Introduction
The phosphoinositide-3 kinase (PI3K)/Akt pathway
participates in many biological processes such as proliferation,
apoptosis, differentiation, metabolism and migration (1). The PI3K signaling cascade is
initiated through activation of receptors with intrinsic tyrosine
kinase activity, which leads to generation of the second messenger
phosphatidylinositol (3,4,5)-trisphosphate (PIP3), acting
on downstream targets such as PI-dependent kinase (PDK1),
integrin-linked kinase (ILK-1) or Akt. Type IA PI3K is a
heterodimer composed of a p85 regulatory subunit encoded by
PIK3R1, PIK3R2 or PIK3R3 and a p110 catalytic
subunit; p110α, p110β or p110δ encoded by PIK3CA,
PIK3CB and PIK3CD, respectively. Deregulation of the
PI3K/Akt pathway is a recurrent feature in numerous human
malignancies with a key role in cancer development, progression and
also in resistance to chemotherapy. Over-activity is commonly
caused by loss of the tumor suppressor gene PTEN(2,3),
oncogenic activation of PIK3CA(4,5)
and/or over-stimulation by various growth factors like IGF-1, EGF
or VEGF (6–8).
Neuroblastoma is a pediatric cancer stemming from
immature precursors of the sympathetic nervous system with tumors
arising in sympathetic ganglia or adrenal gland (9). Neuroblastoma displays high clinical
variability, ranging from more favorable stage 1 tumors to highly
aggressive stage 4 tumors with many times fatal outcome. The
contribution of PI3K/Akt in the carcinogenesis of neuroblastoma is
not fully understood. Mutations in the genes PIK3CA and
PTEN frequently reported in other malignancies, are rare in
neuroblastoma (10,11) although a few mutations have been
reported in PIK3CD(12).
PIK3CD also show lower expression in aggressive
neuroblastomas compared to neuroblastomas with more favorable
biology (13,14). Moreover, further connection to the
PI3K/Akt pathway is seen through Akt, which is found to be
activated in neuroblastoma (15)
in an outcome-correlated manner (16). There are several other markers that
correlate to grade of disease and/or outcome, such as expression of
the different Trk-receptors (17),
degree of neural differentiation (18,19)
or genetic aberrations such as 1p deletion, 11q deletion, gain of
17q and amplification of the oncogene MYCN(20). PI3K signaling has effect on Mycn
protein stability through inactivation of GSK3β and inhibition of
PI3K destabilized Mycn and prevented tumor progression in a murine
model of neuroblastoma (21).
PI3K inhibition is considered to be one of the most
promising targeted therapies for cancer, thus the understanding of
the molecular pathology of the individual tumors will be essential
to match patients with PI3K inhibitors of differing selectivity
profiles. In this study we explored the expression of PI3K/Akt
associated genes and found significant differences at both mRNA and
protein levels between aggressive and favorable neuroblastoma
tumors.
Materials and methods
RNA purification and cDNA
preparation
Fresh frozen tumor samples from patients diagnosed
with neuroblastoma and staged according to the International
Neuroblastoma Staging System Criteria (INSS) and International
Neuroblastoma Risk Group (INRG) were used (Table I). Total-RNA was prepared using
Totally RNA (Ambion, St. Austin, TX) or RNeasy mini kit (Qiagen,
Hilden, Germany) while genomic DNA were removed with DNA-free kit
(Ambion). Purity and integrity of the RNA were assayed with
spectrophotometer and RNA 6000 Nano Bioanalyzer (Agilent, Palo
Alto, CA) before cDNA synthesis using SuperScript™ II Reverse
Transcriptase (Invitrogen, Carlsbad, CA).
 | Table I.Clinical data. |
Table I.
Clinical data.
| Patient | INSS | INRG | Outcome | 1p loss | MNA | 11q loss | Methods
|
|---|
| QPCR | WB | Array |
|---|
| 18E1 | 1 | L | NED | Neg | Neg | Neg | + | | |
| 18E5 | 1 | L | NED | Neg | Neg | Neg | + | | |
| 18E8 | 1 | L | NED | Neg | Neg | Neg | + | | |
| 19R1 | 1 | L | NED | Neg | Neg | Neg | + | | |
| 30R9 | 1 | L | NED | Neg | Neg | Neg | + | | |
| 19R6 | 1 | L | DOD | Pos | Pos | Neg | + | | |
| 17E7 | 2 | L | NED | Neg | Neg | Neg | + | | |
| 10R6 | 2 | L | NED | Neg | Neg | Neg | + | | |
| 14R9 | 2 | L | NED | Pos | Neg | Neg | + | | |
| 25R8 | 2 | L | NED | Neg | Neg | Neg | + | | |
| 27R1 | 2 | L | NED | Neg | Neg | Neg | + | | |
| 33R7 | 2 | L | NED | Neg | Neg | Neg | + | | |
| 8E5 | 3 | L | NED | Neg | Neg | Neg | + | | |
| 16R4 | 3 | L | NED | Neg | Pos | Neg | + | | |
| 34R5 | 3 | L | NED | Neg | Neg | NA | + | | |
| 6E9 | 3 | L | DOD | Pos | Neg | Pos | + | | |
| 13E6 | 3 | L | DOD | Pos | Pos | Pos | + | | |
| 15E1 | 4 | M | NED | Pos | Pos | Neg | + | | |
| 10E6 | 4 | M | NED | Pos | Pos | Neg | + | | |
| 17R3 | 4 | M | NED | Neg | Neg | NA | + | | |
| 25R3 | 4 | M | NED | Neg | Neg | Neg | + | | |
| 29R2 | 4 | M | NED | Pos | Pos | Neg | + | | |
| 32R2 | 4 | M | NED | Pos | Neg | Pos | + | | |
| 40R2 | 4 | M | NED | Neg | Neg | Neg | + | | |
| 4E1 | 4 | M | DOD | Neg | Neg | Pos | + | | |
| 3E2 | 4 | M | DOD | Neg | Neg | Pos | + | | |
| 12E3 | 4 | M | DOD | Pos | Pos | Neg | + | | |
| 16E3 | 4 | M | DOD | Pos | Pos | Neg | + | | |
| 11E4 | 4 | M | DOD | Neg | Neg | Pos | + | | |
| 18E4 | 4 | M | DOD | Pos | Pos | Neg | + | | |
| 13R0 | 4 | M | DOD | Pos | Pos | Neg | + | | |
| 24R3 | 4 | M | DOD | Pos | Pos | NA | + | | |
| 26R8 | 4 | M | DOD | Pos | Pos | NA | + | | |
| 35R2 | NA | L | NED | Neg | Neg | Neg | + | | |
| 14E6 | 1 | L | NED | Neg | Neg | Neg | + | | + |
| 10R7 | 1 | L | NED | Neg | Neg | Neg | + | | + |
| 35R5 | 1 | L | NED | NA | NA | NA | + | | + |
| 35R8 | 1 | L | NED | Neg | Neg | Neg | + | | + |
| 37R6 | 1 | L | NED | Neg | Neg | Neg | + | | + |
| 26R0 | 4 | M | NED | Pos | Pos | Pos | + | | + |
| 25R9 | 2 | L | NED | Neg | Neg | Neg | + | + | + |
| 10R2 | 4 | M | DOD | Pos | Pos | Neg | + | + | + |
| 15R3 | 4 | M | DOD | Pos | Neg | Pos | + | + | + |
| 34R0 | 4 | M | DOD | Neg | Neg | Neg | + | + | + |
| 9R9 | 3 | M | DOD | Pos | Neg | Pos | + | + | |
| 15E7 | 3 | L | DSC | Neg | Neg | Neg | + | + | |
| 15E3 | 3 | L | NED | Neg | Neg | Neg | + | + | |
| 20R9 | 2 | L | NED | Neg | Neg | NA | + | + | |
| 27R7 | 2 | L | NED | Neg | Neg | Neg | + | + | |
| 25R0 | 3 | L | NED | Neg | Neg | Neg | + | + | |
| 17R2 | 4 | M | DOD | Neg | Neg | Pos | + | + | |
| 28R8 | 4 | M | DOD | Neg | Neg | Pos | + | + | |
| 33R5 | 1 | L | NED | Neg | Neg | Neg | | + | |
| 13E8 | 2 | L | NED | Neg | Neg | Neg | | + | |
| 11R4 | 3 | L | DOD | Pos | Pos | Neg | | + | |
| 16E9 | 4 | M | DOD | Neg | Pos | Neg | | + | |
| 10R8 | 3 | L | DOD | Neg | Neg | Pos | | + | |
| 39R1 | 4 | M | NED | Pos | Pos | Neg | | + | + |
| 26R9 | 1 | L | NED | Neg | Neg | Neg | | | + |
| 11E1 | 4 | M | NED | Neg | Neg | Pos | | | + |
| 16E1 | 1 | L | NED | Neg | Neg | Neg | | | + |
| 23R4 | 2 | L | NED | Neg | Neg | Neg | | | + |
| 36R3 | MS | MS | DOD | Neg | Neg | Neg | | | + |
Expression analysis by microarray and
real-time RT-PCR
Four total-RNAs run on Affymetrix HU133A platform as
described previously (46), and
another twelve total-RNAs were run on the Affymetrix HU133plus2
platform by Aros Applied Biotechnology AS (www.arosab.com/). Bioconducter for R 2.9.2 (library
BioC 2.4) was used to perform gcRMA normalisation for each GeneChip
platform set separately. For each probe-set, the maximum expression
values over all samples was determined, and probe-sets that showed
very low or no detectable expression levels were filtered out (max
2log expression <6). For those probe-sets overlapping the two
GeneChip platforms, a probe-specific normalization between the two
platforms was performed based on two individuals run on both
platforms. Next, the mean log2 expression level for each gene
symbol was calculated.
A set of 88 genes with known association to the
PI3K/Akt pathway were selected (Table
II) and a two-sided t-test was performed to identify genes with
significant differential expression when comparing neuroblastoma of
low stage (stage 1, 2 and 4S) (n=10) to stage 4 (n=6). Expression
of identified genes were verified by quantitative real-time PCR
(QPCR) using TaqMan Low Density arrays in a larger set of tumors;
stage 1–2 (n=21), stage 4 (n=22) and stage 3 (n=9). Pooled RNA (40
donors) from normal adrenal gland tissue was used as reference
(Ambion). QPCR was performed using triplicates with pre-designed
primer and probe sets for target genes (PRKCZ: hs.00177051_ml,
EIF4EBP1: hs.00607050_ml, PRKZB1: hs.01030676_ml, PDGFRA:
hs.00183486_ml, PIK3CD: hs.00192399_ml, PIK3R1: hs.00933163_ml,
AKT1: hs.00920503_ml, BAD: hs.00188930_ml, GUSB:
hs.99999908_ml) and ABI PRISM® 7900HT Sequence detection
system (Applied Biosystems). Quantification was performed using the
standard curve method with GUSB (β-glucuronidase) as endogenous
control for normalization of gene expression. The logarithms of
mean expression levels were used in t-tests of microarray and QPCR
data. Expression from microarrays was compared using two-tailed
t-test while expression of genes in the validation-set was compared
using one-tailed t-test. Statistical calculations and boxplots were
made with SPSS ver.18 (SPSS, Chicago, IL) and Excel (Microsoft).
Fold change was calculated by dividing the corresponding values for
stage 4 with that of stage 1 and 2 neuroblastomas. Unsupervised
hierarchal clustering of real-time PCR data from six PI3K pathway
genes and 52 primary neuroblastoma samples. The heat map was based
on Max linkage.
 | Table II.Tested PI3K/Akt associated genes. |
Table II.
Tested PI3K/Akt associated genes.
| Gene | Description | Gene | Description |
|---|
| ADAR | Adenosine
deaminase, RNA-specific isoform a | MAPK1 | Mitogen-activated
protein kinase 1 |
| AKT1 | V-akt murine
thymoma viral oncogene homolog 1 | MAPK14 | Mitogen-activated
protein kinase 14 |
| AKT3 | V-akt murine
thymoma viral oncogene homolog 3 | MAPK3 | Mitogen-activated
protein kinase 3 |
| APC | Adenomatous
polyposis coli | MAPK8 | Mitogen-activated
protein kinase 8 |
| BAD | BCL2-antagonist of
cell death protein | MTCP1 | Mature T-cell
proliferation 1 |
| BTK | Bruton
agammaglobulinemia tyrosine kinase | MYD88 | Myeloid
differentiation primary response gene |
| CASP9 | Caspase 9 isoform
alpha preproprotein | NFKB1 | Nuclear factor
kappa-B, subunit 1 |
| CCND1 | Cyclin D1 | NFKBIA | Nuclear factor of
kappa light polypeptide gene |
| CD14 | CD14 antigen
precursor | NRAS | Neuroblastoma RAS
viral (v-ras) oncogene |
| CDC42 | Small GTP binding
protein CDC42 | PABPC1 | Poly(A) binding
protein, cytoplasmic 1 |
| CDKN1B | Cyclin-dependent
kinase inhibitor 1B | PDGFRA | Platelet-derived
growth factor receptor alpha |
| CTMP | Carboxyl-terminal
modulator protein | PDK1 | 3-phosphoinositide
dependent protein kinase-1 |
| CHUK | Conserved
helix-loop-helix ubiquitous kinase | PDK2 | Pyruvate
dehydrogenase kinase, isozyme 2 |
| CSNK2A1 | Casein kinase II
alpha 1 subunit | PIK3CA |
Phosphoinositide-3-kinase, catalytic,
alpha |
| CTNNB1 | Catenin
(cadherin-associated protein), beta 1 | PIK3CB |
Phosphoinositide-3-kinase, catalytic,
beta |
| CUTL1 | Cut-like homeobox
1 | PIK3CD |
Phosphoinositide-3-kinase, catalytic,
delta |
| EIF2AK2 | Eukaryotic
translation initiation factor 2-alpha | PIK3CG |
Phosphoinositide-3-kinase, catalytic,
gamma |
| EIF4A1 | Eukaryotic
translation initiation factor 4A | PIK3R1 |
Phosphoinositide-3-kinase, regulatory
subunit 1 |
| EIF4B | Eukaryotic
translation initiation factor 4B | PIK3R3 |
Phosphoinositide-3-kinase, regulatory
subunit 3 |
| EIF4E2 | Eukaryotic
translation initiation factor 4E | PP2A | Protein phosphatase
2, catalytic subunit, alpha |
|
EIF4EBP1 | Eukaryotic
translation initiation factor 4E | PRKCA | Protein kinase C,
alpha |
| EIF4G1 | Eukaryotic
translation initiation factor 4 | PRKCB1 | Protein kinase C,
beta isoform 1 |
| ELK1 | ELK1 protein | PRKCZ | Protein kinase C,
zeta |
| FASLG | Tumor necrosis
factor ligand superfamily member 6 | PTEN | Phosphatase and
tensin homolog |
| FKBP1A | FK506-binding
protein 1A | PTK2 | PTK2 protein
tyrosine kinase 2 |
| FOS | C-fos FBJ murine
osteosarcoma viral oncogene | PTPN11 | Protein tyrosine
phosphatase, non-receptor type |
| FOXO1 | Forkhead box
O1 | RAC1 | Ras-related C3
botulinum toxin substrate 1 |
| FOXO3 | Forkhead box
O3A | RAF1 | V-raf-1 murine
leukemia viral oncogene homolog |
| FRAP1
(MTOR) | FK506 binding
protein 12-rapamycin associated | RASA1 | RAS p21 protein
activator 1 |
| GJA1 | Connexin 43 | RBL2 | Retinoblastoma-like
2 (p130) |
| GRB10 | Growth factor
receptor-bound protein 10 | RHEB | Ras homolog
enriched in brain |
| GRB2 | Growth factor
receptor-bound protein 2 | RHOA | Ras homolog gene
family, member A |
| GSK3B | Glycogen synthase
kinase 3 beta | RPS6KA1 | Ribosomal protein
S6 kinase, 90 kDa, polypeptide |
| HRAS | V-Ha-ras Harvey rat
sarcoma viral oncogene | RPS6KB1 | Ribosomal protein
S6 kinase, 70 kDa, polypeptide |
| HSPB1 | Heat shock 27 kDa
protein 1 | SHC1 | SHC (Src homology 2
domain containing) |
| IGF1 | Insulin-like growth
factor 1 i | SOS1 | Son of sevenless
homolog 1 |
| IGF1R | Insulin-like growth
factor 1 receptor | SRF | Serum response
factor |
| ILK | Integrin-linked
kinase | TIRAP | Toll-interleukin 1
receptor domain-containing |
| IRAK1 | Interleukin-1
receptor-associated kinase 1 | TLR4 | Toll-like receptor
4 |
| IRS1 | Insulin receptor
substrate 1 | TOLLIP | Toll interacting
protein |
| ITGB1 | Integrin beta 1
isoform 1B precursor | TSC1 | Tuberous sclerosis
1 protein |
| JUN | Jun oncogene | TSC2 | Tuberous sclerosis
2 |
| KRAS | Ras family small
GTP binding protein K-Ras | WASL | Wiskott-Aldrich
syndrome gene-like protein |
| MAP2K1 | Mitogen-activated
protein kinase kinase 1 | YWHAH | Tyrosine
3-monooxygenase/tryptophan |
Protein isolation, western blot analysis
and antibodies
Fresh frozen neuroblastoma tumors were homogenized
using Tissuelyzer (Qiagen) in RIPA lysis buffer supplemented with
HALT™ Phosphatase and protease inhibitor cocktail (Pierce,
Rockford, IL) while a ready-made protein lysate for normal adrenal
gland (20 pooled donors) was purchased from Clontech (Mountain
View, CA). SDS-PAGE and western blot analysis were carried out
according to standard procedures using 30 μg of total
protein lysate. Immunoblotting was performed with rabbit polyclonal
antibodies against p85α (no. 06–496) (Millipore, Billerica, MA)
4e-bp1 (no. 9452) (Cell Signaling Technology, Danvers, MA) and PKCβ
(sc-209), PKCζ (sc-216), Pdgfrα (sc-338) GAPDH (sc-825778) and
p110δ (sc-7176), from Santa Cruz Biotechnology (Santa Cruz, CA).
Quantification of proteins was performed with the ImageJ software
(available at http://rsb.info.nih.gov/ij). GAPDH was used for
normalization in calculation of relative expression. The logarithms
of expression levels were calculated and the difference between
groups was assessed by a two-tailed independent-samples t-test.
Results
mRNA levels of six PI3K-pathway genes
differs between neuroblastoma stages
Analysis of Affymetrix oligo microarray data on a
panel of neuroblastoma tumors revealed differential expression
between low stage (1, 2 and 4S) and stage 4 patients with
statistical significance (p<0.05) for 8 out of 88 genes
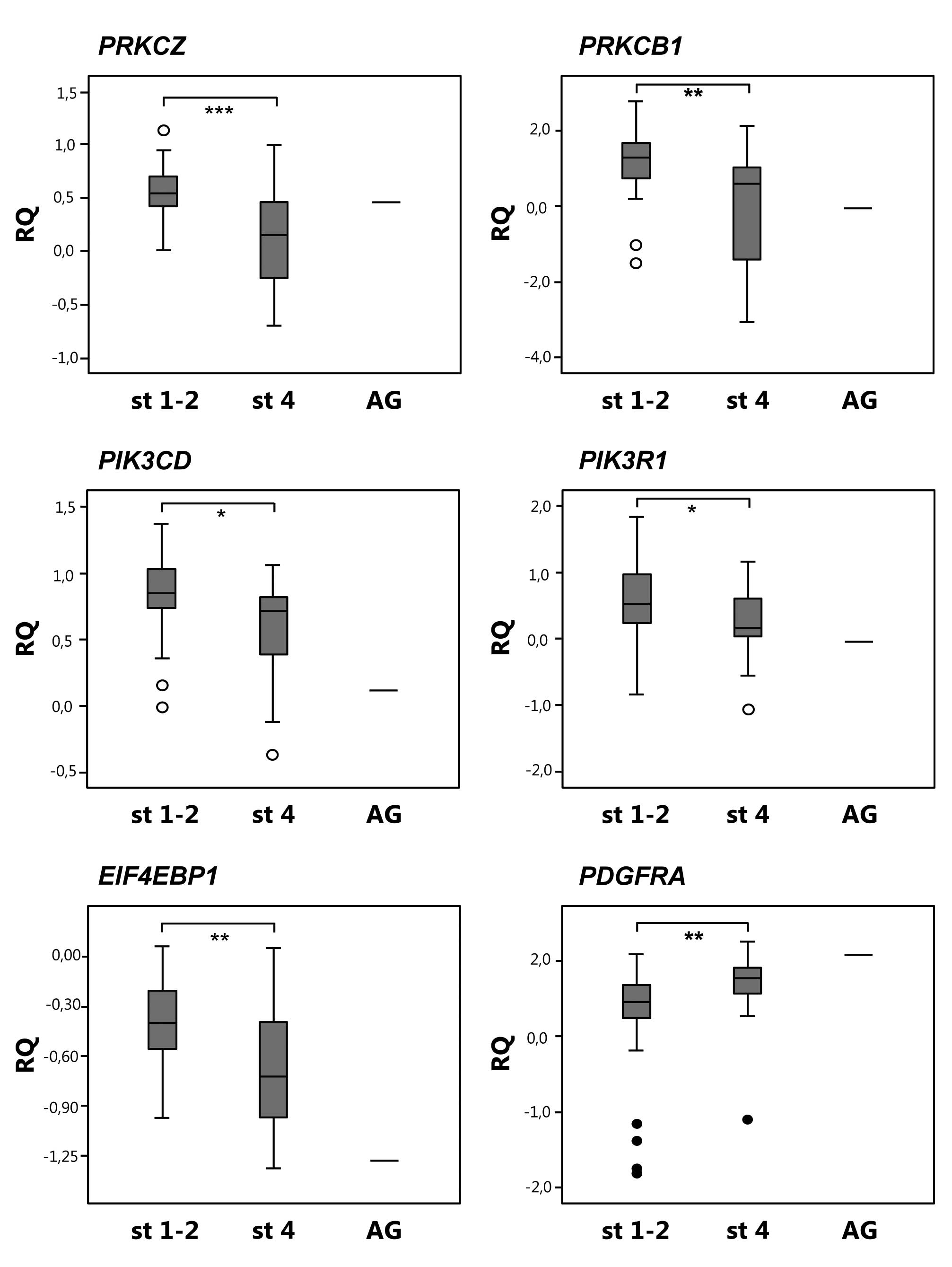
associated with PI3K/Akt signaling (Table III). Expression of these genes were
validated in a larger set of primary neuroblastoma samples using
QPCR and the pattern of expression was confirmed for PRKCZ,
EIF4EBP1, PRKCB1, PIK3CD, PIK3R1, which
showed lower expression in stage 4 compared to stage 1–2 tumors,
and PDGFRA, which showed higher expression in stage 4
compared to stage 1–2 tumors (Fig.
1, Table III).
 | Table III.Results from microarray and QPCR. |
Table III.
Results from microarray and QPCR.
| Gene | Chromosomal
localization | Microarray
| QPCR
|
|---|
| Fold change | P-value* | Fold change | P-value** |
|---|
| PRKCZ | 1p36 | 0.46 | 0.02 | 0.52 | 0.0003 |
|
EIF4EBP1 | 8p12 | 0.60 | 0.02 | 0.64 | 0.006 |
| PRKCB1 | 16p11 | 0.19 | 0.02 | 0.28 | 0.005 |
| PDGFRA | 4q12 | 10.40 | 0.02 | 2.46 | 0.01 |
| PIK3CD | 1p36 | 0.31 | 0.001 | 0.61 | 0.03 |
| PIK3R1 | 5q13 | 0.44 | 0.03 | 0.36 | 0.03 |
| AKT1 | 14q32 | 0.77 | 0.03 | 1.05 | 0.43 |
| BAD | 11q13 | 0.46 | 0.004 | 0.98 | 0.40 |
| GUSB | 7q11 | - | - | - | - |
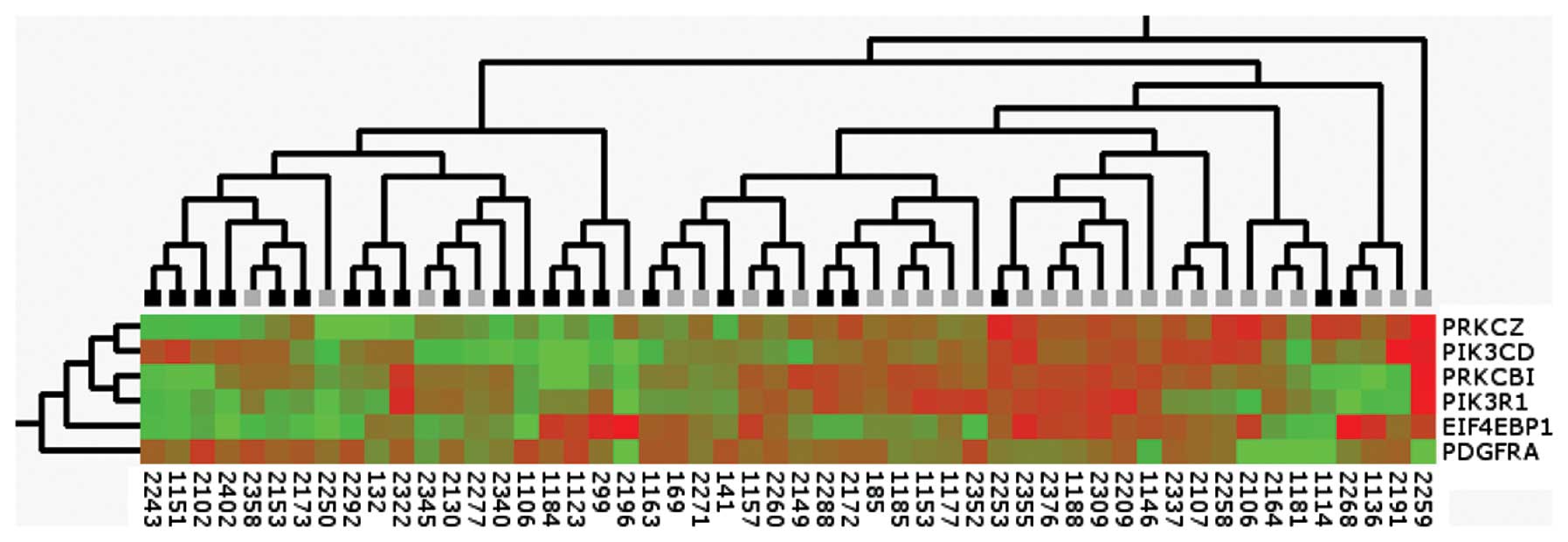
Clustering of six PI3K-pathway genes
Unsupervised hierarchal clustering using Max linkage
of real-time PCR data from PRKCZ, EIF4EBP1,
PRKCB1, PIK3CD, PIK3R1 and PDGFRA in 52
primary tumor samples showed that the expression levels of these
genes cluster neuroblastomas into metastasizing and localized
tumors (Fig. 2).
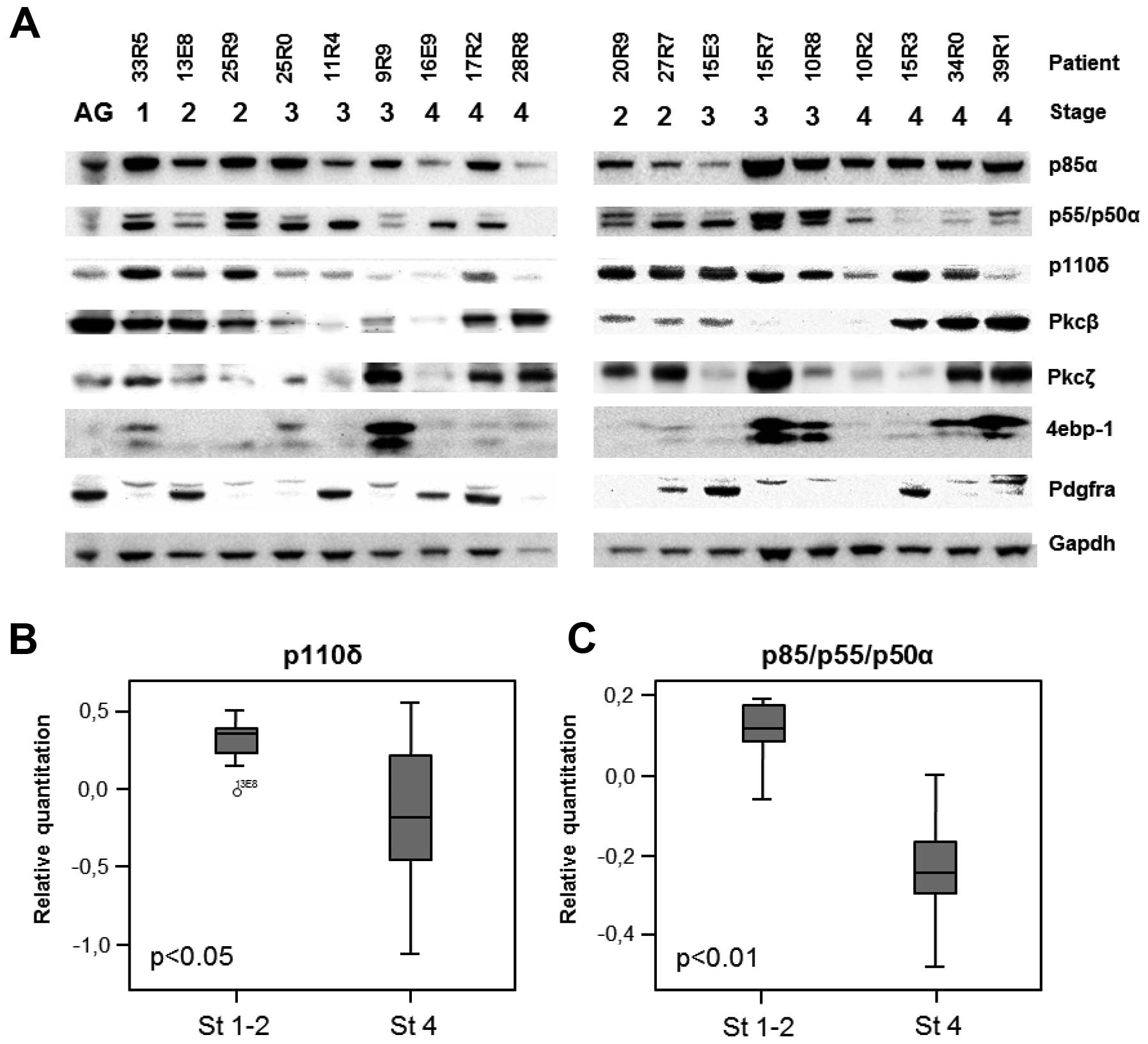
Low p110δ and p85α protein levels in
aggressive neuroblastoma
To further explore the proteins encoded by the
differential expressed genes we performed western blot analysis on
lysates from 18 primary neuroblastoma tumors and normal adrenal
gland. All proteins except 4e-bp1 were detectable in adrenal gland
and to various extents in neuroblastoma tumors (Fig. 3A). p110δ (encoded by PIK3CD)
was detected in all stages, however overall protein levels of p110δ
was significantly lower in stage 4 compared to stage 1–2
neuroblastomas (p=0.04) (Fig. 3B).
The overall protein levels of p85α isomers were significantly lower
in stage 4 compared to stage 1–2 neuroblastoma (p=0.0015) (Fig. 3C). No other proteins encoded by the
genes differently expressed on mRNA-level showed significant
differences in protein levels in these 18 tested neuroblastoma
protein samples.
Discussion
The PI3K/Akt pathway is central for numerous
cellular functions and it is frequently deregulated in human
cancers. This pathway is also suggested to be an important player
in neuroblastoma development and/or progression and we therefore
investigated different actors in PI3K/Akt signaling in primary
tumors through analysis at the mRNA and protein level. Five of 88
investigated genes associated to PI3K/Akt signaling pathway showed
higher levels of mRNA expression in stage 1–2 neuroblastomas
compared to stage 4; EIF4EBP1, PRKCZ, PRKCB1,
PIK3RI and PIK3CD. It is notable that the decreased
expression of PIK3CD and PRKCZ in stage 4
neuroblastoma may be due to their chromosomal localization at 1p36,
a region frequently deleted in stage 4 neuroblastoma.
EIF4EBP1 encodes 4e-bp1, a repressor protein
that inhibits the eukaryotic translation initiation factor 4E
(eIF4E). High expression of EIF4EBP1 in both favorable and
unfavorable neuroblastomas compared to adrenal gland indicates a
general upregulation with higher mRNA levels in stage 1–2 compared
to stage 4 neuroblastoma (Fig. 1).
It is possible that lower expression of EIF4EBP1 mimics the
physiological relevance of phosphorylation of 4e-bp1 since both is
expected to reduce translational inhibition.
The mRNA expression of PRKCB1 and
PRKCZ, encoding PKCβ and PKCζ, respectively, were lower in
stage 4 compared to stage 1–2 (Fig.
1). Members of the PKC family have unique and even opposite
effects on cell growth, survival and differentiation (22–24).
PKCβ stimulates growth and proliferation in neuroblastoma (25) although upregulation of both PKCβ
and PKCζ was noticed under euxanthone-induced differentiation of a
neuroblastoma cell line (26) and
PKCβ activation induced apoptosis in HL60-cells (27). PKCζ participate in negative
regulation of IRS-1 (28) and have
shown proapoptotic functions in ovarian cancer (29). On the other hand, siRNA silencing
of PRKCZ impairs migration and invasion in glioblastoma,
indicating a role in metastasis (30). This suggests different roles of the
PKC isoforms depending on stimuli, and that further effort is
needed to elucidate the functions of PRKCZ and PRKCB1
in neuroblastoma.
PDGFRA encodes a cell surface tyrosine kinase
receptor important in development of the neural crest and has also
been shown to be important in neuroblastoma differentiation
(31,32). Moreover, it has also been found to
be downregulated during neural differentiation (32). We found PDGFRA to be
expressed in all stages even though significantly higher in stage 4
compared to stage 1–2 neuroblastoma, probably explained by the
undifferentiated character of all neuroblastomas, especially stage
4. Since PDGFRA also has been found to be mutated or overexpressed
in cancer and contribute to cancer development by autocrine or
paracrine signaling mechanisms, this could also contribute to the
pathogenesis of neuroblastoma (33).
Pten activity can be modulated by the p85 subunit of
the PI3K (34,35), which also enhances the phosphatase
activity of Pten (36).
Consequently, decreased levels of p85 leads to diminished Pten
activity and hence increased phosphorylation of Akt. In our
material, expression of PIK3R1, encoding three different
p85α isomers, was indeed decreased in stage 4 tumors compared to
stage 1–2 both on mRNA and on protein level (Figs. 1 and 3). In hepatocellular carcinoma
PIK3R1 levels were inversely correlated with grade of
malignancy, consistent with reports of tumor suppressing functions
of p85 (37,38). Besides modulation of Pten, p85
stabilizes and inhibits the p110α isoform (39) and mutations in the SH2-domain of
p85 has been shown to release the inhibitory effect of p110α and
leads to constitutive activation of Akt (40–42).
Both mRNA and protein levels from
PIK3CD/p110δ are decreased in stage 4 neuroblastomas
compared to stage 1–2 as described by us and others previously
(13,14). Signaling through PI3K is required
in neural development (43–46)
and possibly the δ-isoform could be important in neuroblast
differentiation since higher levels of p110δ was detected in stage
1–2 neuroblastoma, commonly expressing more markers of neural
differentiation. However, the contribution of the different p110
isoforms in neural differentiation is not fully understood and
requires further attention.
Although the molecular mechanisms underlying
neuroblastoma are slowly being uncovered, neuroblastoma is still
fatal in many cases. In this study we have detected differential
expression of several members of the PI3K/Akt pathway on mRNA
and/or protein level. Since neuroblastoma is a heterogeneous
disease, tumor initiation and progression could occur through
activation of different signaling pathways. From the present study
we conclude that expression evaluation of a few genes involved in
the PI3K-pathway can predict aggressive disease, and our findings
indicate a stage-dependent involvement of the PI3K-pathway in
neuroblastoma.
Abbreviations:
|
PI3K
|
phosphoinositide-3 kinase;
|
|
QPCR
|
quantitative PCR;
|
|
INRG
|
International Neuroblastoma Risk
Group;
|
|
INSS
|
International Neuroblastoma Staging
System Criteria
|
Acknowledgements
We thank the Sahlgenska Gothenburg
Genomics Core Facility for access to the ABI
PRISM®7900HT System and Grissel Faura for technical
assistance. This study was supported by grants from the Swedish
Cancer Society, the Swedish Children’s Cancer Fund, the Sahlgrenska
University Hospital Foundation, the Assar Gabrielsson Foundation,
Gunvor and Ivan Svensson’s Foundation, Åke Wiberg’s Foundation,
Mary Beves Foundation for research in childhood cancer and
Frimurare Barnhusdirektionen.
References
|
1.
|
Engelman JA, Luo J and Cantley LC: The
evolution of phosphatidylinositol 3-kinases as regulators of growth
and metabolism. Nat Rev Genet. 7:606–619. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Saal LH, Johansson P, Holm K, et al: Poor
prognosis in carcinoma is associated with a gene expression
signature of aberrant PTEN tumor suppressor pathway activity. Proc
Natl Acad Sci USA. 104:7564–7569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Tang JM, He QY, Guo RX and Chang XJ:
Phosphorylated Akt overexpression and loss of PTEN expression in
non-small cell lung cancer confers poor prognosis. Lung Cancer.
51:181–191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Aleskandarany MA, Rakha EA, Ahmed MA, et
al: PIK3CA expression in invasive breast cancer: a biomarker of
poor prognosis. Breast Cancer Res Treat. 122:45–53. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kato S, Iida S, Higuchi T, et al: PIK3CA
mutation is predictive of poor survival in patients with colorectal
cancer. Int J Cancer. 121:1771–1778. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Chapuis N, Tamburini J, Cornillet-Lefebvre
P, et al: Autocrine IGF-1/IGF-1R signaling is responsible for
constitutive PI3K/Akt activation in acute myeloid leukemia:
therapeutic value of neutralizing anti-IGF-1R antibody.
Haematologica. 95:415–423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Muders MH, Zhang H, Wang E, Tindall DJ and
Datta K: Vascular endothelial growth factor-C protects prostate
cancer cells from oxidative stress by the activation of mammalian
target of rapamycin complex-2 and AKT-1. Cancer Res. 69:6042–6048.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Puri N and Salgia R: Synergism of EGFR and
c-Met pathways, cross-talk and inhibition, in non-small cell lung
cancer. J Carcinog. 7:92008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
De Preter K, Vandesompele J, Heimann P, et
al: Human fetal neuroblast and neuroblastoma transcriptome analysis
confirms neuroblast origin and highlights neuroblastoma candidate
genes. Genome Biol. 7:R842006.
|
|
10.
|
Dam V, Morgan BT, Mazanek P and Hogarty
MD: Mutations in PIK3CA are infrequent in neuroblastoma. BMC
Cancer. 6:1772006. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Moritake H, Horii Y, Kuroda H and Sugimoto
T: Analysis of PTEN/MMAC1 alteration in neuroblastoma. Cancer Genet
Cytogenet. 125:151–155. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Caren H, Fransson S, Ejeskar K, Kogner P
and Martinsson T: Genetic and epigenetic changes in the common 1p36
deletion in neuroblastoma tumours. Br J Cancer. 97:1416–1424. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Boller D, Schramm A, Doepfner KT, et al:
Targeting the phosphoinositide 3-kinase isoform p110delta impairs
growth and survival in neuroblastoma cells. Clin Cancer Res.
14:1172–1181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Fransson S, Martinsson T and Ejeskar K:
Neuroblastoma tumors with favorable and unfavorable outcomes:
significant differences in mRNA expression of genes mapped at
1p36.2. Genes Chromosomes Cancer. 46:45–52. 2007. View Article : Google Scholar
|
|
15.
|
Johnsen JI, Segerstrom L, Orrego A, et al:
Inhibitors of mammalian target of rapamycin downregulate MYCN
protein expression and inhibit neuroblastoma growth in vitro and in
vivo. Oncogene. 27:2910–2922. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Opel D, Poremba C, Simon T, Debatin KM and
Fulda S: Activation of Akt predicts poor outcome in neuroblastoma.
Cancer Res. 67:735–745. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Brodeur GM, Minturn JE, Ho R, et al: Trk
receptor expression and inhibition in neuroblastomas. Clin Cancer
Res. 15:3244–3250. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Fredlund E, Ringner M, Maris JM and
Pahlman S: High Myc pathway activity and low stage of neuronal
differentiation associate with poor outcome in neuroblastoma. Proc
Natl Acad Sci USA. 105:14094–14099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Hedborg F, Bjelfman C, Sparen P, Sandstedt
B and Pahlman S: Biochemical evidence for a mature phenotype in
morphologically poorly differentiated neuroblastomas with a
favourable outcome. Eur J Cancer. 31A:435–443. 1995. View Article : Google Scholar
|
|
20.
|
Brodeur GM: Neuroblastoma: biological
insights into a clinical enigma. Nat Rev Cancer. 3:203–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Chesler L, Schlieve C, Goldenberg DD, et
al: Inhibition of phosphatidylinositol 3-kinase destabilizes Mycn
protein and blocks malignant progression in neuroblastoma. Cancer
Res. 66:8139–8146. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Yamamoto M, Acevedo-Duncan M, Chalfant CE,
Patel NA, Watson JE and Cooper DR: The roles of protein kinase C
beta I and beta II in vascular smooth muscle cell proliferation.
Exp Cell Res. 240:349–358. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Borner C, Ueffing M, Jaken S, Parker PJ
and Weinstein IB: Two closely related isoforms of protein kinase C
produce reciprocal effects on the growth of rat fibroblasts.
Possible molecular mechanisms J Biol Chem. 270:78–86.
1995.PubMed/NCBI
|
|
24.
|
Zeidman R, Pettersson L, Sailaja PR, et
al: Novel and classical protein kinase C isoforms have different
functions in proliferation, survival and differentiation of
neuroblastoma cells. Int J Cancer. 81:494–501. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Svensson K, Zeidman R, Troller U, Schultz
A and Larsson C: Protein kinase C beta1 is implicated in the
regulation of neuroblastoma cell growth and proliferation. Cell
Growth Differ. 11:641–648. 2000.PubMed/NCBI
|
|
26.
|
Mak NK, Lung HL, Wong RN, Leung HW, Tsang
HY and Leung KN: Expression of protein kinase C isoforms in
euxanthone-induced differentiation of neuroblastoma cells. Planta
Med. 67:400–405. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Macfarlane DE and Manzel L: Activation of
beta-isozyme of protein kinase C (PKC beta) is necessary and
sufficient for phorbol ester-induced differentiation of HL-60
promyelocytes. Studies with PKC beta-defective PET mutant J Biol
Chem. 269:4327–4331. 1994.PubMed/NCBI
|
|
28.
|
Liu YF, Paz K, Herschkovitz A, et al:
Insulin stimulates PKCzeta-mediated phosphorylation of insulin
receptor substrate-1 (IRS-1). A self-attenuated mechanism to
negatively regulate the function of IRS proteins. J Biol Chem.
276:14459–14465. 2001.
|
|
29.
|
Nazarenko I, Jenny M, Keil J, et al:
Atypical protein kinase C zeta exhibits a proapoptotic function in
ovarian cancer. Mol Cancer Res. 8:919–934. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Guo H, Gu F, Li W, et al: Reduction of
protein kinase C zeta inhibits migration and invasion of human
glioblastoma cells. J Neurochem. 109:203–213. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Mei Y, Wang Z, Zhang L, et al: Regulation
of neuroblastoma differentiation by forkhead transcription factors
FOXO1/3/4 through the receptor tyrosine kinase PDGFRA. Proc Natl
Acad Sci USA. 109:4898–4903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Pahlman S, Johansson I, Westermark B and
Nister M: Platelet-derived growth factor potentiates phorbol
ester-induced neuronal differentiation of human neuroblastoma
cells. Cell Growth Differ. 3:783–790. 1992.
|
|
33.
|
Yu J, Ustach C and Kim HR:
Platelet-derived growth factor signaling and human cancer. J
Biochem Mol Biol. 36:49–59. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Barber DF, Alvarado-Kristensson M,
Gonzalez-Garcia A, Pulido R and Carrera AC: PTEN regulation, a
novel function for the p85 subunit of phosphoinositide 3-kinase.
Sci STKE. 2006.pe492006.PubMed/NCBI
|
|
35.
|
Rabinovsky R, Pochanard P, McNear C, et
al: p85 associates with unphosphorylated PTEN and the
PTEN-associated complex. Mol Cell Biol. 29:5377–5388. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Chagpar RB, Links PH, Pastor MC, et al:
Direct positive regulation of PTEN by the p85 subunit of
phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA.
107:5471–5476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Taniguchi CM, Winnay J, Kondo T, et al:
The phosphoinositide 3-kinase regulatory subunit p85alpha can exert
tumor suppressor properties through negative regulation of growth
factor signaling. Cancer Res. 70:5305–5315. 2010. View Article : Google Scholar
|
|
38.
|
Luo J and Cantley LC: The negative
regulation of phosphoinositide 3-kinase signaling by p85 and it’s
implication in cancer. Cell Cycle. 4:1309–1312. 2005.
|
|
39.
|
Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic
T, Orr GA and Backer JM: Regulation of the p85/p110
phosphatidylinositol 3’-kinase: stabilization and inhibition of the
p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell
Biol. 18:1379–1387. 1998.
|
|
40.
|
Shekar SC, Wu H, Fu Z, et al: Mechanism of
constitutive phosphoinositide 3-kinase activation by oncogenic
mutants of the p85 regulatory subunit. J Biol Chem.
280:27850–27855. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Jimenez C, Jones DR, Rodriguez-Viciana P,
et al: Identification and characterization of a new oncogene
derived from the regulatory subunit of phosphoinositide 3-kinase.
EMBO J. 17:743–753. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Philp AJ, Campbell IG, Leet C, et al: The
phosphatidylinositol 3’-kinase p85alpha gene is an oncogene in
human ovarian and colon tumors. Cancer Res. 61:7426–7429. 2001.
|
|
43.
|
Lopez-Carballo G, Moreno L, Masia S, Perez
P and Barettino D: Activation of the phosphatidylinositol
3-kinase/Akt signaling pathway by retinoic acid is required for
neural differentiation of SH-SY5Y human neuroblastoma cells. J Biol
Chem. 277:25297–25304. 2002. View Article : Google Scholar
|
|
44.
|
Evangelopoulos ME, Weis J and Kruttgen A:
Signalling pathways leading to neuroblastoma differentiation after
serum withdrawal: HDL blocks neuroblastoma differentiation by
inhibition of EGFR. Oncogene. 24:3309–3318. 2005. View Article : Google Scholar
|
|
45.
|
Evangelopoulos ME, Weis J and Kruttgen A:
Mevastatin-induced neurite outgrowth of neuroblastoma cells via
activation of EGFR. J Neurosci Res. 87:2138–2144. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Wilzen A, Nilsson S, Sjoberg RM, Kogner P,
Martinsson T and Abel F: The Phox2 pathway is differentially
expressed in neuroblastoma tumors, but no mutations were found in
the candidate tumor suppressor gene PHOX2A. Int J Oncol.
34:697–705. 2009.
|