Introduction
The Human Genome Project demonstrated that ≥90% of
the human genome is actively transcribed to RNA, but <2% of the
RNA encodes proteins (1,2). Long non-coding RNAs (lncRNAs) are
non-protein-coding RNA molecules longer than 200 nucleotides, which
in the past had been simply dismissed as transcriptional ‘noise’
(3). lncRNAs are highly conserved
throughout mammalian evolution, including in humans. In addition,
lncRNAs have been shown to be aberrantly expressed in various
diseases, including cancer (4). It
has been reported that lncRNAs are associated with a spectrum of
biological processes, such as gene regulation at the
transcriptional and post-transcriptional levels, chromatin
modification and epigenetics, protein activity modulation and
protein localization (5,6). In fact, lncRNAs have been recognized as
hallmarks of the onset and development of various types of cancer
(7,8).
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1), an 8.1-kb lncRNA transcribed from the 52
nuclear-enriched transcript 2, was one of the first
cancer-associated lncRNAs to be identified. In terms of its
association with cancer, MALAT1 has been shown to be oncogenic and
is overexpressed in several types of cancer (9–13). As
regards its function, MALAT1 is localized to nuclear speckles and
has been associated with a number of cancer-related processes, such
as alternative splicing and cell cycle regulation (14–17).
Recent clinical studies have demonstrated that
increased expression of MALAT1 is correlated with poor prognosis in
various types of cancer. Furthermore, MALAT1 in different human
cancers is significantly correlated with certain
clinicopathological characteristics, such as cancer
differentiation, depth of invasion and lymph node metastasis
(9,18–26).
However, a number of these studies are limited by their small size
and single-center design. Therefore, a meta-analysis was performed
to elucidate the prognostic value of MALAT1 in human cancer.
Materials and methods
Search strategy and selection
criteria
The present meta-analysis was performed in line with
the guidelines of the Meta-analysis of Observational Studies in
Epidemiology and Preferred Reporting Items for Systematic Reviews
and Meta-Analyses groups (27,28). The
PubMed/Medline, Web of Science and The Cochrane Library databases
were systematically searched (up to January, 2015) for articles
assessing the prognostic value of MALAT1 in various types of
cancer. The key words and related Medical Subject Headings for
lncRNA, MALAT1, cancer, prognosis, death and survival were used. In
addition, experts were consulted, the reference lists of retrieved
articles were reviewed, and the ‘see related articles’ links were
searched for key publications in PubMed. In addition, the authors
of the articles were contacted if necessary.
The inclusion criteria for the present analysis were
as follows: i) Studies investigating the prognostic role of MALAT1
in patients with various types of cancer; ii) providing enough
information to estimate hazard ratio (HR) and 95% confidence
interval (CI) for overall survival (OS), disease-free survival
(DFS) or recurrence-free survival (RFS); and iii) studies conducted
on adults and published in English. Duplicate studies, non-original
articles and animal experiments were excluded. Two reviewers (J.Y.
and X.Y.Z.) independently scanned all titles and abstracts
identified during the search. In addition, we obtained full-text
reports of articles that indicated or suggested eligibility,
resolving disagreements on exclusion through consensus
adjudication. Finally, a total of 10 studies were included in the
analysis (14,18–26).
Data extraction and quality
assessment
Data were independently extracted by two
investigators (J.Y. and X.Y.Z.) trained to interpret information to
ensure homogeneity in data collection and entry. These data
included name of first author, year of publication, demographic
characteristics of the patients included in the study (number, age,
gender and ethnicity), cancer characteristics (tumor type, tumor
size, differentiation, invasion, lymph node metastasis and stage),
study design (specimen, measuring method, cut-off point defining
high MALAT1 expression and follow-up period) and survival analysis.
HRs were preferentially extracted from multivariate or univariate
analyses; if these were not available, Engauge 4.0 was used to
calculate HRs and corresponding 95% CIs.
Study quality was rated by the Newcastle-Ottawa
Quality Assessment Scale for Cohort Studies (29), in which the quality of the selected
study was determined by 8 questions in 3 domains: selection (0–4
points) and comparability (0–2 points) of the study groups, and
ascertainment of the outcome of interest (0–3 points). Based on
previous recommendations, studies with 5 points were considered to
be of high quality. The two aforementioned investigators
independently evaluated the quality of each study. Disagreements
were resolved by consensus.
Statistical analysis
Estimates of odds ratios (ORs) and HRs were weighted
and pooled using the generic inverse-variance (30). ORs with 95% CIs were used to estimate
the association between MALAT1 expression and clinicopathological
characteristics. Pooled HRs with 95% CIs were used to estimate the
prognostic role of MALAT1 in various cancers. I2 was
calculated as a measure of statistical heterogeneity, with values
of 25, 50 and 75% representing mild, moderate and severe
heterogeneity, respectively. Random-effects models were applied in
cases with significant heterogeneity. Stratified analyses were
conducted to assess potential confounder contribution to
heterogeneity. In addition, publication bias was investigated using
Begg's funnel plots and Egger's linear regression test. Analyses
were performed with STATA statistical software, version 12.0 (Stata
Corporation, College Station, TX, USA). The results were considered
significant at two-sided P-value of 0.05.
Results
Characteristics of included
studies
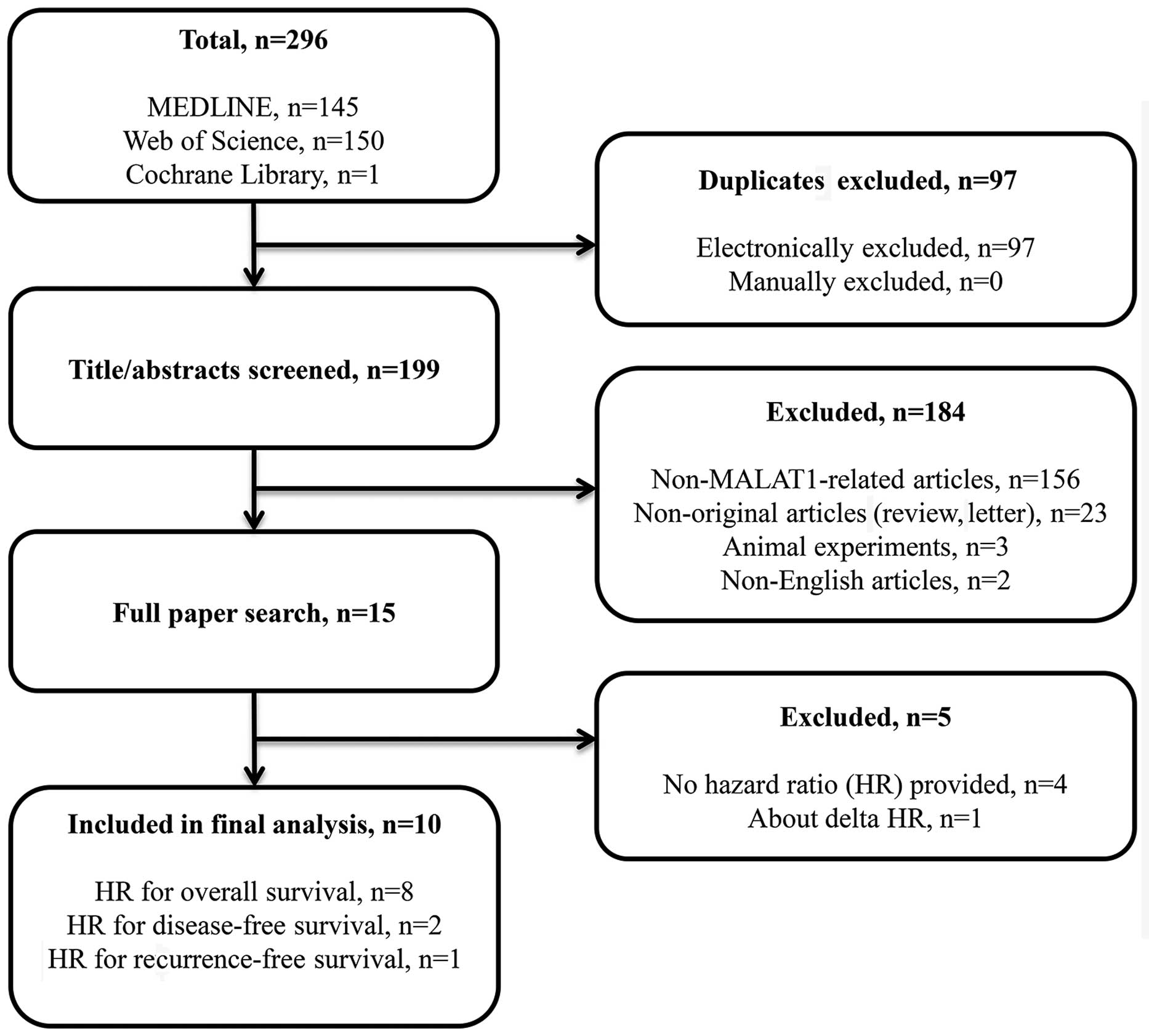
A total of 296 articles were identified during the
electronic search through PubMed/Medline, Web of Science and The
Cochrane Library. Duplicates, animal experiments,
non-MALAT1-related studies, non-English articles, and studies
lacking data on the association of MALAT1 expression with prognosis
were excluded, leaving 10 eligible studies to be included in this
meta-analysis (Fig. 1). The
characteristics of the included studies are summarized in Table I. All the studies were published from
2011 onwards. The participants in 8 studies were Chinese and in the
other 2 studies German and American. Various types of cancers were
recorded, including colorectal, non-small-cell lung and bladder
cancer, pancreatic ductal adenocarcinoma, hepatocellular carcinoma,
pancreatic cancer, clear cell renal cell carcinoma, gastric cancer
and glioma. The expression level of MALAT1 was detected by reverse
transcription quantitative polymerase chain reaction or in
situ hybridization (ISH). The sample size ranged between 45 and
222 patients. There were 8 studies for OS, 2 for DFS and 1 for RFS.
HRs with the corresponding 95% CIs were extracted from univariate
analysis and Kaplan-Meier curves in 10 studies, and from
multivariate analysis in 7 studies. The NOS scores of 90% of the
included studies were ≥5.
 | Table I.Main characteristics of all eligible
studies. |
Table I.
Main characteristics of all eligible
studies.
| Authors, year
(Refs.) | Region | Tumor type | Sample size | Method | Cut-off point | Follow-up
(months) | Outcome measures | Survival
analysis | Method score | Quality |
|---|
| Zheng et al,
2014 (18) | China | Colorectal
cancer | 146 | RT-qPCR | Median | 56.2, 11–72.8 | OS, DFS | U, M | a,b | 6 |
| Schmidt et al,
2011 (9) | Germany | NSCLC | 222 | ISH | 50% | 38 | OS | U, M | a,b | 7 |
| Fan et al,
2014 (19) | China | Bladder cancer | 95 | RT-qPCR | Median | 5–30 | OS | U | b | 4 |
| Liu et al,
2014 (20) | China | Pancreatic ductal
adenocarcinoma | 45 | RT-qPCR | Mean | 1–36 | OS | U, M | a | 7 |
| Lai et al,
2012 (21) | China | Hepatocellular
carcinoma | 112 | RT-qPCR | NR | 1–36 | RFS | U, M | a | 8 |
| Shen et al,
2015 (22) | China | NSCLC | 78 | RT-qPCR | Mean | NR | DFS | U | a | 5 |
| Pang et al,
2014 (23) | China | Pancreatic
cancer | 126 | RT-qPCR | Median | 5–60 | OS | U, M | a | 7 |
| Zhang et al,
2014 (24) | China | Clear-cell renal
cell carcinoma | 106 | RT-qPCR | Mean | 3–60 | OS | U, M | a | 7 |
| Okugawa et
al, 2014 (25) | USA | Gastric cancer | 150 | RT-qPCR | 0.985 | 1–60 | OS | U | a | 7 |
| Ma et al,
2015 (26) | China | Glioma | 118 | RT-qPCR | Median | 5–60 | OS | U, M | a | 7 |
MALAT1 expression and
clinicopathological characteristics
The main results of the association between MALAT1
expression and clinicopathological characteristics are summarized
in Table II. The results
demonstrated that MALAT1 was not associated with
clinicopathological parameters such as age, gender,
diffferentiation, depth of invasion, lymph node metastasis, distant
metastasis or tumor stage, but was significantly associated with
tumor size (HR=2.70, 95%CI: 1.23–5.92, P=0.013).
 | Table II.Main results of the meta-analysis of
the association between MALAT1 and clinicopathological
characteristics. |
Table II.
Main results of the meta-analysis of
the association between MALAT1 and clinicopathological
characteristics.
|
|
| Fixed-effects
model | Random-effects
model |
|
|---|
|
|
|
|
|
|
|---|
| Clinical
parameters | No. of studies | Pooled HR (95%
CI) | P-value | Pooled HR (95%
CI) | P-value | Heterogeneity
(I2) (%) |
|---|
| Age (young vs.
elderly) | 7 | 1.03
(0.77–1.38) | 0.854 | 1.03
(0.76–1.39) | 0.850 | 0.0 |
| Gender (male vs.
female) | 7 | 0.62
(0.85–1.15) | 0.289 | 0.87
(0.57–1.33) | 0.511 | 41.4 |
| Tumor size (small
vs. large) | 6 | 2.46
(1.77–3.41) | 0.000 | 2.70
(1.23–5.92) | 0.013 | 79.8 |
| Differentiation
(high/moderate vs. poor) | 4 | 1.27
(0.80–2.00) | 0.309 | 1.25
(0.79–1.98) | 0.336 | 0.0 |
| Depth of invasion
(T1/2 vs. T3/4) | 3 | 1.88
(1.18–3.01) | 0.008 | 2.16
(0.93–5.02) | 0.075 | 63.7 |
| Lymph node
metastasis (absent vs. present) | 5 | 1.85
(1.30–2.63) | 0.001 | 2.04
(0.94–4.45) | 0.073 | 76.6 |
| Metastasis (absent
vs. present) | 3 | 2.422
(1.09–5.37) | 0.029 | 3.47
(0.36–33.21) | 0.280 | 73.3 |
| Stage (I/II vs.
III/IV) | 4 | 1.24
(0.84–1.84) | 0.277 | 1.54
(0.56–4.29) | 0.407 | 82.3 |
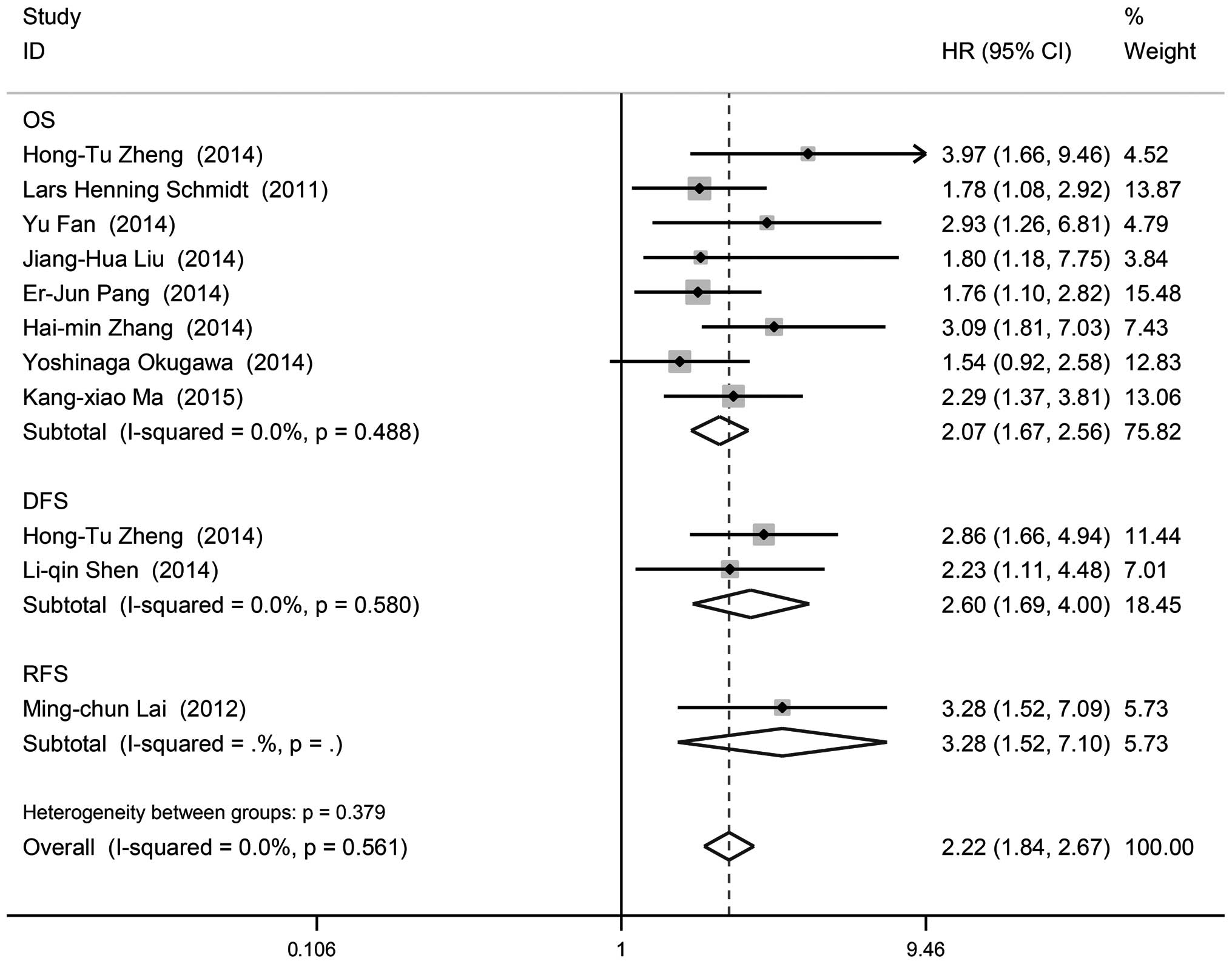
OS
A total of 8 studies including 1,008 patients
reported HRs for OS. Of the 8 eligible studies, one (12.5%)
reported a non-statistically significant HR (i.e., the 95% CIs
crossed 1). A forest plot of all studies is presented in Fig. 2. It was suggested that increased
MALAT1 expression predicted a poor outcome for OS (HR=2.07, 95% CI:
1.67–2.56, P=0.000). There appeared to be no heterogeneity between
the HRs of MALAT1 among these studies (I2=0.0%,
P=0.488). However, subgroup analysis was also conducted to
investigate the association between HRs and these variables,
including type of cancer, region of subjects, sample size, analysis
methods and quality scores (Table
III). Stratified analysis by cancer type indicated a
significant prognostic effect of MALAT1 for digestive (HR=1.86, 95%
CI: 1.37–2.53) and non-digestive system cancers (HR=2.28, 95% CI:
1.70–3.07). Although the differences in HRs between subjects from
China (HR=2.33, 95% CI: 1.79–3.03) and Western countries (HR=1.66,
95% CI: 1.16–2.37) was not statistically significant (P=0.134),
high MALAT1 expression in Chinese subjects was associated with
numerically higher HR values. Moreover, the HRs were significant
for studies including <110 subjects (HR=2.67, 95% CI: 1.69–4.23)
as well as those with >110 subjects (HR=1.93, 95% CI:
1.52–2.45). When different analysis methods were considered, MALAT1
was a strong prognostic marker by univariate (HR=1.83, 95% CI:
1.81–2.85) as well as multivariate (HR=2.15, 95% CI: 1.68–2.73)
analyses. In addition, when performing subgroup analyses stratified
by quality score, increased MALAT1 expression was significantly
associated with poor prognosis in the studies with a score of <7
(HR=3.40, 95% CI: 1.85–6.22), as well as in those with a score of
≥7 (HR=1.93, 95% CI: 1.54–2.42).
 | Table III.Main results of the pooled
analysis. |
Table III.
Main results of the pooled
analysis.
|
|
|
|
|
| Fixed-effects
model | Random-effects
model | Heterogeneity |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Survival | Variables | No. of studies | No. of
patients | Psa | Pooled HR (95%
CI) | Pzb | Pooled HR (95%
CI) | Pzb | I2
(%) | Phc |
|---|
| OS | All | 8 | 1,008 |
| 2.07
(1.67–2.56) | 0 | 2.07
(1.67–2.56) | 0 | 0.00 | 0.488 |
|
| Type of cancer |
|
| 0.345 |
|
|
|
|
|
|
|
|
Digestive | 4 | 467 |
| 1.86
(1.37–2.53) | 0 | 1.89
(1.35–2.65) | 0 | 14.20 | 0.321 |
|
|
Non-digestive | 4 | 541 |
| 2.28
(1.70–3.07) | 0 | 2.28
(1.70–3.07) | 0 | 0.00 | 0.559 |
|
| Region |
|
| 0.134 |
|
|
|
|
|
|
|
|
China | 6 | 636 |
| 2.33
(1.79–3.03) | 0 | 2.33
(1.79–3.03) | 0.005 | 0.00 | 0.542 |
|
| Western
countries | 2 | 372 |
| 1.66
(1.16–2.37) | 0.005 | 1.66
(1.16–2.37) | 0.005 | 0.00 | 0.691 |
|
| Sample size |
|
| 0.221 |
|
|
|
|
|
|
|
|
<110 | 3 | 246 |
| 2.67
(1.69–4.23) | 0 | 2.67
(1.69–4.23) | 0 | 0.00 | 0.638 |
|
|
≥110 | 5 | 762 |
| 1.93
(1.52–2.45) | 0 | 1.93
(1.52–2.46) | 0 | 1.40 | 0.399 |
|
| Analysis
method |
|
| 0.541 |
|
|
|
|
|
|
|
|
Univariate | 2 | 245 |
| 1.83
(1.81–2.85) | 0.007 | 1.94
(1.06–3.55) | 0.032 | 38.50 | 0.202 |
|
|
Multivariate | 6 | 763 |
| 2.15
(1.68–2.73) | 0 | 2.15
(1.68–2.73) | 0 | 0.00 | 0.486 |
|
| Quality score |
|
| 0.087 |
|
|
|
|
|
|
|
|
<7 | 2 | 241 |
| 3.40
(1.85–6.22) | 0 | 3.40
(1.85–6.22) | 0 | 0.00 | 0.623 |
|
| ≥7 | 6 | 767 |
| 1.93
(1.54–2.42) | 0 | 2.07
(1.67–2.56) | 0 | 0.00 | 0.658 |
| DFS | All | 2 | 224 |
| 2.60
(1.69–4.00) | 0 | 2.60
(1.69–4.00) | 0 | 0.00 | 0.58 |
| RFS | All | 1 | 112 |
| 3.28
(1.52–7.09) |
|
|
|
|
|
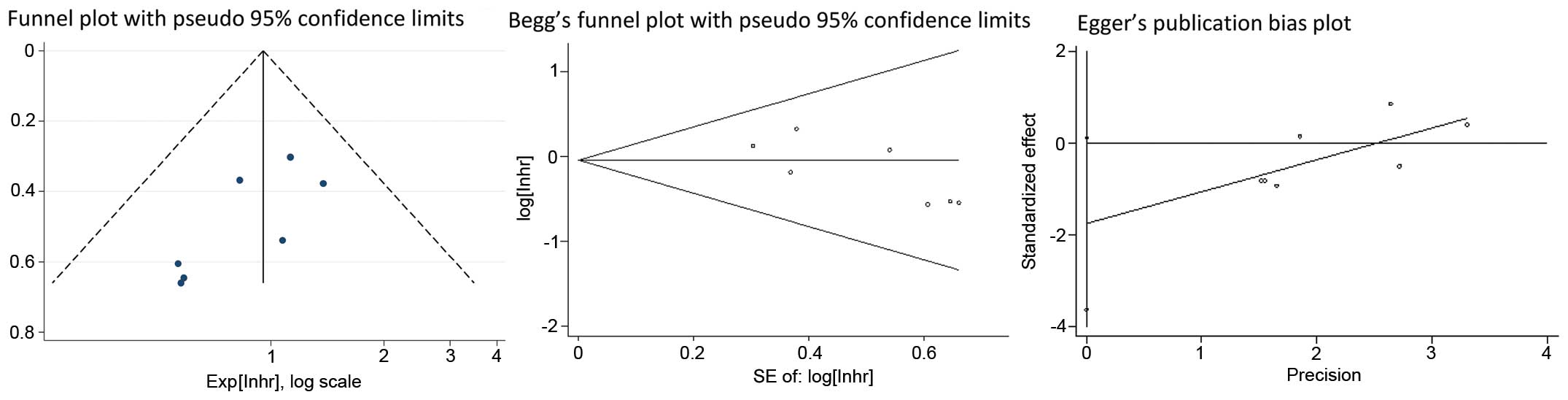
Begg's funnel plots and Egger's linear regression
tests were applied to evaluate publication bias. The shape of the
funnel plot exhibited no significant asymmetry (Fig. 3). Subsequently, Egger's test also
indicated no evidence of publication bias (P=0.174).
DFS and RFS
A total of 2 studies including 224 patients reported
HRs for DFS (Table III). Overall,
upregulation of MALAT1 was associated with an HR for DFS of 2.60
(95% CI: 1.69–4.00). In addition, 1 study including 112 patients
reported HRs for RFS (Table III).
High MALAT1 expression also predicted a poor clinical outcome for
RFS (HR=3.28, 95% CI: 1.52–7.09).
Discussion
As a novel molecular basis, the study of lncRNAs has
focused on their role in cancer pathogenesis and prognosis,
providing a new insight into cancer therapeutic strategy (31,32). This
meta-analysis was conducted to investigate the effect of lncRNA
MALAT1 on tumor prognosis.
A meta-analysis of 10 studies including 1,198 cancer
patients was undertaken. We found that increased MALAT1 expression
was significantly associated with poorer OS, DFS and RFS,
indicating that MALAT1 may be a promising prognostic marker for
cancer patients. Moreover, the significant association was retained
among various cancer type subgroups and across different regions,
sample sizes, analysis methods and quality scores. Our subgroup
analysis by region revealed that the abnormal expression of MALAT1
was strongly correlated with poor prognosis in China, as well as in
Western countries. There was a trend for the association of high
MALAT1 expression with worse OS to be more significant in the more
sensitive and accurate multivariate studies compared with
univariate analysis; however, the magnitude of its effect on OS was
lower in the subgroup with >100 participants and in that with a
quality score of >7. This contradiction indicates that more
studies including more participants are required. In addition, we
identified an association between MALAT1 and clinicopathological
characteristics, which demonstrated that high MALAT1 expression was
significantly correlated with tumor size in the random-effects
model and TNM stage in the fixed-effects model. Both Begg's and
Egger's tests revealed no significant publication bias regarding
the prognostic role of MALAT1 in different types of cancer.
The mechanisms underlying the association of high
MALAT1 expression and poor outcome of cancer patients are poorly
understood. The results of this study may suggest the following
mechanisms as being potentially involved in the prognostic effect
of MALAT1 on carcinogenesis: MALAT1, acting as an oncogene, has
been reported to be involved in the modulation of cellular
processes leading to tumor occurrence, development, metastasis and
drug resistance (13,22,25,33,34).
MALAT1 is retained in the nucleus and controls sequestration of the
paraspeckle proteins PSP1, p54, and factors linked to A-I editing,
which are implicated in mRNA nuclear retention (35); in addition, it has been proven to
participate in the phosphatidylinositol 3-kinase/Akt (36), extracellular signal-regulated
kinase/mitogen-activated protein kinase (37) and Wnt/β-catenin (38) pathways. Downregulating MALAT1 by short
hairpin RNA in the CaSki cervical cancer cell line led to a
decrease of apoptosis-inhibited gene Bcl-2 and Bcl-xL expression
and an increase of apoptosis-promoted proteins caspase-3 and −8,
and Bax (6). Consistently, silencing
of the lncRNA MALAT1 by the microRNA (miR)-101 and miR-217 was
shown to inhibit the proliferation, invasion and migration of
esophageal squamous cell carcinoma cells (39).
Certain limitations in this study should be
acknowledged. First, the number of included studies and the number
of included patients per study was relatively small. Second, only
summarized data rather than individual patient data could be
abstracted. Third, we only included studies reporting HRs and
survival curves; consequently, the excluded publications reporting
on the prognostic value of MALAT1 may lead to bias. Fourth, the
cut-off values of defining the specimens as positive or negative in
terms of MALAT1 expression in cancer patients differed among these
studies. Fifth, certain HRs could not be directly obtained from the
primary studies, requiring calculation or extraction of the HR
estimates from the survival curves. Sixth, non-original English
articles were excluded from our analysis, which may also introduce
bias. Therefore, additional studies with larger sample sizes, high
quality, different ethnic background and same cut-off value are
required to draw a more definitive conclusion.
In conclusion, increased MALAT1 expression is
associated with adverse survival in several types of cancer, and
MALAT1 may serve as an effective prognostic cancer biomarker.
Therefore, investigating the levels of MALAT1 expression in the
clinical setting is a promising approach to identifying patients
who require more intimate care and may enable personalized medical
follow-up.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81470830).
References
|
1
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Human Genome Sequencing Consortium I;
International Human Genome Sequencing Consortium: Finishing the
euchromatic sequence of the human genome. Nature. 431:931–945.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gomes AQ, Nolasco S and Soares H:
Non-coding RNAs: Multi-tasking molecules in the cell. Int J Mol
Sci. 14:16010–16039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmidt LH, Spieker T, Koschmieder S,
Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D,
Marra A, et al: The long noncoding MALAT-1 RNA indicates a poor
prognosis in non-small cell lung cancer and induces migration and
tumor growth. J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: A long non-coding RNA and its important 3 end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.PubMed/NCBI
|
|
11
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hutchinson JN, Ensminger AW, Clemson CM,
Lynch CR, Lawrence JB and Chess A: A screen for nuclear transcripts
identifies two linked noncoding RNAs associated with SC35 splicing
domains. BMC Genomics. 8:392007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miyagawa R, Tano K, Mizuno R, Nakamura Y,
Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al:
Identification of cis- and trans-acting factors involved in the
localization of MALAT-1 noncoding RNA to nuclear speckles. RNA.
18:738–751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang F, Yi F, Han X, Du Q and Liang Z:
MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett.
587:3175–3181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al:
Long noncoding RNA MALAT1 controls cell cycle progression by
regulating the expression of oncogenic transcription factor B-MYB.
PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
19
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shen L, Chen L, Wang Y, Jiang X, Xia H and
Zhuang Z: Long noncoding RNA MALAT1 promotes brain metastasis by
inducing epithelial-mesenchymal transition in lung cancer. J
Neurooncol. 121:101–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 26:2947–2955. 2015. View Article : Google Scholar
|
|
25
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, et
al: Metastasis-associated long non-coding RNA drives gastric cancer
development and promotes peritoneal metastasis. Carcinogenesis.
35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma KX, Wang HJ, Li XR, Li T, Su G, Yang P
and Wu JW: Long noncoding RNA MALAT1 associates with the malignant
status and poor prognosis in glioma. Tumour Biol. 36:3355–3359.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stroup DF, Berlin JA, Morton SC, Olkin I,
Williamson GD, Rennie D, Moher D, Becker BJ, Sipe TA and Thacker
SB: Meta-analysis of observational studies in epidemiology: A
proposal for reporting. Meta-analysis Of Observational Studies in
Epidemiology (MOOSE) group. JAMA. 283:2008–2012. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liberati A, Altman DG, Tetzlaff J, Mulrow
C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J
and Moher D: The PRISMA statement for reporting systematic reviews
and meta-analyses of studies that evaluate health care
interventions: Explanation and elaboration. Ann Intern Med.
151:W65–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oremus M, Oremus C, Hall GB and McKinnon
MC: ECT & Cognition Systematic Review Team: Inter-rater and
test-retest reliability of quality assessments by novice student
raters using the Jadad and Newcastle-Ottawa Scales. BMJ Open.
2:22012. View Article : Google Scholar
|
|
30
|
van der Molen HF, Hoonakker PL, Lehtola
MM, Hsiao H, Haslam RA, Hale AR and Verbeek JH: Writing a Cochrane
systematic review on preventive interventions to improve safety:
The case of the construction industry. Med Lav. 100:258–267.
2009.PubMed/NCBI
|
|
31
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Necsulea A, Soumillon M, Warnefors M,
Liechti A, Daish T, Zeller U, Baker JC, Grützner F and Kaessmann H:
The evolution of lncRNA repertoires and expression patterns in
tetrapods. Nature. 505:635–640. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen H, Xin Y, Zhou L, Huang JM, Tao L,
Cheng L and Tian J: Cisplatin and paclitaxel target significant
long noncoding RNAs in laryngeal squamous cell carcinoma. Med
Oncol. 31:2462014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of long non-coding RNAs on tumorigenesis and
glioma development. Brain Tumor Res Treat. 2:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT1 promotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ji Q, Liu X, Fu X, Zhang L, Sui H, Zhou L,
Sun J, Cai J, Qin J, Ren J, et al: Resveratrol inhibits invasion
and metastasis of colorectal cancer cells via MALAT1 mediated
Wnt/β-catenin signal pathway. PLoS One. 8:e787002013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|