Introduction
microRNAs (miRNAs) are noncoding RNA molecules,
19–24 nucleotides in length, that repress post-transcriptional gene
expression. miRNAs are important in maintaining normal human body
physiological conditions and the abnormal expression of miRNA is
associated with several human diseases, ranging from psychiatric
disorders (1) to various types of
malignant cancer (2,3). In addition, they are important in
regulating host gene expression in virally infected cells and in
types of cancer caused by viral infection (4–7).
miR-122 is a liver specific microRNA and is the most
abundantly expressed type of microRNA in the liver (8,9).
Previous studies suggest that miR-122 is involved in maintaining
the normal function of the liver (10–12).
Esau et al (13)
demonstrated that miR-122 positively regulates lipid metabolism by
reducing lipid-associated protein mRNA and that inhibiting the
expression of miR-122 attenuates liver steatosis in mice fed a
high-fat diet. miR-122 can activate the translation of p53 mRNA
through the suppression of cytoplasmic polyadenylation
element-binding protein and is involved in cellular senescence
(14). miR-122 is also known to be
involved in cholesterol synthesis (13,15).
The hepatitis C virus (HCV) is a positive-sense
single-stranded RNA virus with a 9.6 kb genome that establishes
persistent infections in the liver, eventually leading to cirrhosis
and carcinoma. Notably, miR-122 is used by HCV for triggering viral
replication by repression of heme oxygenase-1 (16) or by interaction with the 5′
untranslated region (UTR) of HCV RNA (17,18).
In addition, miR-122 was found to be dysregulated in different
stages of HCV-infected liver tissues and serum (19,20).
The present study identified target genes of
miR-122. The effect of HCV mRNA overexpression on expression of
target genes was investigated, as well as downstream effects which
reduced cellular immune responses to HCV infection.
Materials and methods
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR analysis was used to determine the relative
expression level of miR-122. Total RNA was extracted from the
tissues using TRIzol (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer’s instructions. The expression
level of miR-122 was detected using TaqMan miRNA RT-qPCR.
Single-stranded cDNA was synthesized using a TaqMan microRNA
Reverse Transcription kit (Applied Biosystems, Foster City, CA,
USA) and then amplified using TaqMan Universal PCR Master mix
(Applied Biosystems) with an miRNA-specific TaqMan MGB probe,
miR-122-5p (Applied Biosystems). U6 snRNA was used for
normalization. Each sample was measured in triplicate and the
experiment was repeated at least three times for the detection of
miR-122.
To detect HCV RNA in plasmid-transfected cells,
total RNA was first subjected to reverse transcription using a
random hexamer as a primer. The cDNA was then subjected to qPCR and
relative mRNA was calculated by normalizing the values of the
indicated genes to that of β-actin.
Cell culture
Huh7 cells (American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s
medium (Invitrogen Life Technologies) containing 10% fetal bovine
serum (HyClone, Logan, UT, USA), 100 IU/ml penicillin and 10 mg/ml
streptomycin (Sigma, St. Louis, MO, USA). All cells were maintained
at 37°C in a 5% CO2 atmosphere.
3′UTR and 5′UTR luciferase reporter
assays
To generate the 3′UTR luciferase reporter, a segment
of 567 bp 3′UTR from STAT3 was cloned into the downstream of the
firefly luciferase gene in a pGL3-control vector (Promega Corp.,
Madison, WI, USA). The miR-122 mimic and miR-122 inhibitor were
synthesized by GenePharma Co., Ltd. (Shanghai, China). A thymidine
kinase promoter-Renilla luciferase reporter plasmid (pRL-TK) vector
containing Renilla luciferase (Promega Corp.) was co-transfected
for data normalization. For the luciferase reporter assays, Huh7
cells were seeded into 48-well plates. Luciferase reporter vectors
were co-transfected with the miR-122 mimic or inhibitor using
lipofectamine 2000 (Invitrogen Life Technologies). After 2 days,
the cells were harvested and assayed using a Dual-Luciferase assay
(Promega Corp.). Each treatment was performed in triplicate in
three independent experiments. The results were expressed as
relative luciferase activity (firefly luciferase/Renilla
luciferase).
To generate the 5′UTR luciferase reporter, wild type
or mutant HCV RNA 5′UTR was cloned into the upstream of the firefly
luciferase gene in pGL3-Basic vector (Promega Corp.). miR-122 mimic
and pGL3-Basic-HCV 5′UTR vectors were co-transfected into Huh7
cells and a pRL-TK vector was used for data normalization.
Western blot analysis
Protein extracts were boiled in sodium dodecyl
sulphate/β-mercaptoethanol sample buffer (Sangon Biotech Co., Ltd,
Shanghai, China) and 20 μg samples were loaded into each lane of
10% polyacrylamide gels. The proteins were separated by
electrophoresis and the proteins in the gels were blotted onto
polyvinylidene difluoride membranes (Amersham Pharmacia Biotech,
St. Albans, UK) by electrophoretic transfer. Antibodies against the
following proteins were used: Monoclonal anti-tubulin (Santa Cruz
Biotechnology, Inc., Austin, TX, USA), anti-β-actin (Santa Cruz
Biotechnology, Inc.) and anti-STAT3 (Cell Signaling Technology,
Inc., Danvers, MA, USA) antibodies (goat anti-human).
Statistical analysis
Data were analyzed using an SPSS statistical
package, version 16 (SPSS, Inc., Chicago, IL, USA) and a two group
independent samples t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-122 represses the expression of STAT3
by targeting the STAT3 mRNA 3′UTR
miRNA is an important post-transcriptional negative
regulator for protein coding genes and may directly target numerous
genes. miR-122 is a liver abundant miRNA, which has several
functions, including the control of lipid metabolism. To expand on
the current knowledge of the function of miR-122 during HCV
infection, the present study searched for new target genes using
bioinformatics tools. Based on the prediction of the online
bioinformatics tool, TargetScan (http://www.targetscan.org/), STAT3 mRNA was identified
as a potential direct target of the miR-122 3′UTR.
To validate whether STAT3 is indeed the target gene
of miR-122, a 567 bp segment of STAT3 3′UTR containing the
predicted miR-122 binding site was cloned downstream in the firefly
luciferase reporter gene, in the pGL3 control vector (designated as
pGL3-STAT3) for the dual luciferase assay (Fig. 1A). Huh7 cells were co-transfected
with pGL3-STAT3 and the miR-122 mimic or inhibitor. Compared with
the miRNA control, the luciferase activity was significantly
suppressed by miR-122, by ~42.1% (P<0.05; Fig. 1B). Furthermore, the luciferase
activity was significantly upregulated by the miR-122 inhibitor
compared with the anti-miR control by ~38.4% (P<0.05). These
results indicated that miR-122 targeted the 3′UTR of STAT3, leading
to a change in firefly luciferase translation.
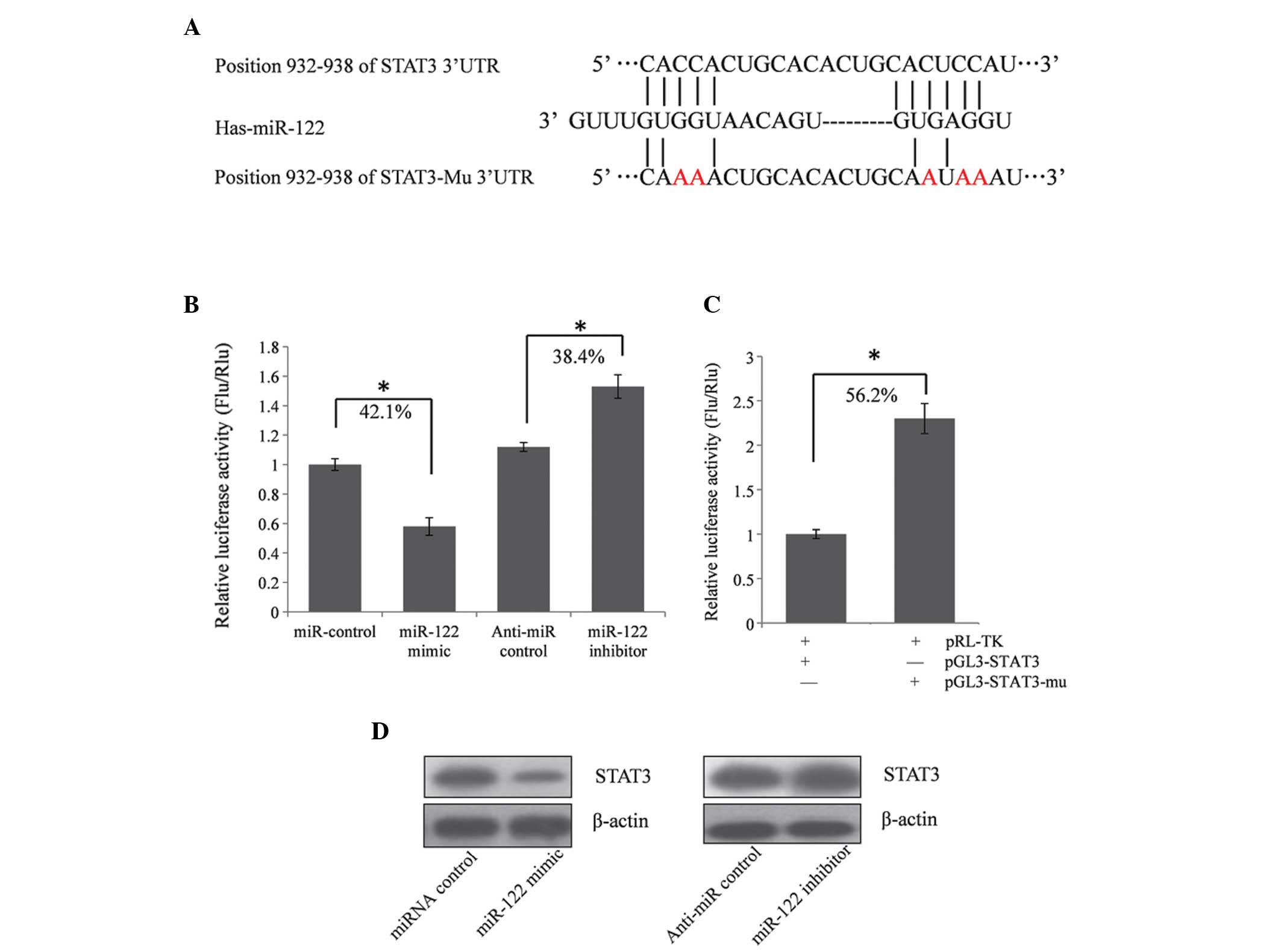 | Figure 1miR-122 suppresses the expression of
STAT3 by targeting the STAT3 mRNA 3′UTR. (A) Predicted miR-122
binding site in STAT3 3′UTR. (B) Confirmation of the target gene of
miR-122. Huh7 cells were co-transfected with the miRNA control,
miR-122 mimic, anti-miR control or miR-122 inhibitor and pGL3-STAT3
for a dual-luciferase assay. PRL-TK containing Rlu was
co-transfected for data normalization. (C) Mutation analysis of the
miR-122 binding sites. When five nucleotides of the miR-122 binding
site were mutated (pGL3-STAT3-Mu), the luciferase activity was
significantly increased compared with the wild type STAT3. (D)
STAT3 protein level in the miR-122 mimic or inhibitor-treated Huh7
cells was detected by western blot analysis. *P<0.05
vs. controls. miR, microRNA; STAT3, signal transducer and activator
of transcription 3; 3′UTR, 3′ untranslated region; PRL-TK,
thymidine kinase promoter-Renilla luciferase reporter plasmid; mu,
mutant; Rlu, Renilla luciferase; Flu, firefly luciferase. |
A seed sequence mutation clone was also used to
further confirm the binding site for miR-122 (Fig. 1A). The vector containing a putative
miR-122 binding region in the 3′UTR of STAT3 with five mutant
nucleotides (designated as pGL3-STAT3-Mu) was used and a wild type
STAT3 vector was used as a control. The histogram in Fig. 1C shows that the enzyme activity was
reduced by ~56.2% in cells transfected with pGL3-STAT3 compared
with pGL3-STAT3-Mu (P<0.05). These data indicated that miR-122
may have suppressed the expression of STAT3 by binding to the seed
sequence at the 3′UTR of STAT3 and that STAT3 may be a direct
target gene of miR-122.
miR-122 inhibits endogenous STAT3
expression in Huh7 cells
Although STAT3 was identified as a target gene for
miR-122, it remains to be elucidated whether miR-122 is able to
regulate endogenous expression of STAT. Huh7 cells were transfected
with either miR-122 mimics or an miR-122 inhibitor to establish
whether the dysregulation of miR-122 expression affected the
endogenous expression of STAT3. Compared with the corresponding
control, the protein level of STAT3 was significantly suppressed by
the miR-122 mimics and upregulated by the miR-122 inhibitor
(Fig. 1D).
HCV RNA overexpression sponges miR-122
and rescues the expression of STAT3 in Huh7 cells
‘miRNA sponges’ are miRNA competitive inhibitors,
which are expressed from strong promoters, containing multiple,
tandem binding sites to one or several miRNAs of interest. When
vectors encoding these sponges are transiently transfected into
cultured cells, sponges can derepress miRNA targets (21).
To examine whether HCV RNA can protect the
expression of miR-122 target genes by interacting with miR-122, the
experiment revealing the ability of miR-122 to target HCV mRNA
5′UTR was repeated (Fig. 2A). As
shown in Fig. 2B, overexpression
of miR-122 enhanced the expression of luciferase compared with the
scramble miR-control. When three nucleotides of the miR-122 target
sites were changed, the relative firefly luciferase activity was
significantly reduced. Transient transfection of the vector
encoding 9658 bp HCV genotype 1b RNA into Huh7 cells was performed
and RT-qPCR was used to determine the expression of miR-122 24 h
after transfection. The present study demonstrated that expression
of miR-122 partially reduced compared with the control, however,
the difference was not significant (Fig. 2C). Subsequently, the expression of
STAT3 was assessed by western blot analysis. Notably, the
expression of STAT3 was upregulated by overexpression of HCV RNA
compared with the empty vector and the miR-122 target site mutant
vector (Fig. 2D), suggesting that
HCV RNA protected the expression of STAT3 by absorbing miR-122.
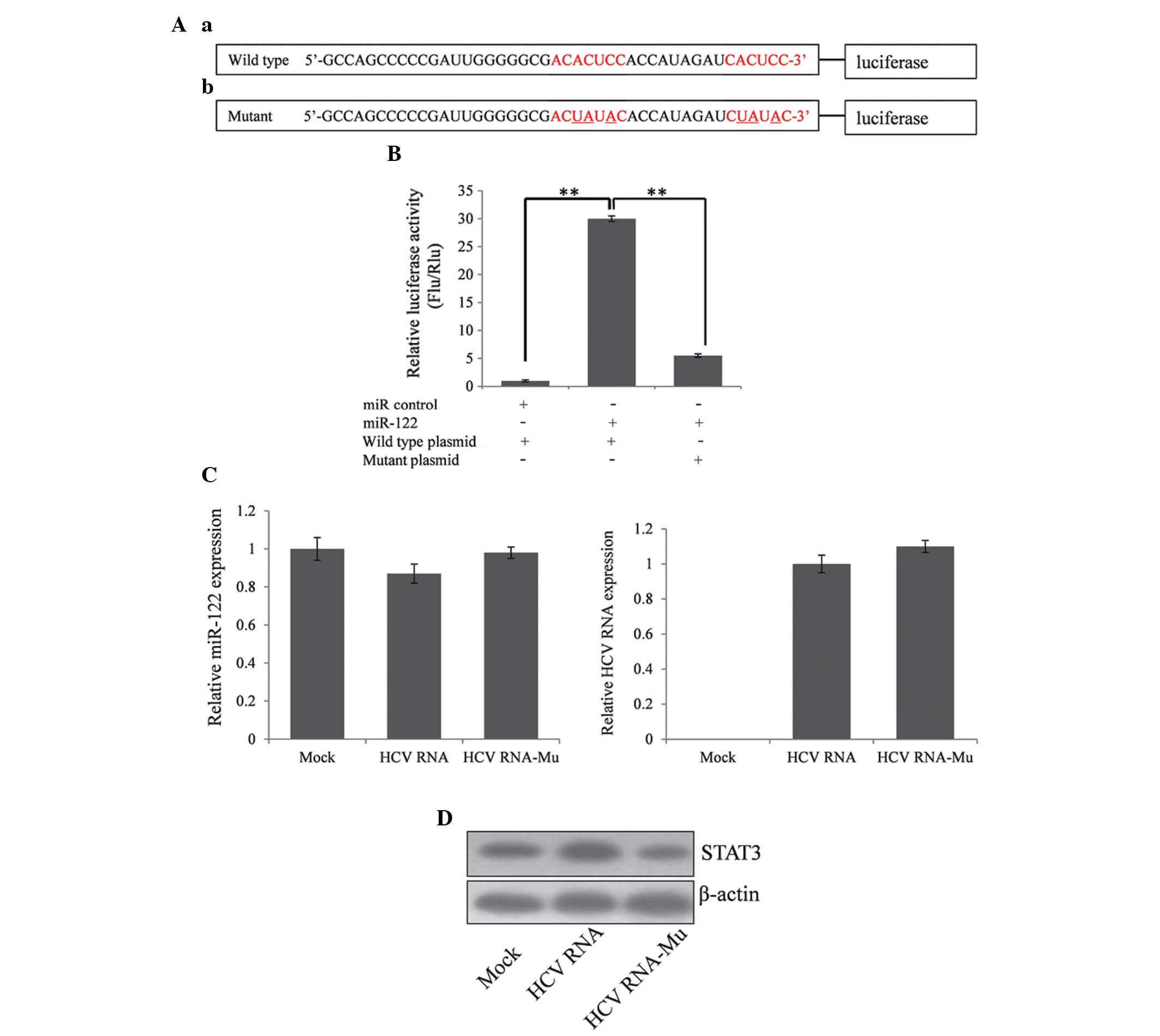 | Figure 2HCV mRNA represses the expression of
polyinosinic-polycytidylic acid-stimulated type I interferon by
upregulating STAT3. (Aa) Wild type and miR-122 target site mutant
HCV mRNA 5′UTR were added prior to the firefly luciferase coding
sequence of the pGL3-basic vector. (Ab) A dual luciferase assay was
used to detect the interaction between the miR-122 and the HCV
5′UTR. miR-122 enhanced luciferase activity by targeting the HCV
5′UTR. (B) When the binding sites were mutated, the relative
luciferase activity was significantly reduced. (C) miR-122
expression reduced when the wild type HCV mRNA was overexpressed,
however, the difference was not significant. (D) The expression of
STAT3 was upregulated by wild type HCV mRNA rather than the miR-122
target site mutant HCV mRNA, thus, HCV mRNA protects the expression
of STAT3 against inhibition from miR-122. *P<0.05,
**P<0.01 vs. cells co-transfected with miR112 and
wild-type plasmid. HCV, hepatitis C virus; miR, microRNA; 5′UTR,
untranslated region; STAT3, signal transducer and activator of
transcription 3; Mu, mutant. |
HCV RNA overexpression inhibits the
expression of IFN by rescuing STAT3
STAT3 is considered to be a negative regulator of
the type I IFN-mediated antiviral response. STAT3 knockdown or
knockout cells exhibit enhanced gene expression and antiviral
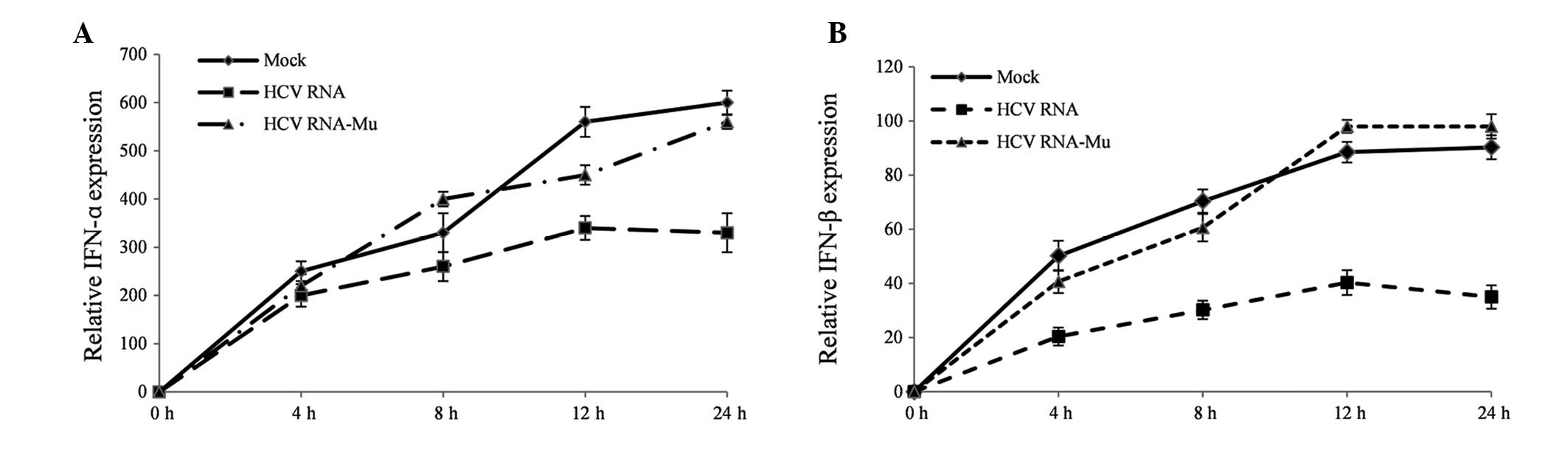
activity in response to IFN-α and IFN-β (22). To examine the response of HCV RNA
overexpression on the generation of type I IFN, the present study
detected the expression of IFN-α and IFN-β using RT-qPCR at four
time points following plasmid and polyinosinic-polycytidylic acid
transfection. HCV RNA repressed the expression of IFN-α (Fig. 3A) and IFN-β (Fig. 3B), however, the miR-122 target site
mutant HCV RNA did not, suggesting that HCV RNA repressed the cell
anti-viral response by absorbing miR-122 directly.
Discussion
HCV is a positive-sense single-stranded RNA virus.
At present, >170 million individuals worldwide are chronically
infected with HCV, and cirrhosis and hepatocellular carcinoma
induced by HCV infection are life-threatening diseases (23,24).
miR-122 is a liver-specific miRNA and its ability to promote,
rather than inhibit, HCV RNA function has rendered miR-122 an
attractive therapeutic target. In the present study, STAT3 was
initially confirmed as a direct target gene of miR-122. STAT3 is a
signaling mediator of the interleukin (IL)-6 and IL-10 family
members and other cytokines, including leptin and granulocyte
colony-stimulating factor. Previous studies have identified a
negative effect of STAT3 in the type I IFN response (22,25).
Ho and Ivashkiv (22) demonstrated
that the overexpression of STAT3 in THP-1 cells downregulated IFN-α
activated, STAT1-dependent genes, including interferon regulatory
factor 1, CXC chemokine ligand (CXCL)9 and CXCL10. In addition
STAT3 knockdown resulted in an increase in the expression of the
same genes. Using gain-of-function and loss-of-function approaches,
Wang et al (26)
demonstrated that STAT3 negatively regulates the type I
IFN-mediated response. Previous studies have also demonstrated that
hepatic and circulating levels of miR-122 in HCV-infected patients
were altered, indicating that HCV may regulate the host antiviral
response by disturbing the expression of miR-122 (19,27,28).
The production of cell lines and transgenic
organisms with continuous miRNA loss of function is enabled by a
method termed the miRNA ‘sponge’. The sponge mRNA usually contains
multiple target sites complementary to the miRNA of interest. To
determine whether the HCV mRNA can act as an miR-122 sponge
molecule, the HCV mRNA was overexpressed in Huh7 cells and the
expression of miR-122 and STAT3 was detected. Although the
expression of miR-122 was not significantly altered, the expression
of STAT3 was rescued by HCV mRNA. These results indicated that HCV
mRNA protected the miR-122 target genes by sponging miR-122.
In conclusion, the present study revealed another
aspect of the association between HCV and miR-122. Previous studies
have demonstrated that the expression of miR-122 enhances the
propagation of HCV through genetic interaction with the 5′UTR of
the HCV genome (29,30). miR-122 is a liver specific miRNA,
the function of which is mainly associated with the maintenance of
normal liver physiology. Therefore, it was hypothesized that
miR-122 may be involved in the antiviral response of the liver. The
present study initially predicted and confirmed that STAT3 is a
target gene of miR-122. Subsequently, the wild type HCV genome RNA
was found to protect the expression of STAT3 by absorbing miR-122,
which functions as an RNA sponge. Subsequently, it was demonstrated
that the HCV genome RNA inhibited the expression of IFN-α and IFN-β
by upregulating the expression of STAT3, indicating that HCV RNA
may act as an miR-122 sponge molecule that protects genes that are
of benefit to the virus. The present study provided a further
understanding of the complex roles of miR-122 in the interaction
between HCV and host factors and supported the use of an miR-122
inhibition strategy in the treatment of HCV infection.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant nos. 81171643 and 30872230).
References
|
1
|
Maes OC, Chertkow HM, Wang E and Schipper
HM: MicroRNA: Implications for Alzheimer disease and other human
CNS disorders. Curr Genomics. 10:154–168. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu J, Li Y, Wang F, et al: Suppressed
miR-424 expression via upregulation of target gene Chk1 contributes
to the progression of cervical cancer. Oncogene. 32:976–987. 2013.
View Article : Google Scholar
|
|
3
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singaravelu R, Chen R, Lyn RK, et al:
Hepatitis C virus induced up-regulation of microRNA-27: A novel
mechanism for hepatic steatosis. Hepatology. 59:98–108. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang J, Wang F, Argyris E, et al:
Cellular microRNAs contribute to HIV-1 latency in resting primary
CD4+ T lymphocytes. Nat Med. 13:1241–1247. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S, Qiu L, Yan X, et al: Loss of
microRNA 122 expression in patients with hepatitis B enhances
hepatitis B virus replication through cyclin G(1)-modulated P53
activity. Hepatology. 55:730–741. 2012. View Article : Google Scholar
|
|
7
|
Lajer CB, Garnaes E, Friis-Hansen L, et
al: The role of miRNAs in human papilloma virus (HPV)-associated
cancers: bridging between HPV-related head and neck cancer and
cervical cancer. Br J Cancer. 106:1526–1534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang J, Provost P and Taylor JM:
Resistance of human hepatitis delta virus RNAs to dicer activity. J
Virol. 77:11910–11917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jopling CL, Schütz S and Sarnow P:
Position-dependent function for a tandem microRNA miR-122-binding
site located in the hepatitis C virus RNA genome. Cell Host
Microbe. 4:77–85. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Castoldi M, Vujic Spasic M, Altamura S, et
al: The liver-specific microRNA miR-122 controls systemic iron
homeostasis in mice. J Clin Invest. 121:1386–1396. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esau C, Davis S, Murray SF, et al: miR-122
regulation of lipid metabolism revealed by in vivo antisense
targeting. Cell Metab. 3:87–98. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Burns DM, D’Ambrogio A, Nottrott S and
Richter JD: CPEB and two poly(A) polymerases control miR-122
stability and p53 mRNA translation. Nature. 473:105–108. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wen J and Friedman JR: miR-122 regulates
hepatic lipid metabolism and tumor suppression. J Clin Invest.
122:2773–2776. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shan Y, Zheng J, Lambrecht RW and
Bonkovsky HL: Reciprocal effects of micro-RNA-122 on expression of
heme oxygenase-1 and hepatitis C virus genes in human hepatocytes.
Gastroenterology. 133:1166–1174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jopling CL, Yi M, Lancaster AM, Lemon SM
and Sarnow P: Modulation of hepatitis C virus RNA abundance by a
liver-specific MicroRNA. Science. 309:1577–1581. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krutzfeldt J, Rajewsky N, Braich R, et al:
Silencing of microRNAs in vivo with ‘antagomirs’. Nature.
438:685–689. 2005. View Article : Google Scholar
|
|
19
|
Trebicka J, Anadol E, Elfimova N, et al:
Hepatic and serum levels of miR-122 after chronic HCV-induced
fibrosis. J Hepatol. 58:234–239. 2013. View Article : Google Scholar
|
|
20
|
Choi Y, Dienes HP and Krawczynski K:
Kinetics of miR-122 expression in the liver during acute HCV
infection. PloS One. 8:e765012013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ho HH and Ivashkiv LB: Role of STAT3 in
type I interferon responses. Negative regulation of STAT1-dependent
inflammatory gene activation. J Biol Chem. 281:14111–14118. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge D, Fellay J, Thompson AJ, et al:
Genetic variation in IL28B predicts hepatitis C treatment-induced
viral clearance. Nat. 461:399–401. 2009. View Article : Google Scholar
|
|
24
|
Ohata K, Hamasaki K, Toriyama K, et al:
Hepatic steatosis is a risk factor for hepatocellular carcinoma in
patients with chronic hepatitis C virus infection. Cancer.
97:3036–3043. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Minegishi Y, Saito M, Tsuchiya S, et al:
Dominant-negative mutations in the DNA-binding domain of STAT3
cause hyper-IgE syndrome. Nature. 448:1058–1062. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang WB, Levy DE and Lee CK: STAT3
negatively regulates type I IFN-mediated antiviral response. J
Immunol. 187:2578–2585. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu L, Fan H, Jin W, et al:
miR-122-induced down-regulation of HO-1 negatively affects
miR-122-mediated suppression of HBV. Biochem Bioph Res Co.
398:771–777. 2010. View Article : Google Scholar
|
|
28
|
Jopling CL: Regulation of hepatitis C
virus by microRNA-122. Biochem Soc Trans. 36(Pt 6): 1220–1223.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mortimer SA and Doudna JA: Unconventional
miR-122 binding stabilizes the HCV genome by forming a trimolecular
RNA structure. Nucleic Acids Res. 41:4230–4240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kambara H, Fukuhara T, Shiokawa M, et al:
Establishment of a novel permissive cell line for the propagation
of hepatitis C virus by expression of microRNA miR122. J Virol.
86:1382–1393. 2012. View Article : Google Scholar :
|