% in the neoadjuvant setting and
2.1–4.9% in the recurrence setting (3,4). Given
the high prevalence of BRAF and KRAS mutations in ovarian serous
borderline tumors, testing the effect of inhibitors targeting the
MAPK pathway has gained increasing attention. For the case of
serous ovarian tumors, preclinical studies demonstrating a profound
effect of MAPK pathway inhibition with the compound CI-1040 on
tumor cells carrying BRAF or KRAS mutations indicate that this
approach may have clinical potential (11,12).
The MEK inhibitor selumetinib has been used in a phase II treatment
trial for serous ovarian tumors (29).
Acknowledgements
This study was supported by Grants-in-Aid for
Scientific Research, the Japan Society for the Promotion of Science
(JSPS) (nos. 24592097 and 25293303). The authors thank Mr. Yoichi
Tani, who was responsible for immunostaining.
References
|
1
|
Kurman RJ and Shih IeM: Molecular
pathogenesis and extraovarian origin of epithelial ovarian cancer -
shifting the paradigm. Hum Pathol. 42:918–931. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crispens MA, Bodurka D, Deavers M, et al:
Response and survival in patients with progressive or recurrent
serous ovarian tumors of low malignant potential. Obstet Gynecol.
99:3–10. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gershenson DM, Sun CC, Bodurka D, et al:
Recurrent low-grade serous ovarian carcinoma is relatively
chemoresistant. Gynecol Oncol. 114:48–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmeler KM, Sun CC, Bodurka DC, et al:
Neoadjuvant chemotherapy for low-grade serous carcinoma of the
ovary or peritoneum. Gynecol Oncol. 108:510–514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shvartsman HS, Sun CC, Bodurka DC, et al:
Comparison of the clinical behavior of newly diagnosed stages II–IV
low-grade serous carcinoma of the ovary with that of serous ovarian
tumors of low malignant potential that recur as low-grade serous
carcinoma. Gynecol Oncol. 105:625–629. 2007.PubMed/NCBI
|
|
6
|
Singer G, Oldt R III, Cohen Y, et al:
Mutations in BRAF and KRAS characterize the development of
low-grade ovarian serous carcinoma. J Natl Cancer Inst. 95:484–486.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mayr D, Hirschmann A, Lohrs U and Diebold
J: KRAS and BRAF mutations in ovarian tumors: a comprehensive study
of invasive carcinomas, borderline tumors and extraovarian
implants. Gynecol Oncol. 103:883–887. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sieben NL, Macropoulos P, Roemen GM, et
al: In ovarian neoplasms, BRAF, but not KRAS, mutations are
restricted to low-grade serous tumors. J Pathol. 202:336–340. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones S, Wang TL, Kurman RJ, et al:
Low-grade serous carcinomas of the ovary contain very few point
mutations. J Pathol. 226:413–420. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bollag G, Hirth P, Tsai J, et al: Clinical
efficacy of a RAF inhibitor needs broad target blockade in
BRAF-mutant melanoma. Nature. 467:596–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakayama N, Nakayama K, Yeasmin S, et al:
KRAS or BRAF mutation status is s useful predictor of sensitivity
to MEK inhibition in ovarian cancer. Br J Cancer. 99:2020–2028.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pohl G, Ho CL, Kurman RJ, et al:
Inactivation of the mitogen-activated protein kinase pathway as a
potential target-based therapy in ovarian serous tumors with KRAS
or BRAF mutations. Cancer Res. 65:1994–2000. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Didelot A, Le Corre D, Luscan A, et al:
Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR
mutation detection in clinical formalin fixed paraffin embedded
samples. Exp Mol Pathol. 92:275–280. 2012. View Article : Google Scholar
|
|
14
|
Capper D, Preusser M, Habel A, et al:
Assessment of BRAF V600E mutation status by immunohistochemistry
with a mutation-specific monoclonal antibody. Acta Neuropathol.
122:11–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Capper D, Berghoff AS, Magerle M, et al:
Immunohistochemical testing of BRAF V600E status in 1,120 tumor
tissue samples of patients with brain metastases. Acta Neuropathol.
123:223–233. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sasaki H, Shimizu S, Tani Y, et al:
Usefulness of immunohistochemistry for the detection of the BRAF
V600E mutation in Japanese lung adenocarcinoma. Lung Cancer.
82:51–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bösmüller H, Fischer A, Pham DL, et al:
Detection of the BRAF V600E mutation in serous ovarian tumors: a
comparative analysis of immunohistochemistry with a
mutation-specific monoclonal antibody and allele-specific PCR. Hum
Pathol. 44:329–335. 2013.PubMed/NCBI
|
|
18
|
Bustin SA, Benes V, Garson JA, et al: The
MIQE guidelines: minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lamy A, Blanchard F, Le Pessort F, et al:
Metastatic colorectal cancer KRAS genotyping in routine practice:
results and pitfalls. Mod Pathol. 24:1090–1100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anderson S, Bloom KJ, Vallera DU, et al:
Multisite analytic performance studies of a real-time polymerase
chain reaction assay for the detection of BRAF V600E mutations in
formalin-fixed, paraffin-embedded tissue specimens of malignant
melanoma. Arch Pathol Lab Med. 136:1385–1391. 2012. View Article : Google Scholar
|
|
21
|
Yancovitz M, Litterman A, Yoon J, et al:
Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in
primary and metastatic melanoma. PLos One. 7:e293362012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang T, Zhuge J and Zhang WW: Sensitive
detection of BRAF V600E mutation by Amplification Refractory
Mutation System (ARMS)-PCR. Biomark Res. 1:32013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brevet M, Arcita M and Ladanyi M:
Assessment of EGFR mutation status in lung adenocarcinoma by
immunohistochemistry using antibodies specific to the two major
forms of mutant EGFR. J Mol Diagn. 12:169–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sahm F, Capper D, Meyer J, et al:
Immunohistochemical analysis of 1844 human epithelial and
haemotopoietic tumours and sarcomas for IDH1R132H mutation.
Histopathology. 58:1167–1172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Raab S: The cost-effectiveness of
immunohistochemistry. Arch Pathol Lab Med. 124:1185–1191.
2000.PubMed/NCBI
|
|
26
|
Rossle M, Sigg M, Ruschoff JH, et al:
Ultra-deep sequencing confirms immunohistochemistry as a highly
sensitive and specific method for detecting BRAF V600E mutations in
colorectal carcinoma. Virchows Arch. 463:623–631. 2013. View Article : Google Scholar
|
|
27
|
Grisham RN, Iyer G, Garg K, et al: BRAF
mutation is associated with early stage disease and improved
outcome in patients with low-grade serous ovarian cancer. Cancer.
119:548–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong KK, Tsang YT, Deavers MT, et al: BRAF
mutation is rare in advanced-stage low-grade ovarian serous
carcinomas. Am J Pathol. 177:1611–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Farley J, Brady WE, Vathipadiekal V, et
al: Selumetinib in women with recurrent low-grade serous carcinoma
of the ovary or peritoneum: an open-label, single-arm, phase 2
study. Lancet Oncol. 14:134–140. 2013. View Article : Google Scholar : PubMed/NCBI
|