Cancer tissue is a sophisticated construct of both
malignant tumor cells and nonmalignant host stromal cells. In 1889,
Paget (1) proposed a novel
concept, the ‘seed and soil’ hypothesis, postulating that the
congenial microenvironment (the ‘soil’) is prerequisite for the
progression of tumor cells (the ‘seeds’). Tumor cells are
disseminated throughout the body via the blood stream, but only in
congenial ‘soil’ can metastases develop. In the past, cancer
research primarily focused on neoplastic cells. This led to a rapid
progression of knowledge pertaining to the genetic and epigenetic
changes they undergo and elucidation of their signaling pathways in
tumor cells (2,3). Despite the advancement of knowledge
in the malignant transformation of tumor cells, existing therapies
remain relatively ineffective for most types of cancer. Hertenstein
et al (2), Sala-Torra et
al (5) and Xiao et al
(6), respectively, reported that
leukemia patients suffered from donor cell leukemia (DCL) following
allogeneic hematopoietic stem cell transplantation (allo-HSCT). In
Xiao et al’s study (6), the
patient in question as well as his donor-sister had the CCAAT
enhancer binding protein α genetic abnormality, however, leukemia
did not manifest in the patient’s sister. Other studies have also
demonstrated that cells containing abnormal genetic changes only
lead to tumor formation in a congenial microenvironment (7–10).
The results of the aforementioned studies revealed
that genetic abnormality in tumor cells alone is not sufficient to
produce cancer cells with malignant characteristics. The tumor
microenvironment may be a necessity in the inception of malignant
tumors and is increasingly being recognized to have a vital role in
the progression of solid tumors and hematological malignancies
(11–14). Cancer-associated fibroblasts (CAFs)
are the most ubiquitous element of tumor stroma and are found in
numerous types of cancer, including breast (15,16),
NSCLC (17), colorectal (18–20),
liver (21) and prostate cancer
(22). The exact origin and
specific markers of CAFs remain to be elucidated. Contemporary
knowledge suggests that CAFs may be derived from: i) Local
resident fibroblasts that undergo education by tumor cell-secreted
cytokines; ii) bone marrow-derived mesenchymal stem cells
(BMMSCs); iii) cancer cells undergoing
epithelial-mesenchymal transition (EMT); iv) endothelial
cells undergoing endothelial-to-mesenchymal transition (EndoMT);
and v) other mechanisms (23–25).
Fibroblast activation protein α (FAPα) is an important surface
marker of CAFs. Aggregated data revealed that the elimination of
FAPα led to stunted tumor growth and progression and stimulated the
immune system to enhance the effects of tumor vaccination (26–30).
In the present review, the current knowledge
regarding the role of FAPα in the interaction between cancer cells
and the tumor microenvironment, as well as its biological and
therapeutic implications, were summarized.
FAPα, expressed in activated stromal fibroblasts and
remodeling tissue, is a type II cell-surface-bound transmembrane
glycoprotein with Mr 95,000. It consists of 760 amino acids, most
of which possess a hydrolytic area exposed laterally of the
plasmalemma. ~20 amino acids are anchored in the plasma membrane,
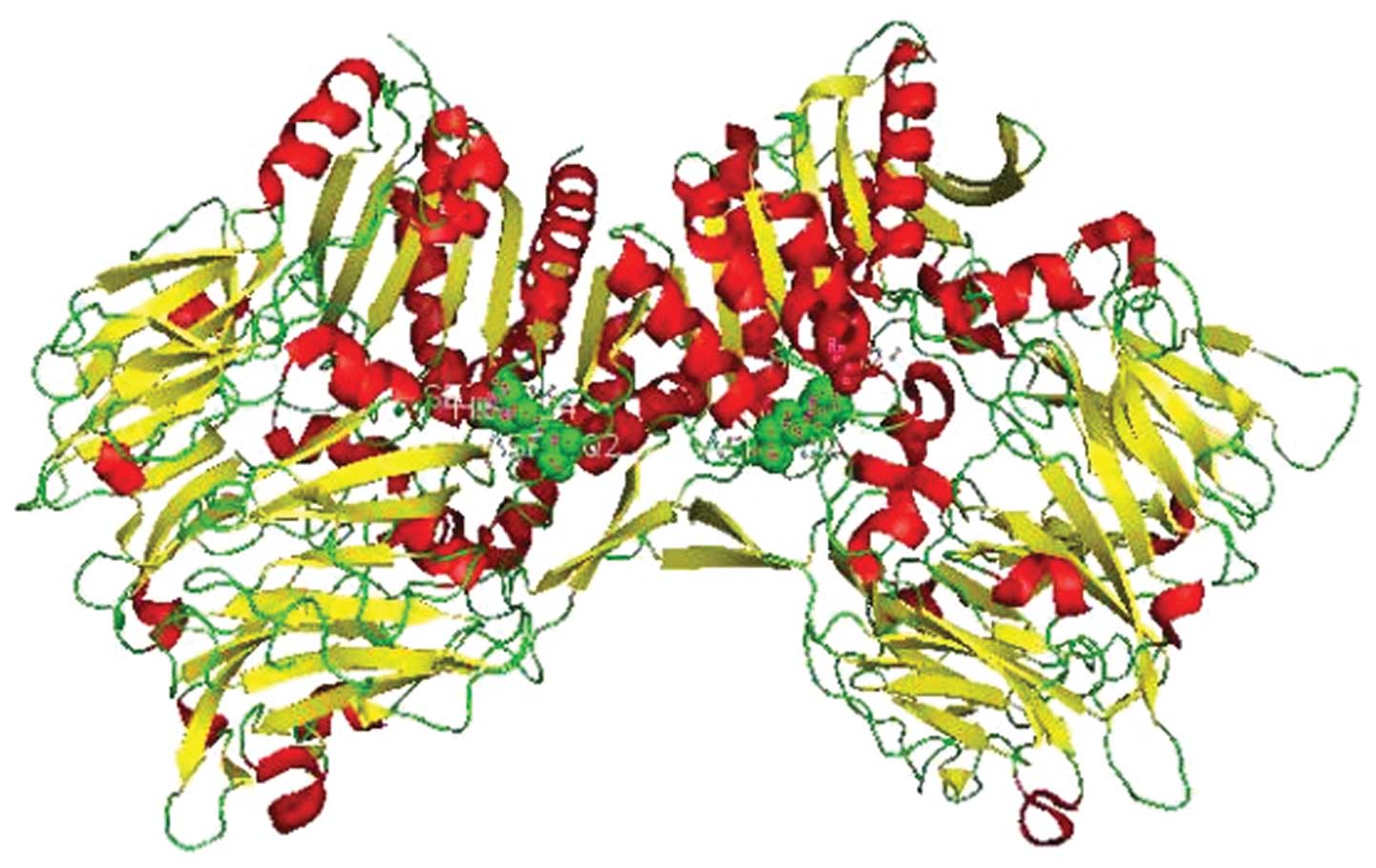
and 6 amino acids are located in the cytoplasm (42). The conserved catalytic triad of
FAPα is comprised of serine (S624), aspartate (D702) and histidine
(H734) (42,43) (Fig.
1). FAPα is a member of the peptidase S9b family, a serine
prolyl oligopeptidase subfamily, with post-prolyl peptidase
activities able to cleave proteins and peptides following proline
residues at the penultimate and P1 positions (44). In addition to FAPα (EC=3.4.21),
this S9b serine peptidase family includes dipeptidyl peptidase 4
(DPP4, also termed CD26, which is identical to FAPβ, EC=3.4.14.5),
dipeptidyl aminopeptidase-like protein 6 (also named DPPX or DPP6),
DPP8 (EC=3.4.14.5), DPP9 (EC=3.4.14.5) and DPP10, and has been
implicated in diabetes, cancer and inflammatory diseases (45–47)
(additional information is available at: http://www.uniprot.org; http://enzyme.expasy.org). FAPα shares 48% amino acid
sequence identity with DPP4 (35).
FAPα and DPP4 are able to form homodimer FAPα/FAPα or heterodimer
FAPα/DPP4 complexes to execute functions. The FAPα monomer is
inactive, therefore dimerization is prerequisite for its catalytic
function (43,48,49).
FAPα and DPP4 are encoded by genes on human chromosomes 2q23 and
2q24.3, respectively (41,50). DPP8 and DPP9 are localized to
chromosomes 15q22 and 19p13.3, respectively (51). DPP6 is encoded by a gene on human
chromosome 7 (41,52) and DPP10 is encoded by a gene
localized to chromosome 2 (2q12.3–2q14.2) (47). Murine FAPα shares 89%
amino-acid-sequence identity with human FAPα (37). A promoter element of FAPα, early
growth response 1 (EGR1), has been described (53).
Approximately 90% of reactive stromal fibroblasts of
epithelial tumors, but not malignant tumor cells, overexpress FAPα
(31,54). Immunohistochemical analysis using
formalin-fixed and paraffin-embedded sections disclosed expression
of FAPα in infiltrating ductal carcinomas (IDC) (55). The data indicated that the majority
of stromal fibroblasts of epithelial tumors and certain malignant
tumor cells are characterized by an overexpression of FAPα
(Table I).
Further to overexpression in the cells and tissues
mentioned above and summarized in Table I, FAPα is also expressed in certain
benign diseases and normal tissues.
In normal tissue, cultured fibroblasts, but not
resting fibroblasts, have a strong expression of FAPα. The cultural
conditions may mimic the ‘wounds that do not heal’ state. Another
important normal cell type that expresses FAPα is bone marrow
mesenchymal stem cells (BMMSCs) (61,62).
In the light of present knowledge, it is difficult to make a clear
distinction between BMMSCs and fibroblasts. It appears that MSCs
and fibroblasts share properties beyond those previously understood
and that MSCs may in fact be fibroblasts’ new ‘clothes’ (63). Therefore, BMMSCs may be regarded as
cultured fibroblasts. Table I
lists FAPα-expressing tissues.
FAPα expression may be elevated under the influence
of an altered tumor microenvironment or inflammation. In
vitro FAPα expression was observed in fibroblasts and
melanocytes cultured in fibroblast growth factor (FGF) and phorbol
ester (33). Treatment of FB20
cells with human transforming growth factor-βl (TGF-β1),
12-o-tetradecanoyl phorbol-13-acetate (TPA), retinol or
retinoic acid for 24–48 h increased FAPα expression in the cells
(34). FAPα expression in CD
strictured myofibroblasts under the stimulation of 10 ng/ml tumor
necrosis factor α (TNF-α) or TGF-β1 for 48 h was significantly
increased (59). TNF-α, produced
by macrophages, was also able to induce FAPα expression in cultured
human aortic smooth muscle cells (60). Further to the cytokines and
chemical substances which induce FAPα expression, physical
stimulants, including ultraviolet radiation, also induce
upregulation of FAPα expression in fibroblasts, melanocytes and
primary melanoma cells to facilitate invasion and migration of the
cells (69).
HEK293 cells and MDA-MB-231 human mammary
adenocarcinoma cells were transfected with FAPα cDNA to
constitutively express FAPα, and subsequently xenografted into SCID
mice. The transfected cells were more likely to develop
subcutaneous tumors and demonstrated enhanced tumor growth
(70,71) as well as increased microvessel
density (71), compared with
mock-transfected cells. Antibodies that neutralized FAPα attenuated
the tumor growth rate (70). The
human breast cancer cell lines MDA-MB-435 and MDA-MB-436, stably
transfected with anti-sense oligonucleotides of FAPα, demonstrated
slower proliferation than their FAPα-expressing counterparts in
serum-free medium but not in serum-containing medium, indicating
that breast cancer cells with high FAPα expression levels may be
independent from exogenous serum factors for growth (72). Planting FAPα-silenced SKOV3 cells
in a xenograft mouse model resulted in significantly decreased
tumor growth (73). This is
consistent with the observation that the elimination of
FAPα-expressing cells led to stunted tumor growth and enhanced
anti-tumor immune response in a mouse model (30). Radioimmunotherapy with novel
internalizing antibody ESC11 delayed growth of established tumors
and extended survival of mice (74). Mutation at the site of
Ser624→Ala624 of FAPα resulted in
~100,000-fold decrease in DPP activity and attenuated tumor growth
when HEK293 cells transfected with enzymatic mutant (S624A) FAPα
were inoculated subcutaneously into a CB17-SCID mouse (27). FAPα was upregulated in bone marrow
mesenchymal stem cells and osteoclasts when co-cultured with
myeloma cells and supported myeloma cell survival (61). Inhibition of FAPα with PT-100
(Val-boro-Pro) influenced the expression of adhesion molecules in
osteoclasts and reduced myeloma growth and bone disease (75). In a mouse model, inhibition of FAPα
with PT-100 resulted in an antitumor effect implicating
tumor-specific cytotoxic T lymphocytes, protection of immunological
memory, augmented antitumor activity of antibody-increasing
cytokines [interleukin (IL)-1, IL-6, interferon, granulocyte-colony
stimulating factor] and chemokines (76). Taken together, these studies
indicated that FAPα is a tumor promoter.
Tumor immunotherapy is important for eradicating
tumors with minimal residual disease. Tumor-associated antigens are
able to spontaneously elicit a CD8(+) T-cell response (77). However, the results of therapeutic
vaccination with such antigens in inhibiting tumor growth have been
relatively ineffective. This may be associated with the
immunosuppressive effect of the stromal cells surrounding tumors. A
study demonstrated that depleting FAPα-expressing cells in a
transgenic mouse elicited antitumor immunity, and thus indicated
that FAPα-expressing cells are an immune-suppressive component of
the tumor microenvironment (30).
This result is in accordance with evidence that an oral DNA vaccine
targeting FAPα is able to suppress primary breast carcinoma growth
and metastasis (28). This process
may be associated with a shift in the immune microenvironment from
expression of T helper cells (Th)2 to Th1
(78).
While numerous studies demonstrated that FAPα was a
tumor suppressor, in 1993, Rettig et al (33) observed that FAPα expression in
melanocytes was downregulated once they transformed into malignant
cells and acquired tumorigenic potential. Analysis of human skin
lesions, detected by immunohistochemical analysis, indicated that
FAPα was expressed in only a fraction of melanocytic nevi and
expression was scarce in both primary and metastatic melanoma
lesions (79). By hybridizing
normal fibroblasts with tumorigenic and nontumorigenic HeLa cells,
Tsujimoto et al (80)
identified FAPα as a potential inhibitor of tumorigenesis. All
these observations were consistent with Brown et al’s
(81) discovery that Xenopus
laevis demonstrate a marked expression of FAPα whilst
reabsorbing tadpole tails during amphibian metamorphosis. This
indicated that FAPα was a pro-apoptotic factor involved in tissue
remodeling. FAPα also enhanced apoptosis in the mouse B16 melanoma
cell line independent of DPP4 and its enzymatic activity (82).
Recently, a study using transgenic mice revealed
that FAPα(+) cells may have important functions in maintaining
normal muscle mass and hematopoiesis, and their expression in
normal tissues may have an important role in the paraneoplastic
syndromes of cachexia and anemia (83). Niedermeyer et al (84) found that, in vivo,
homozygous FAPα-deficient mice generated from homologous
recombination in the embryonic stem cell line R1 were fertile and
exhibited no overt developmental defects or general changes in
cancer susceptibility. Therefore, the function of FAPα may vary
between tumor contexts and require further study.
FAPα not only has an important role in regulating
tumor behavior, but also influences CAF behavior. Silencing FAPα
with short interfering RNA transfected using a lentiviral vector
inhibited growth and resulted in cell cycle arrest at the
G2 and S phases of cancer-associated fibroblasts in
vitro (73).
Tissue remodeling is important in development, wound
healing, chronic inflammation, fibrosis and cancer. It is
understood that an active stroma is essential for cancer cell
invasion and metastasis (85).
Invasion and metastasis of malignant cancer cells requires the
degradation of the extracellular matrix (ECM). FAPα displays DPP
and gelatinolytic activity as proved by gelatin zymography and can
cleave native ECM proteins, including collagen I, collagen IV,
fibronectin, laminin and gelatin (38–40,49,86).
These enzyme activities depend on the mutation at position
Ser624, which abrogates the DPP and collagenase activity
of FAPα (49). These enzymatic
activities indicate that FAPα may have a prominent role in tumor
invasion, metastasis and angiogenesis (86–88).
Clinical observation revealed that the overexpression of FAPα by
ductal carcinomas is congruent with the invasion and metastasis of
infiltrating ductal carcinomas (IDC) of the breast (55). Using an in vivo-like
three-dimensional matrix system, Lee et al (89) observed that FAPα remodeled the ECM
and increased the invasive capability and metastasis of pancreatic
tumors, mediated by β1-integrin/focal adhesion kinase.
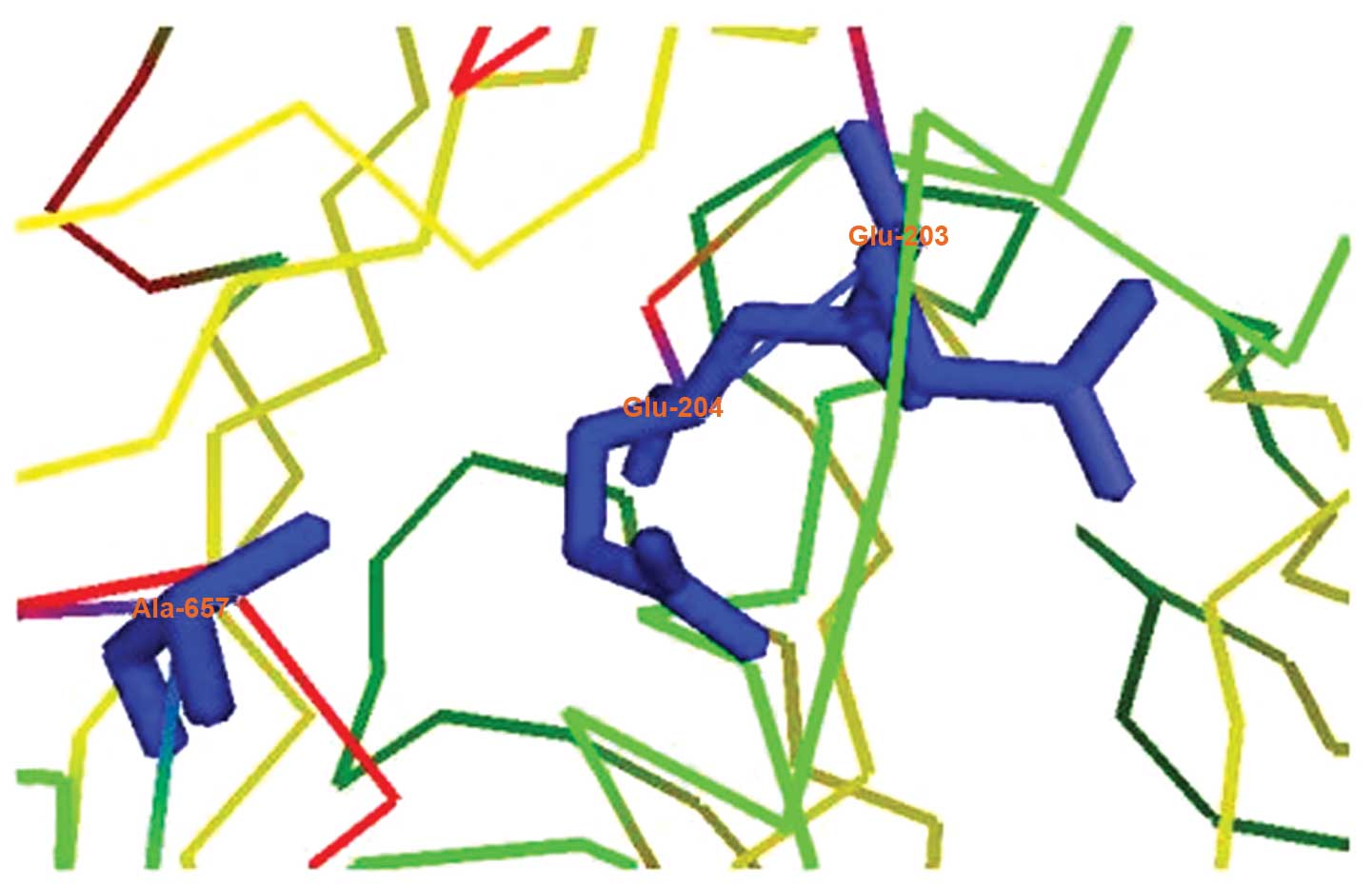
In addition to DPP activity, FAPα also demonstrates
endopeptidase activity due to the presence of Ala657,
which leads to decreased acidity in the active site of the FAPα Glu
motif (E203–E204; Fig. 2)
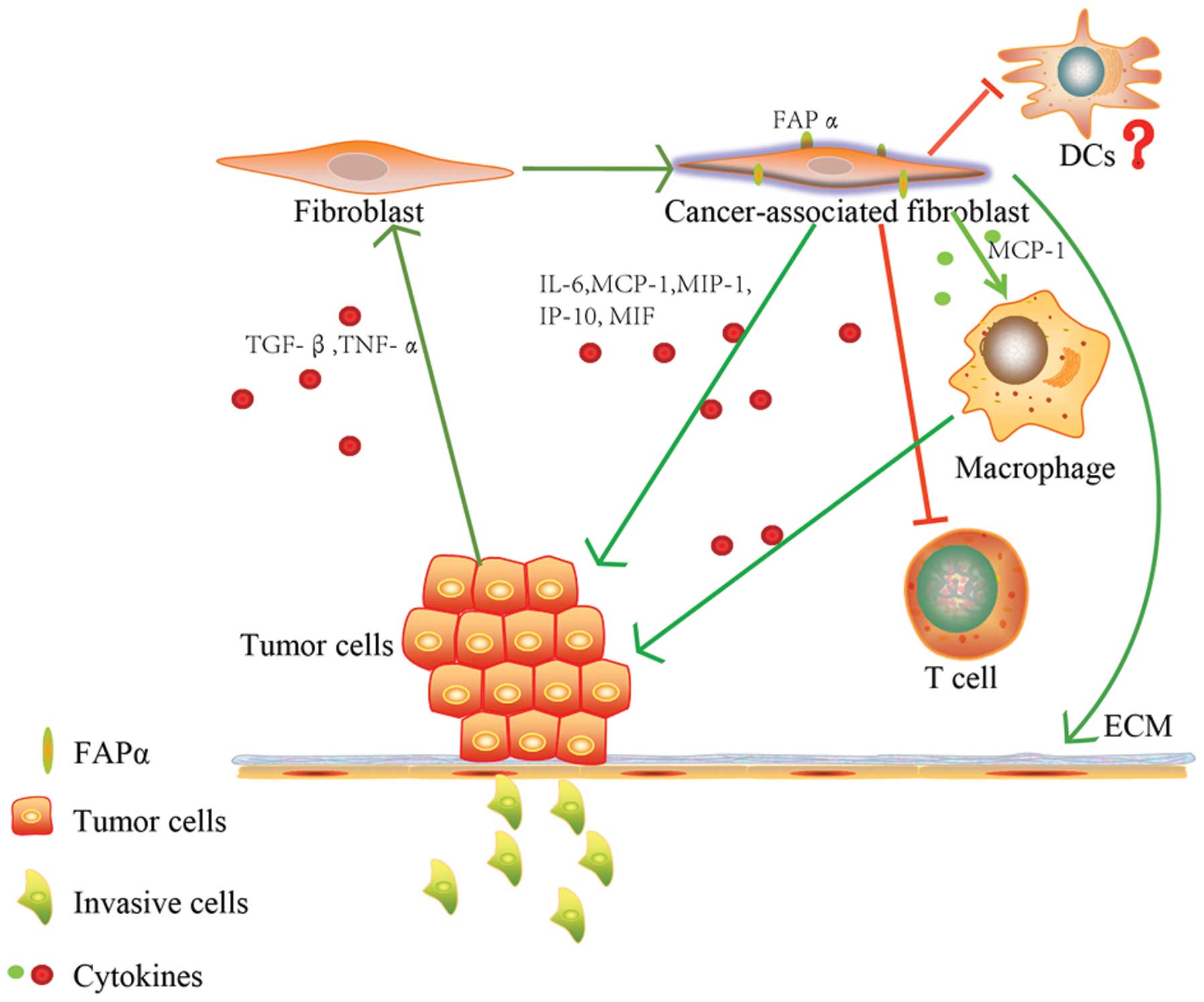
(43,49). Fig.
3 summarizes the intricate interaction of tumor cells with FAPα
in the tumor microenvironment.
The role of FAPα is controversial as it remains
associated with tumor promotion and inhibition; therefore, the
clinical significance of FAPα expression requires further study.
Using immunohistochemical analysis, Wikberg et al (90) found that FAPα was expressed by
stromal fibroblasts in 85–90% of colorectal cancers and that
increased FAPα expression in the cancer center, but not in the
outlying regions, was associated with microsatellite instability,
high CpG island methylator phenotype and poor prognosis. FAPα
expression in pancreatic adenocarcinoma is associated with
desmoplasia and a worse prognosis (64,92).
Henry et al (93) reported
that patients with colon cancer who had high levels of stromal FAPα
expression were more likely to demonstrate progression of disease,
latent occurrence or recurrence of metastases and poor prognosis.
FAPα is also involved in tumor re-growth and recurrence and high
FAPα expression is correlated with poor prognosis in rectal cancer
following chemoradiotherapy (94).
Conversely, Ariga et al (95), discovered that higher expression of
FAPα in the mesenchyme of invasive ductal carcinoma of breast
cancer is associated with longer overall and disease-free
survival.
To date, numerous endogenous substrates of FAPα have
remained to be elucidated. In 2004, Lee et al (96) discovered and purified a proteinase
from human plasma, antiplasmin-cleaving enzyme (APCE), which is
capable of cleaving the Pro12-Asn13 bond of Met-α2-antiplasmin
(α2-AP) to yield Asn-α2-AP. Subsequently, this APCE was identified
as a soluble form of FAPα (97).
In addition to α2-AP, gelatin and collagen, further substrates have
been identified. Recently, neuropeptide Y, B-type natriuretic
peptide, peptide YY, incretins, substance P, glucagon-like
peptide-1 and glucose-dependent insulinotropic peptide were
identified as substrates of FAPα (98). A study indicated that α2-AP was not
a robust substrate of FAPα in vitro, but a novel substrate,
Spry2 (also called Sprouty2, a member of the Sprouty family) was
identified (99).
The general and abundant expression of FAPα in the
stroma of tumors makes it a potential target for the diagnosis and
therapy of numerous carcinomas. A phase I clinical study was
executed and indicated that FAPα was highly expressed by reactive
stromal fibroblasts in >95% of primary and metastatic tumors in
patients with colorectal carcinomas (36). A phase I open-label study
demonstrated that a humanized antibody (sibrotuzumab), directed
against human FAPα expressed by advanced or metastatic
FAPα-positive cancer, may be administered safely. However, the
study did not indicate sibrotuzumab efficacy for the treatment of
FAPα-positive cancer (100). In
2003, an early phase II trial of sibrotuzumab in patients with
metastatic colorectal cancer revealed that progressive disease was
evident in 15 out of 17 evaluable patients (101). T cells, engineered with
FAPα-reactive chimeric antigen receptors and stimulated with FAPα
or FAPα-expressing cell lines, degranulated and produced effector
cytokines (102). However,
adoptive transfer of FAPα-reactive T cells into mice infected with
various tumors, mediated weak antitumor effects (102). FAPα-specific redirected T cells
for the treatment of FAPα-positive malignant pleural mesothelioma
are currently subject to clinical trials (103).
The tumor stroma has been increasingly recognized as
a vital participant in tumorigenesis, drug-resistance,
angiogenesis, invasion and metastasis in numerous types of cancer.
FAPα is highly expressed in CAFs and is important in mediating
their function. Ubiquitous expression by the majority of the stroma
of epithelial tumors makes FAPα an ideal target for cancer therapy.
Since the discovery of FAPα, it has been studied extensively.
However, though a large amount of promising results were observed
in vitro, clinical application of FAPα-targeting has thus
far remained ineffective. In view of the complexity of its
functions, FAPα requires further study.
The authors would like to thank their colleagues at
the Department of Hematology, The First Affiliated Hospital,
Zhejiang University, Hangzhou, China. The present review was
supported by the Major Research Plan of the National Natural
Science Foundation of China (no. 91029740) and the National Natural
Science Foundation of China (no. 81071936).
|
1
|
Paget S: The distribution of secondary
growths in cancer of the breast. 1889. Cancer Metastasis Rev.
8:98–101. 1989.PubMed/NCBI
|
|
2
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
3
|
Bhowmick NA, Neilson EG and Moses HL:
Stromal fibroblasts in cancer initiation and progression. Nature.
432:332–337. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hertenstein B, Hambach L, Bacigalupo A,
Schmitz N, McCann S, Slavin S, Gratwohl A, Ferrant A, Elmaagacli A,
Schwertfeger R, et al: Chronic Leukaemia Working Party of the
European Group for Blood and Marrow Transplantation: Development of
leukemia in donor cells after allogeneic stem cell transplantation
- a survey of the European Group for Blood and Marrow
Transplantation (EBMT). Haematologica. 90:969–975. 2005.PubMed/NCBI
|
|
5
|
Sala-Torra O, Hanna C, Loken MR, Flowers
ME, Maris M, Ladne PA, Mason JR, Senitzer D, Rodriguez R, Forman
SJ, et al: Evidence of donor-derived hematologic malignancies after
hematopoietic stem cell transplantation. Biol Blood Marrow
Transplant. 12:511–517. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao H, Shi J, Luo Y, Tan Y, He J, Xie W,
Zhang L, Wang Y, Liu L, Wu K, et al: First report of multiple CEBPA
mutations contributing to donor origin of leukemia relapse after
allogeneic hematopoietic stem cell transplantation. Blood.
117:5257–5260. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Raaijmakers MH, Mukherjee S, Guo S, Zhang
S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian
RP, Scadden EO, et al: Bone progenitor dysfunction induces
myelodysplasia and secondary leukaemia. Nature. 464:852–857. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ross FM, Chiecchio L, Dagrada G, Protheroe
RK, Stockley DM, Harrison CJ, Cross NC, Szubert AJ, Drayson MT and
Morgan GJ: UK Myeloma Forum: The t(14;20) is a poor prognostic
factor in myeloma but is associated with long-term stable disease
in monoclonal gammopathies of undetermined significance.
Haematologica. 95:1221–1225. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Walkley CR, Olsen GH, Dworkin S, Fabb SA,
Swann J, McArthur GA, Westmoreland SV, Chambon P, Scadden DT and
Purton LE: A microenvironment-induced myeloproliferative syndrome
caused by retinoic acid receptor gamma deficiency. Cell.
129:1097–1110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Walkley CR, Shea JM, Sims NA, Purton LE
and Orkin SH: Rb regulates interactions between hematopoietic stem
cells and their bone marrow microenvironment. Cell. 129:1081–1095.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sounni NE and Noel A: Targeting the tumor
microenvironment for cancer therapy. Clin Chem. 59:85–93. 2013.
View Article : Google Scholar
|
|
12
|
Zhang J and Liu J: Tumor stroma as targets
for cancer therapy. Pharmacol Ther. 137:200–215. 2013. View Article : Google Scholar :
|
|
13
|
Zhang B, Li M, McDonald T, Holyoake TL,
Moon RT, Campana D, Shultz L and Bhatia R: Microenvironmental
protection of CML stem and progenitor cells from tyrosine kinase
inhibitors through N-cadherin and Wnt-β-catenin signaling. Blood.
121:1824–1838. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Konopleva MY and Jordan CT: Leukemia stem
cells and microenvironment: biology and therapeutic targeting. J
Clin Oncol. 29:591–599. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eck SM, Côté AL, Winkelman WD and
Brinckerhoff CE: CXCR4 and matrix metalloproteinase-1 are elevated
in breast carcinoma-associated fibroblasts and in normal mammary
fibroblasts exposed to factors secreted by breast cancer cells. Mol
Cancer Res. 7:1033–1044. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao MQ, Kim BG, Kang S, Choi YP, Park H,
Kang KS and Cho NH: Stromal fibroblasts from the interface zone of
human breast carcinomas induce an epithelial-mesenchymal
transition-like state in breast cancer cells in vitro. J Cell Sci.
123(Pt 20): 3507–3514. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hellevik T, Pettersen I, Berg V, Winberg
JO, Moe BT, Bartnes K, Paulssen RH, Busund LT, Bremnes R, Chalmers
A and Martinez-Zubiaurre I: Cancer-associated fibroblasts from
human NSCLC survive ablative doses of radiation but their invasive
capacity is reduced. Radiat Oncol. 7:592012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berdiel-Acer M, Bohem ME, López-Doriga A,
Vidal A, Salazar R, Martínez-Iniesta M, Santos C, Sanjuan X,
Villanueva A and Molleví DG: Hepatic carcinoma-associated
fibroblasts promote an adaptative response in colorectal cancer
cells that inhibit proliferation and apoptosis: nonresistant cells
die by nonapoptotic cell death. Neoplasia. 13:931–946.
2011.PubMed/NCBI
|
|
19
|
Mueller L, Goumas FA, Himpel S, Brilloff
S, Rogiers X and Broering DC: Imatinib mesylate inhibits
proliferation and modulates cytokine expression of human
cancer-associated stromal fibroblasts from colorectal metastases.
Cancer Lett. 250:329–338. 2007. View Article : Google Scholar
|
|
20
|
Henriksson ML, Edin S, Dahlin AM,
Oldenborg PA, Öberg Å, Van Guelpen B, Rutegård J, Stenling R and
Palmqvist R: Colorectal cancer cells activate adjacent fibroblasts
resulting in FGF1/FGFR3 signaling and increased invasion. Am J
Pathol. 178:1387–1394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin ZY, Chuang YH and Chuang WL:
Cancer-associated fibroblasts up-regulate CCL2, CCL26, IL6 and
LOXL2 genes related to promotion of cancer progression in
hepatocellular carcinoma cells. Biomed Pharmacother. 66:525–529.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
True LD, Zhang H, Ye M, Huang CY, Nelson
PS, von Haller PD, Tjoelker LW, Kim JS, Qian WJ, Smith RD, et al:
CD90/THY1 is overexpressed in prostate cancer-associated
fibroblasts and could serve as a cancer biomarker. Mod Pathol.
23:1346–1356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mao Y, Keller ET, Garfield DH, Shen K and
Wang J: Stromal cells in tumor microenvironment and breast cancer.
Cancer Metastasis Rev. 32:303–315. 2013. View Article : Google Scholar
|
|
24
|
Mishra PJ, Humeniuk R, Medina DJ, Medina
DJ, Alexe G, Mesirov JP, Ganesan S, Glod JW and Banerjee D:
Carcinoma-associated fibroblast-like differentiation of human
mesenchymal stem cells. Cancer Res. 68:4331–4339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lecomte J, Masset A, Blacher S, Maertens
L, Gothot A, Delgaudine M, Bruyère F, Carnet O, Paupert J, Illemann
M, et al: Bone marrow-derived myofibroblasts are the providers of
pro-invasive matrix metalloproteinase 13 in primary tumor.
Neoplasia. 14:943–951. 2012.PubMed/NCBI
|
|
26
|
Fassnacht M, Lee J, Milazzo C, Boczkowski
D, Su Z, Nair S and Gilboa E: Induction of CD4(+) and CD8(+) T-cell
responses to the human stromal antigen, fibroblast activation
protein: implication for cancer immunotherapy. Clin Cancer Res.
11:5566–5571. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng JD, Valianou M, Canutescu AA, Jaffe
EK, Lee HO, Wang H, Lai JH, Bachovchin WW and Weiner LM: Abrogation
of fibroblast activation protein enzymatic activity attenuates
tumor growth. Mol Cancer Ther. 4:351–360. 2005.PubMed/NCBI
|
|
28
|
Loeffler M, Kruger JA, Niethammer AG and
Reisfeld RA: Targeting tumor-associated fibroblasts improves cancer
chemotherapy by increasing intratumoral drug uptake. J Clin Invest.
116:1955–1962. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Santos AM, Jung J, Aziz N, Kissil JL and
Puré E: Targeting fibroblast activation protein inhibits tumor
stromagenesis and growth in mice. J Clin Invest. 119:3613–3625.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kraman M, Bambrough PJ, Arnold JN, Roberts
EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA and Fearon DT:
Suppression of antitumor immunity by stromal cells expressing
fibroblast activation protein-alpha. Science. 330:827–830. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rettig WJ, Chesa PG, Beresford HR,
Feickert HJ, Jennings MT, Cohen J, Oettgen HF and Old LJ:
Differential expression of cell surface antigens and glial
fibrillary acidic protein in human astrocytoma subsets. Cancer Res.
46(12 Pt 2): 6406–6412. 1986.PubMed/NCBI
|
|
32
|
Rettig WJ, Garin-Chesa P, Beresford HR,
Oettgen HF, Melamed MR and Old LJ: Cell-surface glycoproteins of
human sarcomas: differential expression in normal and malignant
tissues and cultured cells. Proc Natl Acad Sci USA. 85:3110–3114.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rettig WJ, Garin-Chesa P, Healey JH, Su
SL, Ozer HL, Schwab M, Albino AP and Old LJ: Regulation and
heteromeric structure of the fibroblast activation protein in
normal and transformed cells of mesenchymal and neuroectodermal
origin. Cancer Res. 53:3327–3335. 1993.PubMed/NCBI
|
|
34
|
Rettig WJ, Su SL, Fortunato SR, Scanlan
MJ, Raj BK, Garin-Chesa P, Healey JH and Old LJ: Fibroblast
activation protein: purification, epitope mapping and induction by
growth factors. Int J Cancer. 58:385–392. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Scanlan MJ, Raj BK, Calvo B, Garin-Chesa
P, Sanz-Moncasi MP, Healey JH, Old LJ and Rettig WJ: Molecular
cloning of fibroblast activation protein alpha, a member of the
serine protease family selectively expressed in stromal fibroblasts
of epithelial cancers. Proc Natl Acad Sci USA. 91:5657–5661. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Welt S, Divgi CR, Scott AM, Garin-Chesa P,
Finn RD, Graham M, Carswell EA, Cohen A, Larson SM, Old LJ, et al:
Antibody targeting in metastatic colon cancer: a phase I study of
monoclonal antibody F19 against a cell-surface protein of reactive
tumor stromal fibroblasts. J Clin Oncol. 12:1193–1203.
1994.PubMed/NCBI
|
|
37
|
Niedermeyer J, Scanlan MJ, Garin-Chesa P,
Daiber C, Fiebig HH, Old LJ, Rettig WJ and Schnapp A: Mouse
fibroblast activation protein: molecular cloning, alternative
splicing and expression in the reactive stroma of epithelial
cancers. Int J Cancer. 71:383–389. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Aoyama A and Chen WT: A 170-kDa
membrane-bound protease is associated with the expression of
invasiveness by human malignant melanoma cells. Proc Natl Acad Sci
USA. 87:8296–8300. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Monsky WL, Lin CY, Aoyama A, Kelly T,
Akiyama SK, Mueller SC and Chen WT: A potential marker protease of
invasiveness, seprase, is localized on invadopodia of human
malignant melanoma cells. Cancer Res. 54:5702–5710. 1994.PubMed/NCBI
|
|
40
|
Piñeiro-Sánchez ML, Goldstein LA, Dodt J,
Howard L, Yeh Y, Tran H, Argraves WS and Chen WT: Identification of
the 170-kDa melanoma membrane-bound gelatinase (seprase) as a
serine integral membrane protease. J Biol Chem. 272:7595–7601.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mathew S, Scanlan MJ, Mohan Raj BK, Murty
VV, Garin-Chesa P, Old LJ, Rettig WJ and Chaganti RS: The gene for
fibroblast activation protein alpha (FAP), a putative cell
surface-bound serine protease expressed in cancer stroma and wound
healing, maps to chromosome band 2q23. Genomics. 25:335–337. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kelly T: Fibroblast activation
protein-alpha and dipeptidyl peptidase IV (CD26): cell-surface
proteases that activate cell signaling and are potential targets
for cancer therapy. Drug Resist Updat. 8:51–58. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Aertgeerts K, Levin I, Shi L, Snell GP,
Jennings A, Prasad GS, Zhang Y, Kraus ML, Salakian S, Sridhar V, et
al: Structural and kinetic analysis of the substrate specificity of
human fibroblast activation protein alpha. J Biol Chem.
280:19441–19444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rosenblum JS and Kozarich JW: Prolyl
peptidases: a serine protease subfamily with high potential for
drug discovery. Curr Opin Chem Biol. 7:496–504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yazbeck R, Howarth GS and Abbott CA:
Dipeptidyl peptidase inhibitors, an emerging drug class for
inflammatory disease? Trends Pharmacol Sci. 30:600–607. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sedo A and Malík R: Dipeptidyl peptidase
IV-like molecules: homologous proteins or homologous activities?
Biochim Biophys Acta. 1550:107–116. 2001. View Article : Google Scholar
|
|
47
|
Qi SY, Riviere PJ, Trojnar J, Junien JL
and Akinsanya KO: Cloning and characterization of dipeptidyl
peptidase 10, a new member of an emerging subgroup of serine
proteases. Biochem J. 373(Pt 1): 179–189. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ghersi G, Zhao Q, Salamone M, Yeh Y,
Zucker S and Chen WT: The protease complex consisting of dipeptidyl
peptidase IV and seprase plays a role in the migration and invasion
of human endothelial cells in collagenous matrices. Cancer Res.
66:4652–4661. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Park JE, Lenter MC, Zimmermann RN,
Garin-Chesa P, Old LJ and Rettig WJ: Fibroblast activation protein,
a dual specificity serine protease expressed in reactive human
tumor stromal fibroblasts. J Biol Chem. 274:36505–36512. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abbott CA, Baker E, Sutherland GR and
McCaughan GW: Genomic organization, exact localization, and tissue
expression of the human CD26 (dipeptidyl peptidase IV) gene.
Immunogenetics. 40:331–338. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Abbott CA, Yu DM, Woollatt E, Sutherland
GR, McCaughan GW and Gorrell MD: Cloning, expression and
chromosomal localization of a novel human dipeptidyl peptidase
(DPP) IV homolog, DPP8. Eur J Biochem. 267:6140–6150. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yokotani N, Doi K, Wenthold RJ and Wada K:
Non-conservation of a catalytic residue in a dipeptidyl
aminopeptidase IV-related protein encoded by a gene on human
chromosome 7. Hum Mol Genet. 2:1037–1039. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang J, Valianou M and Cheng JD:
Identification and characterization of the promoter of fibroblast
activation protein. Front Biosci (Elite Ed). 2:1154–1163. 2010.
View Article : Google Scholar :
|
|
54
|
Garin-Chesa P, Old LJ and Rettig WJ: Cell
surface glycoprotein of reactive stromal fibroblasts as a potential
antibody target in human epithelial cancers. Proc Natl Acad Sci U S
A. 87:7235–7239. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kelly T, Kechelava S, Rozypal TL, West KW
and Korourian S: Seprase, a membrane-bound protease, is
overexpressed by invasive ductal carcinoma cells of human breast
cancers. Mod Pathol. 11:855–863. 1998.PubMed/NCBI
|
|
56
|
Levy MT, McCaughan GW, Abbott CA, Park JE,
Cunningham AM, Müller E, Rettig WJ and Gorrell MD: Fibroblast
activation protein: a cell surface dipeptidyl peptidase and
gelatinase expressed by stellate cells at the tissue remodelling
interface in human cirrhosis. Hepatology. 29:1768–1778. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Acharya PS, Zukas A, Chandan V,
Katzenstein AL and Puré E: Fibroblast activation protein: a serine
protease expressed at the remodeling interface in idiopathic
pulmonary fibrosis. Hum Pathol. 37:352–360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bauer S, Jendro MC, Wadle A, Kleber S,
Stenner F, Dinser R, Reich A, Faccin E, Gödde S, Dinges H, et al:
Fibroblast activation protein is expressed by rheumatoid
myofibroblast-like synoviocytes. Arthritis Res Ther. 8:R1712006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rovedatti L, Di Sabatino A, Knowles CH,
Sengupta N, Biancheri P, Corazza GR and MacDonald TT: Fibroblast
activation protein expression in Crohn’s disease strictures.
Inflamm Bowel Dis. 17:1251–1253. 2011. View Article : Google Scholar
|
|
60
|
Brokopp CE, Schoenauer R, Richards P,
Bauer S, Lohmann C, Emmert MY, Weber B, Winnik S, Aikawa E, Graves
K, et al: Fibroblast activation protein is induced by inflammation
and degrades type I collagen in thin-cap fibroatheromata. Eur Heart
J. 32:2713–2722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ge Y, Zhan F, Barlogie B, Epstein J,
Shaughnessy J Jr and Yaccoby S: Fibroblast activation protein (FAP)
is upregulated in myelomatous bone and supports myeloma cell
survival. Br J Haematol. 133:83–92. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Bae S, Park CW, Son HK, Ju HK, Paik D,
Jeon CJ, Koh GY, Kim J and Kim H: Fibroblast activation protein
alpha identifies mesenchymal stromal cells from human bone marrow.
Br J Haematol. 142:827–830. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Haniffa MA, Collin MP, Buckley CD and
Dazzi F: Mesenchymal stem cells: the fibroblasts’ new clothes?
Haematologica. 94:258–263. 2009. View Article : Google Scholar :
|
|
64
|
Shi M, Yu DH, Chen Y, Zhao CY, Zhang J,
Liu QH, Ni CR and Zhu MH: Expression of fibroblast activation
protein in human pancreatic adenocarcinoma and its
clinicopathological significance. World J Gastroenterol.
18:840–846. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Iwasa S, Okada K, Chen WT, Jin X, Yamane
T, Ooi A and Mitsumata M: Increased expression of seprase, a
membrane-type serine protease, is associated with lymph node
metastasis in human colorectal cancer. Cancer Lett. 227:229–236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mori Y, Kono K, Matsumoto Y, Fujii H,
Yamane T, Mitsumata M and Chen WT: The expression of a type II
transmembrane serine protease (Seprase) in human gastric carcinoma.
Oncology. 67:411–419. 2004. View Article : Google Scholar
|
|
67
|
Jin X, Iwasa S, Okada K, Mitsumata M and
Ooi A: Expression patterns of seprase, a membrane serine protease,
in cervical carcinoma and cervical intraepithelial neoplasm.
Anticancer Res. 23:3195–3198. 2003.PubMed/NCBI
|
|
68
|
Mentlein R, Hattermann K, Hemion C,
Jungbluth AA and Held-Feindt J: Expression and role of the cell
surface protease seprase/fibroblast activation protein-α (FAP-α) in
astroglial tumors. Biol Chem. 392:199–207. 2011. View Article : Google Scholar
|
|
69
|
Wäster P, Rosdahl I, Gilmore BF and
Seifert O: Ultraviolet exposure of melanoma cells induces
fibroblast activation protein-α in fibroblasts: Implications for
melanoma invasion. Int J Oncol. 39:193–202. 2011.
|
|
70
|
Cheng JD, Dunbrack RL Jr, Valianou M,
Rogatko A, Alpaugh RK and Weiner LM: Promotion of tumor growth by
murine fibroblast activation protein, a serine protease, in an
animal model. Cancer Res. 62:4767–4772. 2002.PubMed/NCBI
|
|
71
|
Huang Y, Wang S and Kelly T: Seprase
promotes rapid tumor growth and increased microvessel density in a
mouse model of human breast cancer. Cancer Res. 64:2712–2716. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Goodman JD, Rozypal TL and Kelly T:
Seprase, a membrane-bound protease, alleviates the serum growth
requirement of human breast cancer cells. Clin Exp Metastasis.
20:459–470. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lai D, Ma L and Wang F: Fibroblast
activation protein regulates tumor-associated fibroblasts and
epithelial ovarian cancer cells. Int J Oncol. 41:541–550.
2012.PubMed/NCBI
|
|
74
|
Fischer E, Chaitanya K, Wüest T, Wadle A,
Scott AM, van den Broek M, Schibli R, Bauer S and Renner C:
Radioimmunotherapy of fibroblast activation protein positive tumors
by rapidly internalizing antibodies. Clin Cancer Res. 18:6208–6218.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Pennisi A, Li X, Ling W, Khan S, Gaddy D,
Suva LJ, Barlogie B, Shaughnessy JD, Aziz N and Yaccoby S:
Inhibitor of DASH proteases affects expression of adhesion
molecules in osteoclasts and reduces myeloma growth and bone
disease. Br J Haematol. 145:775–787. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Adams S, Miller GT, Jesson MI, Watanabe T,
Jones B and Wallner BP: PT-100, a small molecule dipeptidyl
peptidase inhibitor, has potent antitumor effects and augments
antibody-mediated cytotoxicity via a novel immune mechanism. Cancer
Res. 64:5471–5480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
van der Bruggen P, Traversari C, Chomez P,
Lurquin C, De Plaen E, Van den Eynde B, Knuth A and Boon T: A gene
encoding an antigen recognized by cytolytic T lymphocytes on a
human melanoma. Science. 254:1643–1647. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liao D, Luo Y, Markowitz D, Xiang R and
Reisfeld RA: Cancer associated fibroblasts promote tumor growth and
metastasis by modulating the tumor immune microenvironment in a 4T1
murine breast cancer model. PLoS One. 4:e79652009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Huber MA, Kraut N, Park JE, Schubert RD,
Rettig WJ, Peter RU and Garin-Chesa P: Fibroblast activation
protein: differential expression and serine protease activity in
reactive stromal fibroblasts of melanocytic skin tumors. J Invest
Dermatol. 120:182–188. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tsujimoto H, Nishizuka S, Redpath JL and
Stanbridge EJ: Differential gene expression in tumorigenic and
nontumorigenic HeLa × normal human fibroblast hybrid cells. Mol
Carcinog. 26:298–304. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Brown DD, Wang Z, Furlow JD, Kanamori A,
Schwartzman RA, Remo BF and Pinder A: The thyroid hormone-induced
tail resorption program during Xenopus laevis metamorphosis. Proc
Natl Acad Sci USA. 93:1924–1929. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ramirez-Montagut T, Blachere NE,
Sviderskaya EV, Bennett DC, Rettig WJ, Garin-Chesa P and Houghton
AN: FAPalpha, a surface peptidase expressed during wound healing,
is a tumor suppressor. Oncogene. 23:5435–5446. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Roberts EW, Deonarine A, Jones JO, Denton
AE, Feig C, Lyons SK, Espeli M, Kraman M, McKenna B, Wells RJ, et
al: Depletion of stromal cells expressing fibroblast activation
protein-α from skeletal muscle and bone marrow results in cachexia
and anemia. J Exp Med. 210:1137–1151. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Niedermeyer J, Kriz M, Hilberg F,
Garin-Chesa P, Bamberger U, Lenter MC, Park J, Viertel B, Püschner
H, Mauz M, Rettig WJ and Schnapp A: Targeted disruption of mouse
fibroblast activation protein. Mol Cell Biol. 20:1089–1094. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Jacob M, Chang L and Puré E: Fibroblast
activation protein in remodeling tissues. Curr Mol Med.
12:1220–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ghersi G, Dong H, Goldstein LA, Yeh Y,
Hakkinen L, Larjava HS and Chen WT: Regulation of fibroblast
migration on collagenous matrix by a cell surface peptidase
complex. J Biol Chem. 277:29231–29241. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chen WT and Kelly T: Seprase complexes in
cellular invasiveness. Cancer Metastasis Rev. 22:259–269. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
O’Brien P and O’Connor BF: Seprase: an
overview of an important matrix serine protease. Biochim Biophys
Acta. 1784:1130–1145. 2008. View Article : Google Scholar
|
|
89
|
Lee HO, Mullins SR, Franco-Barraza J,
Valianou M, Cukierman E and Cheng JD: FAP-overexpressing
fibroblasts produce an extracellular matrix that enhances invasive
velocity and directionality of pancreatic cancer cells. BMC Cancer.
11:2452011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang XM, Yu DM, McCaughan GW and Gorrell
MD: Fibroblast activation protein increases apoptosis, cell
adhesion, and migration by the LX-2 human stellate cell line.
Hepatology. 42:935–945. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wikberg ML, Edin S, Lundberg IV, Van
Guelpen B, Dahlin AM, Rutegård J, Stenling R, Oberg A and Palmqvist
R: High intratumoral expression of fibroblast activation protein
(FAP) in colon cancer is associated with poorer patient prognosis.
Tumour Biol. 34:1013–1020. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Cohen SJ, Alpaugh RK, Palazzo I, Meropol
NJ, Rogatko A, Xu Z, Hoffman JP, Weiner LM and Cheng JD: Fibroblast
activation protein and its relationship to clinical outcome in
pancreatic adenocarcinoma. Pancreas. 37:154–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Henry LR, Lee HO, Lee JS, Klein-Szanto A,
Watts P, Ross EA, Chen WT and Cheng JD: Clinical implications of
fibroblast activation protein in patients with colon cancer. Clin
Cancer Res. 13:1736–1741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Saigusa S, Toiyama Y, Tanaka K, Yokoe T,
Okugawa Y, Fujikawa H, Matsusita K, Kawamura M, Inoue Y, Miki C and
Kusunoki M: Cancer-associated fibroblasts correlate with poor
prognosis in rectal cancer after chemoradiotherapy. Int J Oncol.
38:655–663. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ariga N, Sato E, Ohuchi N, Nagura H and
Ohtani H: Stromal expression of fibroblast activation
protein/seprase, a cell membrane serine proteinase and gelatinase,
is associated with longer survival in patients with invasive ductal
carcinoma of breast. Int J Cancer. 95:67–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lee KN, Jackson KW, Christiansen VJ, Chung
KH and McKee PA: A novel plasma proteinase potentiates
alpha2-antiplasmin inhibition of fibrin digestion. Blood.
103:3783–3788. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lee KN, Jackson KW, Christiansen VJ, Lee
CS, Chun JG and McKee PA: Antiplasmin-cleaving enzyme is a soluble
form of fibroblast activation protein. Blood. 107:1397–1404. 2006.
View Article : Google Scholar
|
|
98
|
Keane FM, Nadvi NA, Yao TW and Gorrell MD:
Neuropeptide Y, B-type natriuretic peptide, substance P and peptide
YY are novel substrates of fibroblast activation protein-α. FEBS J.
278:1316–1332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Huang CH, Suen CS, Lin CT, Chien CH, Lee
HY, Chung KM, Tsai TY, Jiaang WT, Hwang MJ and Chen X:
Cleavage-site specificity of prolyl endopeptidase FAP investigated
with a full-length protein substrate. J Biochem. 149:685–692. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Scott AM, Wiseman G, Welt S, Adjei A, Lee
FT, Hopkins W, Divgi CR, Hanson LH, Mitchell P, Gansen DN, et al: A
Phase I dose-escalation study of sibrotuzumab in patients with
advanced or metastatic fibroblast activation protein-positive
cancer. Clin Cancer Res. 9:1639–1647. 2003.PubMed/NCBI
|
|
101
|
Hofheinz RD, al-Batran SE, Hartmann F,
Hartung G, Jäger D, Renner C, Tanswell P, Kunz U, Amelsberg A,
Kuthan H and Stehle G: Stromal antigen targeting by a humanised
monoclonal antibody: an early phase II trial of sibrotuzumab in
patients with metastatic colorectal cancer. Onkologie. 26:44–48.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Tran E, Chinnasamy D, Yu Z, Morgan RA, Lee
CC, Restifo NP and Rosenberg SA: Immune targeting of fibroblast
activation protein triggers recognition of multipotent bone marrow
stromal cells and cachexia. J Exp Med. 210:1125–1135. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Petrausch U, Schuberth PC, Hagedorn C,
Soltermann A, Tomaszek S, Stahel R, Weder W and Renner C:
Re-directed T cells for the treatment of fibroblast activation
protein (FAP)-positive malignant pleural mesothelioma (FAPME-1).
BMC Cancer. 12:6152012. View Article : Google Scholar : PubMed/NCBI
|