Introduction
Approximately, 239,000 new cases of ovarian cancer
and 152,000 deaths due to this disease were reported worldwide
(1). The frequency of clear cell
carcinoma (CCC) is thought to be 5%-10% of all epithelial ovarian
cancers in Western countries, but it is higher (>20%) in Japan.
Ovarian clear cell carcinoma (OCCC) is resistant to platinum
chemotherapy and it is characterized by poor prognosis. Therefore,
novel strategy to overcome OCCC is required for a more effective
outcome.
The transcription factor HNF1 homeobox β (HNF-1β) is
upregulated in endometriosis and OCCC, suggesting that it might be
a key molecule in endometriosis-associated CCC (2). We previously reported that HNF-1β
promotes G2 phase cell cycle arrest and survival in human CCC cell
lines through up-regulation of the phosphorylation of Chk1 (p-Chk1)
protein in response to a genotoxic stress (3). Moreover, we reported that HNF-1β
overexpressing cells survive by persistent Chk1 activation,
facilitated by USP28-mediated Claspin stabilization (4). Therefore, therapy targeting the
HNF-1β-USP28-Claspin pathway could be a novel targeted molecular
therapy for HNF-1β overexpressing CCC. However, pharmacological
inhibition of HNF-1β or Chk1 could cause several adverse effects,
because they show comparable abundance in numerous organs such as
the kidney, liver, pancreas, and digestive tract. While some study
reported potential targets of HNF-1β (5,6), to
further investigate the potential role of HNF-1β, we conducted
small interfering RNA (siRNA) library screening, through which the
effects of gene silencing on biological phenotypes can be
systematically explored. Our results are expected to provide
insights into the molecular mechanisms underlying the
HNF-1β-mediated cell survival in OCCC.
Materials and methods
Cell lines
All cells were maintained in humidified incubator at
37˚C with 5% CO2. These cells were maintained in
Dulbecco's modified Eagle's medium/Ham's F-12 with L-Glutamine and
Phenol Red containing 10% fetal bovine serum and 100 U/ml
penicillin and streptomycin, and used at sub confluent status.
TOV-21G and ES2 cell lines were obtained from American Type Culture
Collection. Among these CCC lines, TOV-21G shows HNF-1β
overexpression, while ES2 is negative for HNF-1β expression.
siRNA library screening
We carried out siRNA library screening of human cell
cycle regulation-related genes (G-003205; Dharmacon). TOV-21G cell
line was grown in 6-well plate at a concentration of
4.0x105 cells per well and si-HNF-1β (M-007921-01;
Dharmacon) or si-control (D-001210-02; Dharmacon) was reverse
transfected rapidly at 5 nM according to manufacturer's recommended
protocol. At 24 h after transfection, HNF-1β knockdown and control
cells were plated in three wells of 96-well plate, respectively, at
a concentration of 5,000 cells per well. In each of the three wells
of HNF-1β knockdown and control cells, we transfected 5 nM of
siRNAs for screening. After 48 h, we measured cell viability by MTT
assay (Cell Proliferation kit I; Roche) according to the
recommended protocol. For each 96-well plate, we transfected
si-control as negative control, and si-PLK1 (M-003290-01;
Dharmacon) as positive control. Candidates were extracted as
follows. Firstly, difference in cell viability between TOV-21G
(si-control) and TOV-21G (si-HNF-1β) was considered to be an effect
of HNF-1β interference on cells. Secondly, the cell viability of
negative control group was verified to show normal distribution,
and we corrected the test results based on difference with negative
control.
Western blotting
TOV-21G and ES2 cells were grown in 6-well dish
(4.0x105 cells per well) and si-glycogen synthase
kinase-3β (si-GSK-3β; D-003010-09; Dharmacon) and si-control were
reverse transfected at 5 nM according to manufacturer's recommended
protocol. TOV-21G cells were grown in 6-well dish
(2.0x105 cells per well) and si-GSK-3β, si-HNF-1β
(M-007921-01 or D-009721-02; Dharmacon) and si-control were reverse
transfected at 5 or 20 nM according to manufacturer's recommended
protocol. Then, we extracted protein at 48 and 72 h after
transfection. Samples were applied to Mini-PROTEAN® TGX™
Gels 4-15%, and transferred by Trans-Blot® Turbo™
Transfer Pack (Bio-Rad Laboratories, Inc.). Protein extraction from
nuclear and cytoplasma separately were conducted using NE-PER
Nuclear and Cytoplasmic Extraction reagents (78833; Thermo Fisher
Scientific, Inc.) according to manufacturer's recommended protocol.
The following antibodies were used for western blotting: primary
antibodies against HNF-1β (ab187744, diluted 1:10,000; Abcam),
GSK-3β (#12456, diluted 1:1,000; Cell Signaling Technology, Inc.),
phospho-GSK-3α/β (#9331, diluted 1:1,000; Cell Signaling
Technology, Inc.), NFκB (#8242, diluted 1:1,000, Cell Signaling
Technology, Inc.), phospho-NFκB (#3033, diluted 1:1,000; Cell
Signaling Technology, Inc.), phospho-Chk1 (#2349, diluted 1:10,000;
Cell Signaling Technology, Inc.) and actin (sc-8432, diluted
1:5,000; Santa Cruz Biotechnology, Inc.). Horseradish
peroxidase-conjugated secondary antibodies against mouse (sc-2005,
1:10,000; Santa Cruz Biotechnology, Inc.) and rabbit (sc-2004,
diluted 1:10,000; Santa Cruz Biotechnology, Inc.) were used.
quantitative polymerase chain reaction
(qPCR)
RNA extraction from TOV-21G and ES2 cells were
performed at 24 and 48 h after transfection (5 nM) by Taq Man Gene
Expression Cells-to-CT™ kit (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. PCR was performed
on StepOnePlus™ Real Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with 4 µl of cDNA, 10 µl of TaqMan Gene
Expression Master Mix (4369016; Applied Biosystems; Thermo Fisher
Scientific, Inc.), 1 µl of GSK-3β or GAPDH TaqMan Gene Expression
Assay (Hs01047719_m1 or Hs99999905_m1; Applied Biosystems; Thermo
Fisher Scientific, Inc.) and 5 µl of nuclease-free water
(B-003000-WB-100; Dharmacon) by 2-∆∆Cq method (7).
Cell cycle analysis
TOV-21G cells were grown in 6-well dish
(2.0x105 cells per well) and si-GSK-3β and si-control
were reverse transfected at 5 nM according to manufacturer's
recommended protocol. Then, the cells were harvested for 48 h and
washed in phosphate-buffered saline (PBS) before fixation in cold
70% ethanol which were added drop wise to the pellet while
vortexing. Cells were fixed for 30 min at 4˚C. Fixed cells were
washed twice in PBS and centrifuged by 3,000 x g for 5 min. Cells
were incubated with 950 µl from 10 mg/ml of a ribonuclease
(313-01461; Nippon Gene Co., Ltd) and 50 µl from 1 mg/ml of a
propidium iodide (P378; Dojindo Laboratories). A BD FACSCalibur™
(BD Biosciences) flow cytometer was used to analyze the cell
population for cell cycle changes.
Apoptosis assay
TOV-21G cells were seeded into 6-well plates at a
concentration of 2.0x105 cells per well and the cells
were then treated with si-GSK-3β and si-control. Harvest the cells
after the incubation period for 48 h and wash in cold PBS.
Re-centrifuge the washed cells, discard the supernatant and
resuspend the cells in 1X Annexin-binding buffer (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Determine the cell
density and dilute in 1X Annexin-binding buffer to
1.0x105 cells/100 µl. Add 5 µl Alexa Fluor®
488 Annexin V (Applied Biosystems; Thermo Fisher Scientific, Inc.)
and propidium iodide to adjust its final concentration as 2.5
µg/ml. Incubate the cells at room temperature for 15 min. After the
incubation period, add 400 µl of 1X Annexin-binding buffer, mix
gently and keep the samples on ice. As soon as possible, analyze
the stained cells by BD FACSCalibur™ to assess the percentage
changes in early and late apoptosis.
IncuCyte ZOOM™ image capture and
analyses for cell growth
Cell proliferation was studied using the IncuCyte
ZOOM™ Live-Cell Imaging system (Essen BioScience) as previously
described for kinetic monitoring of proliferation and cytotoxicity
of cultured cells (8). IncuCyte
image assays quantify how rapidly the proportion of the area
covered by cells increases with time as a function of cell
proliferation rate (8). TOV-21G
cells were seeded into 6-well plates at a concentration of
2.0x105 cells per well and all cells were then treated
by 42 µM bleomycin at 0 h. The AR-A014418 (20 µM) or vehicle
(dimethyl sulfoxide) group were transferred to the IncuCyte ZOOM™
apparatus, and incubations continued over 72 h, with images
collected every three hours. All images were analyzed focused on
confluence (%).
In vivo assay
All animal experiments were conducted according to
Guidelines for Proper Conduct of Animal Experiments (June 1, 2006;
Science Council of Japan). And this study was approved by the
animal ethics committee of Nara Medical University (reference no.
12594). To generate murine subcutaneous tumors, 4.5x106
TOV-21G cells in 200 µl of PBS were injected subcutaneously into
the neck of the dorsal midline in 5- to 6-week-old athymic nude
mice (SLC) under maintenance of 2% after 5% introduction of inhaled
isoflurane (Pfizer Inc.). Ten days after the injection, from the
point of tumor palpable, we separated the mice into two groups:
inhibitor group and control group (n=5 for each), and
intraperitoneally injected AR-A014418 (S7435; Shelleck Chemicals)
at a dose of 1 mg/kg or PBS with the same amount of dimethyl
sulfoxide every day for one week. Physical method (cervical
dislocation) was applied to conduct sacrifice of the mice.
Statistical analysis
Data were assessed whether they present normal
distribution by Shapiro-Wilk analysis. In normal distribution,
t-test was applied and presented as mean ± SD. In case of variables
that did not present normal distribution, Mann-Whitney U test were
applied and expressed as median ± SD. Analyses were performed by
SPSS version 25.0 (IBM Corp.). All statistical analysis was
performed at least twice. Two-sided P<0.05 was considered to
indicate a statistically significant difference.
Results
GSK-3β is a key gene regulating HNF-1β
overexpression in CCC
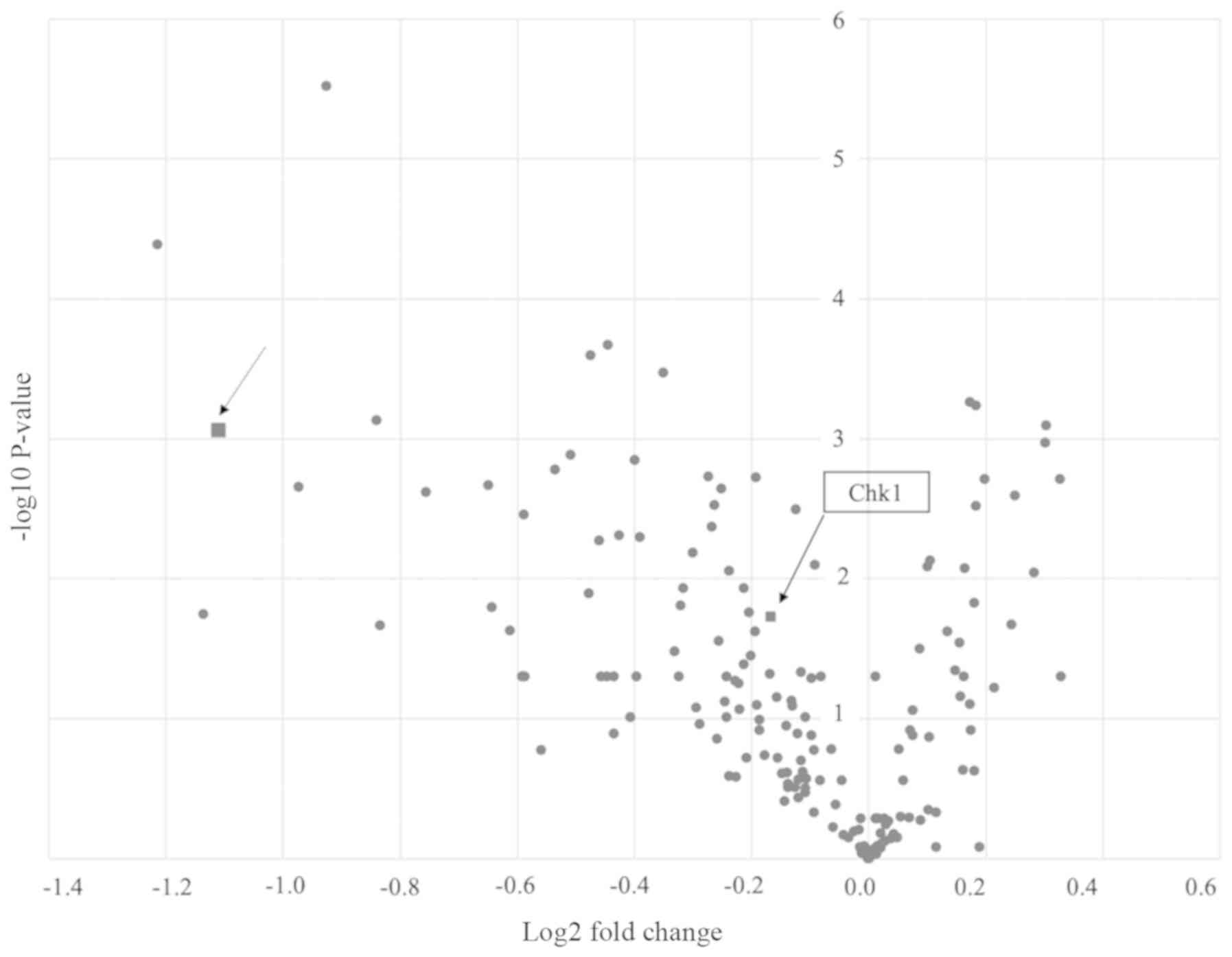
The siRNA library screening was conducted by
focusing on the cell survival rate among 169 cell cycle-related
genes. We prepared HNF-1β knockdown and control cell line using
TOV-21G (HNF-1β overexpressing OCCC cell line), and transfected
siRNA library to both cell lines. Our hypothesis was as follows: if
there would be a candidate gene related to HNF-1β signaling, the
wild type cell line (with HNF-1β overexpression) would show low
viability. TOV-21G si-control cell line showed significant reduced
rate of cell survival compared with TOV-21G si-HNF-1β cells
(46.31±8.60 vs. 100.00±5.82, P=0.001; Fig. 1). As a result, GSK-3β was extracted
as the candidate gene. As each siRNA library consisted of four
different sequences, we determined the most effective sequence
(5'-GAAGUCAGCUAUACAGACA-3') by MTT assay.
GSK-3β plays important role in growth
of HNF-1β overexpressing cells
To determine whether GSK-3β has a selective effect
in cell lines with or without HNF-1β, we assessed the effect of
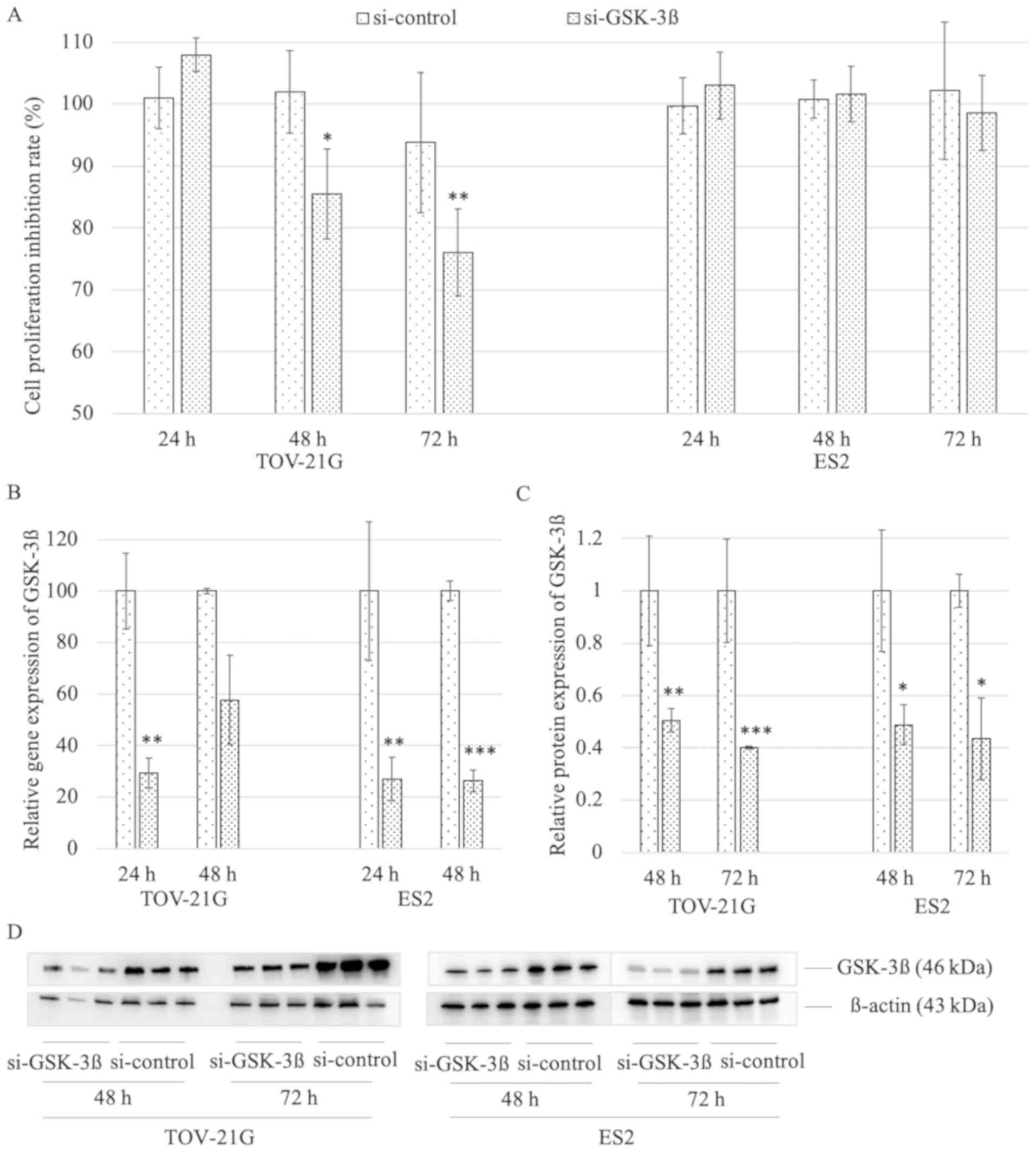
GSK-3β RNA interference by MTT assay. In TOV-21G (HNF-1β
overexpression), si-GSK-3β group showed significantly decreased
cell proliferation at 48 and 72 h to si-control group (85.47±7.24
vs. 101.93±6.65, P=0.015; 76.03±7.02 vs. 93.79±11.29, P=0.009;
respectively). In contrast, the si-GSK-3β in ES2 (HNF-1β negative)
cells did not show differentiation neither at 48 and 72 h
(101.61±4.44 vs. 100.79±3.09; 98.61±6.08 vs. 102.16±11.09)
(Fig. 2A). To confirm the
interference of GSK-3β, we further assessed the relative GSK-3β
mRNA and protein expression levels between GSK-3β knockdown group
and control group by RT-PCR and western blotting, respectively. As
a result, GSK-3β knockdown group was confirmed to sufficiently
suppress mRNA expression of GSK-3β (Fig. 2B), and protein levels of GSK-3β were
also decreased in GSK-3β knockdown group compared to control group
(Fig. 2C and D).
Interference of GSK-3β expression
affects cell cycle
We assessed the effect of interference of GSK-3β on
cell cycle and apoptosis in TOV-21G cells. Knockdown of GSK-3β
resulted in sub-G1 phase accumulation and S phase reduction
compared with the control group (10.84±1.66 vs. 3.60±0.45, P=0.002;
16.64±0.06 vs. 24.21±0.56, P<0.001, respectively) (Fig 3A). However, in the apoptosis assay,
no significant difference was observed between the si-GSK-3β and
control group (Fig. 3B).
HNF-1β regulates expression of GSK-3β
and phosphorylation of NFκB
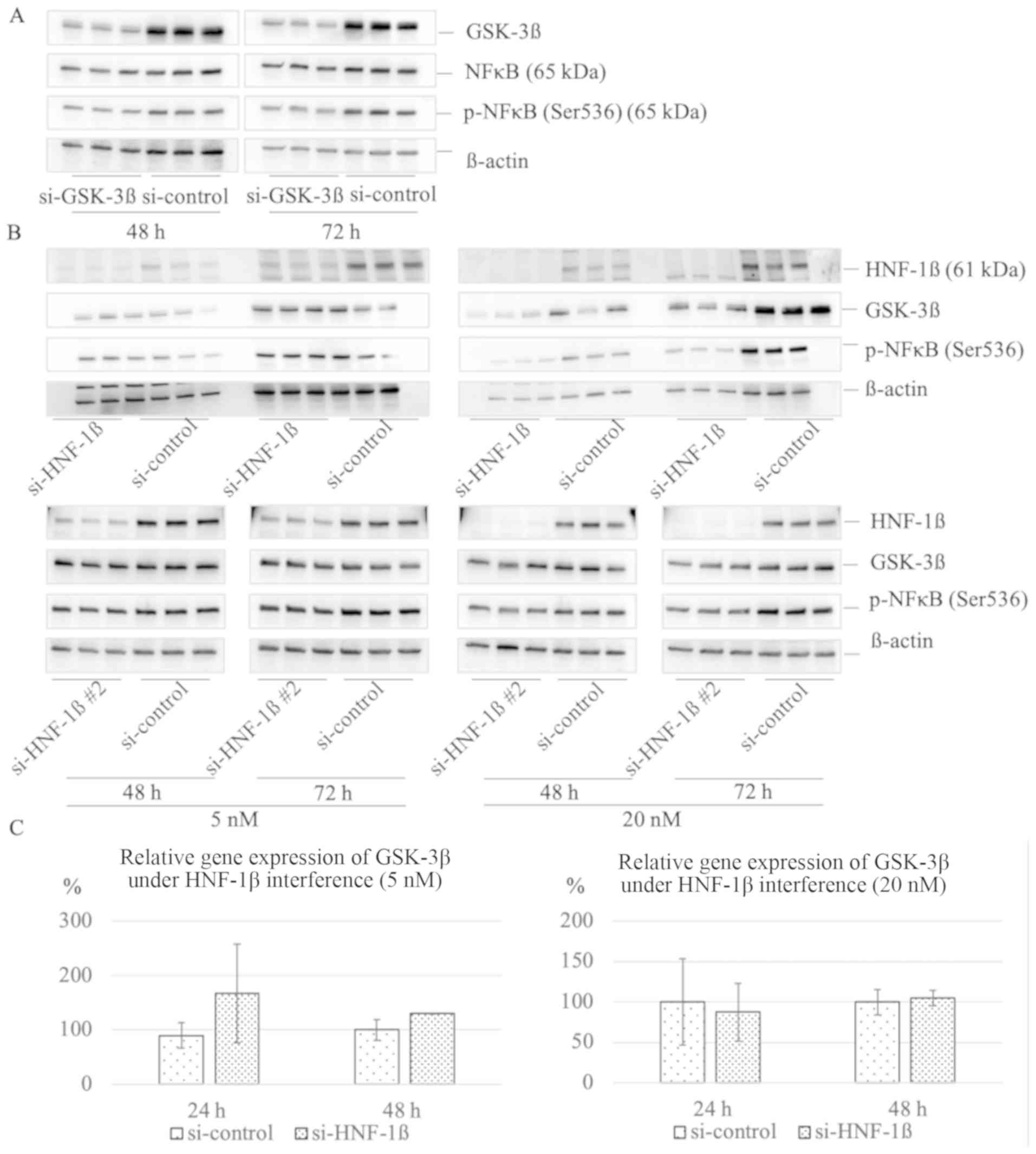
Previous studies have shown that GSK-3β regulates
serine 536 phosphorylation of NFκB subunit (9,10). We
confirmed effective reduction of GSK-3β expression at 48 h and 72 h
(0.60±0.043 vs. 1.00±0.11, P=0.004; 0.43±0.068 vs. 1.00±0.021,
P<0.001, respectively). At 48 h after transfection of 5 nM
si-GSK-3β, there was no significant reduction in the levels of
phosphorylated NFκB subunit (p-NFκB) compared with the si-control
group (0.97±0.11 vs. 1.00±0.15, P=0.801); however, at 72 h, a
significant reduction of p-NFκB (0.64±0.015 vs. 1.00±0.034,
P<0.001) was observed (Fig. 4A).
To assess whether HNF-1β have an effect on GSK-3β protein
expression and phosphorylation of NFκB subunit, we transfected 5 or
20 nM of si-HNF-1β or si-HNF-1β #2 into TOV-21G. At the lower
concentration, si-HNF-1β #2 (5 nM) reduced phosphorylation of NFκB
at 72 h (0.80±0.033 vs. 1.00±0.019, P=0.001). At the higher
concentration of si-HNF-1β or si-HNF-1β #2 (20nM), the expression
of GSK-3β at 72 h was significantly suppressed (0.63±0.13 vs.
1.00±0.14, P=0.029; 0.64±0.079 vs. 1.00±0.14, P=0.018,
respectively), and phosphorylation of NFκB was significantly
reduced at 48 h (0.29±0.18 vs. 1.00±0.25, P=0.015; 0.70±0.14 vs.
1.00±0.045, P=0.021, respectively) and 72 h (0.37±0.025 vs.
1.00±0.16, P=0.002, 0.59±0.035 vs. 1.00±0.074, P=0.001;
respectively) compared with the control group (Fig. 4B). We investigated whether HNF-1β
directly regulate GSK-3β messenger RNA, but HNF-1β does not promote
production of GSK-3β mRNA (Fig.
4C)
HNF-1β plays important role on
regulating GSK-3β activity
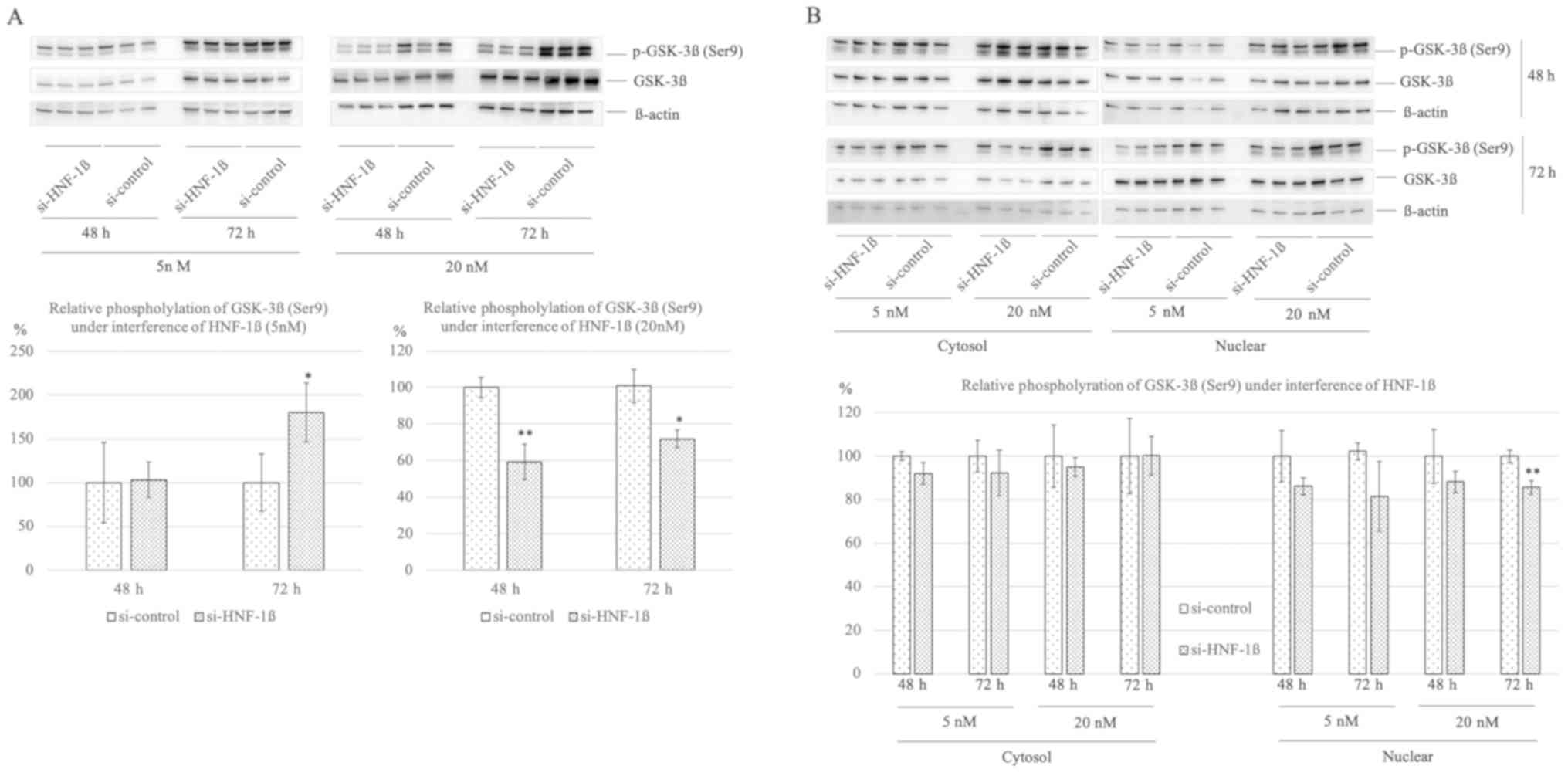
To investigate GSK-3β protein activity and
stability, relative phosphorylation of GSK-3β (Ser 9) was assessed
under lower and higher concentration of HNF-1β interference at 48
and 72 h. Under lower interference of HNF-1β, relative GSK-3β
phosphorylation increased compared to si-control at 72 h
(180.27±33.81 vs. 100.00±32.61, P=0.042). Under higher interference
of HNF-1β, relative GSK-3β phosphorylation significantly decreased
compared to si-control at both times (59.24±9.75 vs. 100.00±5.64,
P=0.003; 69.08±4.92 vs. 100.00±9.17, P=0.049; Fig. 5A). Furthermore, we assessed whether
phosphorylated GSK-3β (Ser 9) distribution differ in the cytosol or
nuclear under lower and higher HNF-1β interference. There were
trends to decrease relative GSK-3β phosphorylation in cytosol and
nuclear. Especially, under higher interference of HNF-1β at 72 h,
significant reduction of relative GSK-3β phosphorylation was
confirmed compared to si-control (85.65±3.17 vs. 100.00±2.91,
P=0.004; Fig. 5B).
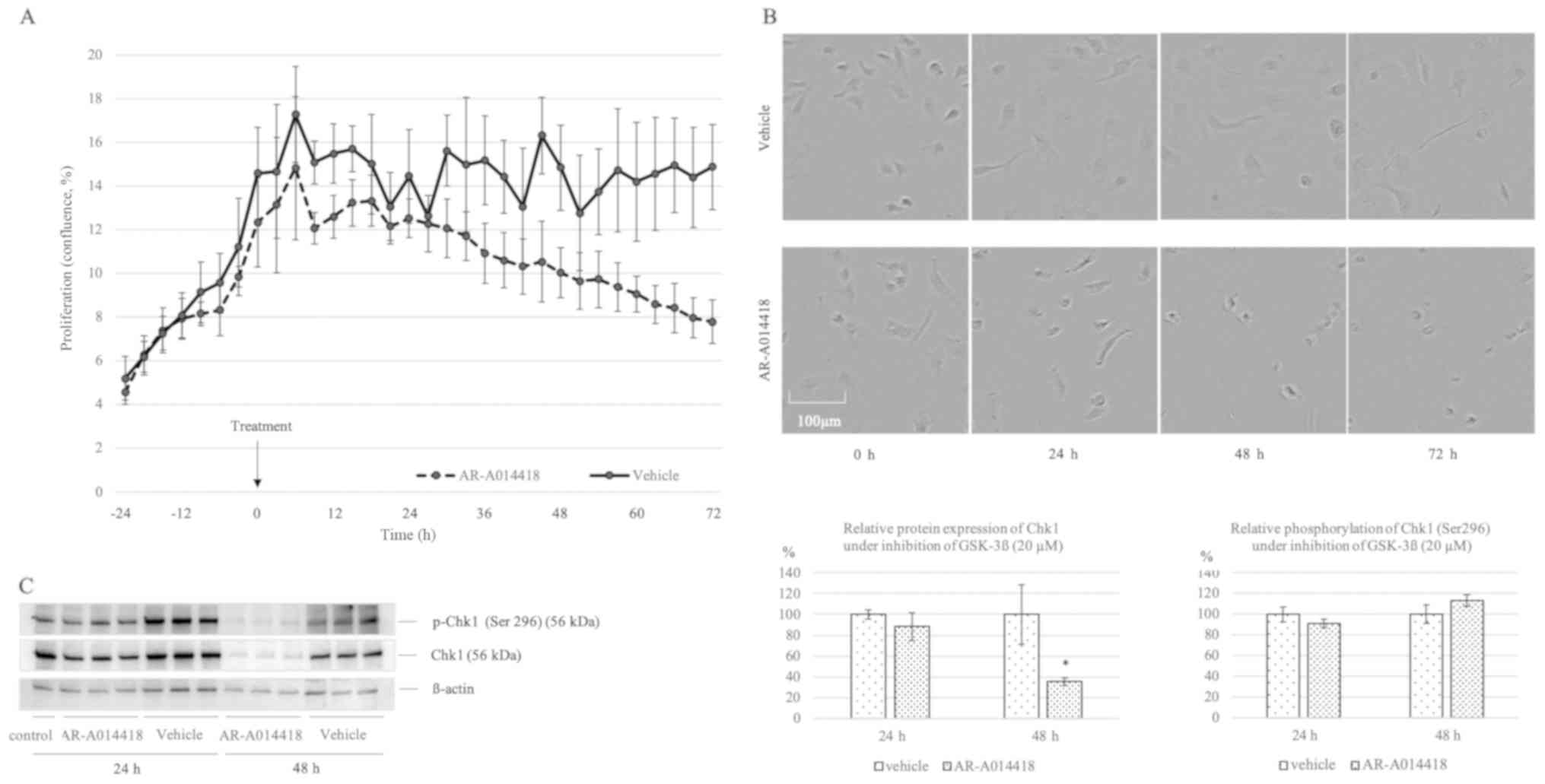
To determine whether GSK-3β inhibitor AR-A014418 is
cytotoxic to TOV-21G cells in vitro, cell proliferation was
determined after exposure to AR-A014418 by the IncuCyte ZOOM™
Live-Cell Imaging system. TOV-21G cells were grown in 6-well dish
(2.0x105 cells per well) treated by 42 µM bleomycin at 0
h. The AR-A014418 or vehicle group were measured confluence (%)
every 3 h by IncuCyte Zoom (Fig.
6A). IncuCyte Zoom imaging suggested that TOV-21G cells treated
by AR-A014418 lose its adherence or migration activity (Fig. 6B). We assessed whether GSK-3β act as
a downstream target to promote Chk1 activation as phosphorylation.
Protein expression of Chk1 and relative phosphorylation of Chk1
upon treatment of GSK-3β inhibitor after bleomycin stimulation were
assessed. Although relative Chk1 phosphorylation showed no
differentiation, protein expression of Chk1 decreased in inhibitor
AR-A014418 group compared to vehicle group at 48 h (35.29±3.63 vs.
100.00±28.42, P=0.017; Fig.
6C).
HNF-1β expression level affects the
efficacy of GSK-3β inhibitor
Cells were treated with two structurally distinct
pharmacological inhibitors of GSK-3β (AR-A014418 and SB-216763) to
determine whether GSK-3β is downstream of the HNF-1β signaling
pathway by cell viability. Consistent with the western blotting
result, lower knockdown of HNF-1β with a low siRNA concentration (5
nM) did not yield a significant difference between the si-HNF-1β
and si-control groups. However, under higher concentration (20 nM),
the knockdown of HNF-1β significantly rescued the effect of both
inhibitors (applied at 40 µM) (72.43±5.69 vs. 39.53±2.86,
P<0.001; 67.86±9.73 vs. 30.89±4.12, P<0.001, respectively)
(Fig. 7).
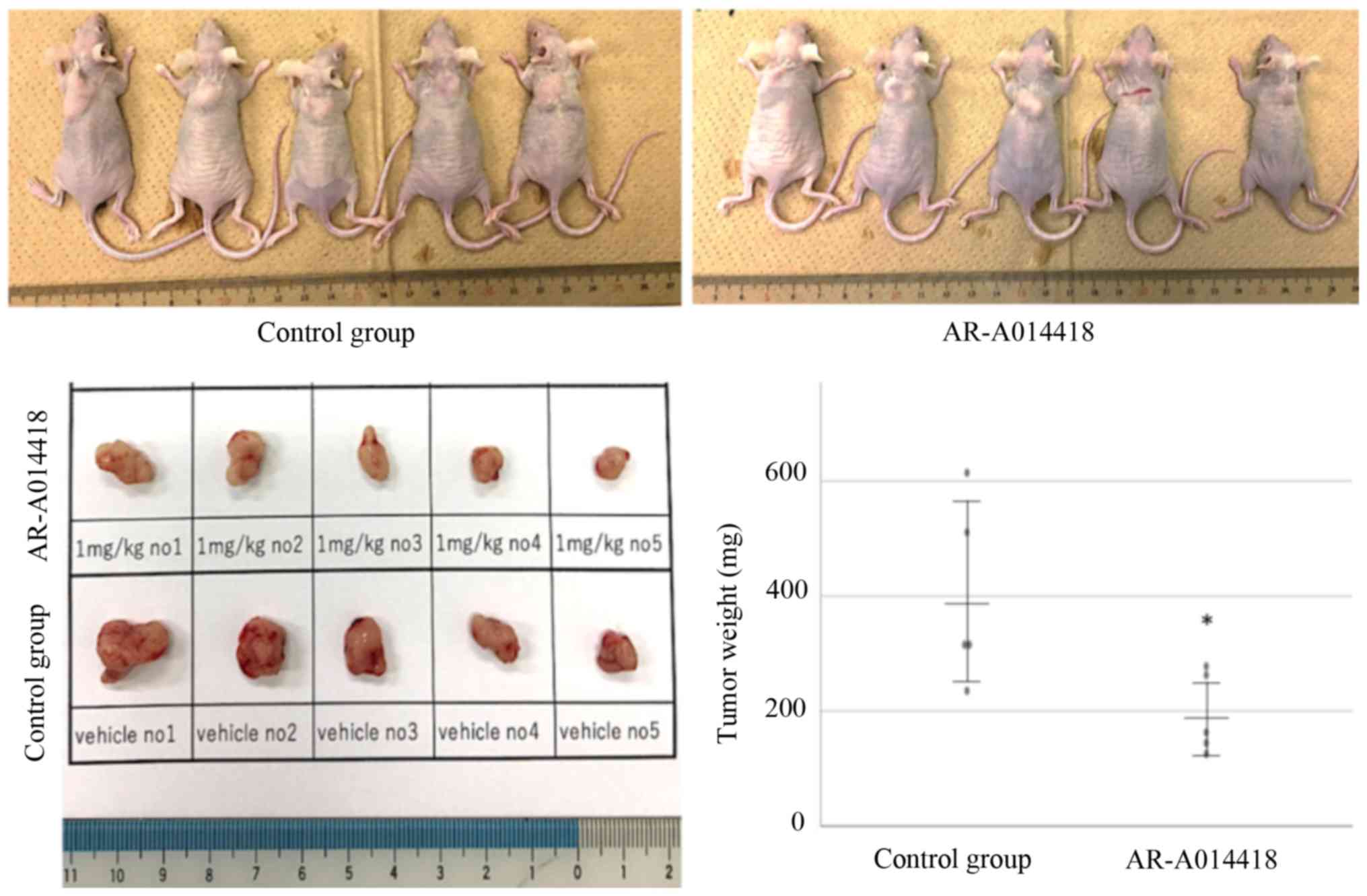
AR-A014418 inhibits tumor growth in
xenograft mouse model
To determine whether the GSK-3β inhibitor shows a
suppressive effect on tumor growth, we conducted an in vivo
assay using a xenograft mouse model. To make murine subcutaneous
tumors, 4.5x106 TOV-21G cells in 200 µl of PBS were
injected subcutaneously into the neck of the dorsal midline in 5-
to 6-week-old athymic nude mice (SLC). After tumor palpable point,
the inhibitor (AR-A014418) and vehicle were administered
intraperitoneally for 7 days. At the beginning and end of
administration, mice weight did not show significant
differentiation between inhibitor-treated group and vehicle group
(15.99±1.30 vs. 16.54±2.10, P=0.632; 17.27±1.38 vs. 16.39±1.94,
P=0.428, respectively). The inhibitor-treated group showed
significantly decreased tumor growth compared with the control
group (194.48±70.28 vs. 398.30±157.80, P=0.042; Fig. 8).
Discussion
A high concentration of free iron due to repeated
hemorrhage and inflammation is frequently detected in ovarian
endometriotic cysts; this condition leads to carcinogenesis through
iron-induced persistent oxidative stress and DNA damage (11-13).
Interestingly, iron-induced reactive oxygen species (ROS) signaling
promotes survival of endometriotic cell, possibly by activating the
detoxification and anti-apoptotic pathways via overexpression of
HNF-1β (3,4,14,15).
Given that some ROS act as a messenger in the TNF-α and okadaic
acid-induced post-translational activation of NFκB/p65(16), it is suggested that the persistent
oxidative stress in endometriotic cysts may serve as an activator
of NFκB signaling and HNF-1β expression.
Our previous study showed that HNF-1β is
overexpressed in OCCC (3). This
transcription factor is associated with cell survival and cell
cycle arrest at the G2 phase, concomitant with accumulation of
p-Chk1, a key regulator of the cell cycle arrest after a genotoxic
stress (3). We further identified
an ubiquitin hydrolase, USP28, as a candidate downstream target of
HNF-1β and as a stabilizer of Claspin protein, which is a binding
partner of Chk1. Taken together, HNF-1β regulates Claspin protein
stability and Chk1 protein activation, leading to the G2 cell cycle
arrest in response to DNA damage, by controlling the
USP28-dependent ubiquitin-proteasome pathway (4).
In the current study, we identified GSK-3β as a key
gene in HNF-1β overexpressing OCCC. Among several candidate genes,
GSK-3β was chosen to be a desirable target by following reasons: i)
small molecular inhibitor targeted HNF-1β is not exist. ii) GSK-3β
is reported not only to play an important role in cancer
progression, but also associated with several neuro degenerative
disease, including Parkinson's disease (5), Alzheimer's disease (17-21),
Huntington's disease (22).
Moreover in the several clinical trials using GSK-3β inhibitor
targeted Alzheimer's disease, advanced solid tumors and acute
myelogenous leukemia inhibitors of GSK-3β revealed its safety
(23-25).
GSK-3β has been previously reported to play an essential role in
maintaining constitutive NFκB reporter activity and expression of
NFκB target genes in pancreatic cancer cells (10). Moreover, the two GSK-3β isoforms
function to regulate constitutive NFκB activity in Panc-1 and
MiaPaCa-2 cells (9). We confirmed
that GSK-3β activates NFκB by phosphorylating it in the TOV-21G
OCCC cell line. We further showed that GSK-3β is a downstream
target of HNF-1β based on the following three observations: i) the
effect of GSK-3β interference on cell proliferation was observed
only in the background of HNF-1β overexpression. ii) Based on our
western blotting analysis, HNF-1β could regulate both the
expression of GSK-3β and phosphorylation of NFκB. iii) Strong
silencing of HNF-1β increased the IC50 of two
structurally distinct pharmacological inhibitors of GSK-3β
(AR-A014418 and SB-216763). Together, these results support that
GSK-3β is a candidate downstream target of HNF-1β.
Previous study reported that NFκB modulates
transcriptional upregulation of HNF-1β through alteration in bcl-2
expression (26). In the current
study, the stronger silencing of HNF-1β resulted in a more dramatic
change in the phosphorylation of NFκB rather than in the protein
expression NF-κB. This suggests that the role of GSK-3β is mainly
to phosphorylate NFκB to activate downstream signaling. To our best
knowledge, there are no reports regarding auto phosphorylation of
NFκB, suggesting that GSK-3β might be crucial for activation of
NFκB.
Our study has some limitations. Firstly, we
evaluated the effectiveness of GSK-3β inhibition only using two
types of OCCC cell lines, namely TOV-21G (HNF-1β overexpression)
and ES2 (HNF-1β negative). To validate these results and identify
more specific effects on HNF-1β overexpression, further studies
should be conducted using knocking or knockout HNF-1β cell lines.
Secondly, because HNF-1β does not promote production of GSK-3β mRNA
(Fig. 4C), there could be
involvement of some kind of proteasome degradation-related protein,
such as members of the USP family, as identified in our previous
study. Therefore, to investigate the detailed mechanism, further
screening focusing on the USP family is needed.
In conclusion, this study showed for the first time
that GSK-3β is a target gene of HNF-1β. In addition, our findings
reveal the novel pathway of HNF-1β-GSK-3β-p-NFκB axis in response
to DNA damage. Targeting this pathway may represent a putative,
novel, anticancer strategy in OCCC.
Acknowledgements
Not applicable.
Funding
The current study was supported by JSPS KAKENHI
(grant no. JP16K11150) and the Kanzawa Medical Research Foundation
(grant no. 30-28).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contribution
NK and HK deigned the current study. NK and AM
collected data from PubMed and performed the experiments. AM, SM
and YT conducted in vivo experiments. NK wrote and proofread
the manuscript. All authors read and approved the manuscript
Ethics approval and consent to
participate
The present study was approved by the Animal Ethics
Committee of Nara Medical University (reference no. 12594).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Ervik M,
Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D and
Bray F (eds): GLOBOCAN 2012: Estimated Cancer Incidence, Mortality
and Prevalence Worldwide in 2012. IARC CancerBase No. 11.
International Agency for Research on Cancer, Lyon, 2013.
|
|
2
|
Tsuchiya A, Sakamoto M, Yasuda J, Chuma M,
Ohta T, Ohki M, Yasugi T, Taketani Y and Hirohashi S: Expression
profiling in ovarian clear cell carcinoma: Identification of
hepatocyte nuclear factor-1 beta as a molecular marker and a
possible molecular target for therapy of ovarian clear cell
carcinoma. Am J Pathol. 163:2503–2512. 2003.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shigetomi H, Sudo T, Shimada K, Uekuri C,
Tsuji Y, Kanayama S, Naruse K, Yamada Y, Konishi N and Kobayashi H:
Inhibition of cell death and induction of G2 arrest accumulation in
human ovarian clear cells by HNF-1β transcription factor:
Chemosensitivity is regulated by checkpoint kinase CHK1. Int J
Gynecol Cancer. 24:838–843. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ito F, Yoshimoto C, Yamada Y, Sudo T and
Kobayashi H: The HNF-1β-USP28-Claspin pathway upregulates DNA
damage-induced Chk1 activation in ovarian clear cell carcinoma.
Oncotarget. 9:17512–17522. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Senkel S, Lucas B, Klein-Hitpass L and
Ryffel GU: Identification of target genes of the transcription
factor HNF1beta and HNF1alpha in a human embryonic kidney cell
line. Biochim Biophys Acta. 1731:179–190. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tanaka T, Tomaru Y, Nomura Y, Miura H,
Suzuki M and Hayashizaki Y: Comprehensive search for
HNF-1beta-regulated genes in mouse hepatoma cells perturbed by
transcription regulatory factor-targeted RNAi. Nucleic Acids Res.
32:2740–2750. 2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2 (Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Johnston ST, Shah ET, Chopin LK, Sean
McElwain DL and Simpson MJ: Estimating cell diffusivity and cell
proliferation rate by interpreting IncuCyte ZOOM™ assay data using
the Fisher-Kolmogorov model. BMC Syst Biol. 9(38)2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wilson W III and Baldwin AS: Maintenance
of constitutive IkappaB kinase activity by glycogen synthase
kinase-3α/β in pancreatic cancer. Cancer Res. 68:8156–8163.
2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Ougolkov AV, Fernandez-Zapico ME, Savoy
DN, Urrutia RA and Billadeau DD: Glycogen synthase kinase-3beta
participates in nuclear factor kappaB-mediated gene transcription
and cell survival in pancreatic cancer cells. Cancer Res.
65:2076–2081. 2005.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kajihara H, Yamada Y, Kanayama S, Furukawa
N, Noguchi T, Haruta S, Yoshida S, Sado T, Oi H and Kobayashi H:
Clear cell carcinoma of the ovary: Potential pathogenic mechanisms
(Review). Oncol Rep. 23:1193–1203. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yamaguchi K, Mandai M, Toyokuni S,
Hamanishi J, Higuchi T, Takakura K and Fujii S: Contents of
endometriotic cysts, especially the high concentration of free
iron, are a possible cause of carcinogenesis in the cysts through
the iron-induced persistent oxidative stress. Clin Cancer Res.
14:32–40. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Niiro E, Kawahara N, Yamada Y, Yoshimoto
C, Shimada K, Sudo T and Kobayashi H: Immunohistochemical
expression of CD44v9 and 8-OHdG in ovarian endometrioma and the
benign endometriotic lesions adjacent to clear cell carcinoma. J
Obstet Gynaecol Res. 45:2260–2266, Epub ahead of print.
2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yamada Y, Shigetomi H, Onogi A, Haruta S,
Kawaguchi R, Yoshida S, Furukawa N, Nagai A, Tanase Y, Tsunemi T,
et al: Redox-active iron-induced oxidative stress in the
pathogenesis of clear cell carcinoma of the ovary. Int J Gynecol
Cancer. 21:1200–1207. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Shigetomi H, Higashiura Y, Kajihara H and
Kobayashi H: A potential link of oxidative stress and cell cycle
regulation for development of endometriosis. Gynecol Endocrinol.
28:897–902. 2012.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Schmidt KN, Amstad P, Cerutti P and
Baeuerle PA: The roles of hydrogen peroxide and superoxide as
messengers in the activation of transcription factor NF-κ B. Chem
Biol. 2:13–22. 1995.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Y, Huang N, Yan F, Jin H, Zhou S,
Shi J and Jin F: Diabetes mellitus and Alzheimer's disease: GSK-3β
as a potential link. Behav Brain Res. 339:57–65. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Llorens-Martín M, Jurado J, Hernández F
and Avila J: GSK-3β, a pivotal kinase in Alzheimer disease. Front
Mol Neurosci. 7(46)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Llorens-Martín M, Blazquez-Llorca L,
Benavides-Piccione R, Rabano A, Hernandez F, Avila J and DeFelipe
J: Selective alterations of neurons and circuits related to early
memory loss in Alzheimer's disease. Front Neuroanat.
8(38)2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Llorens-Martín M, Fuster-Matanzo A,
Teixeira CM, Jurado-Arjona J, Ulloa F, Defelipe J, Rábano A,
Hernández F, Soriano E and Avila J: Alzheimer disease-like cellular
phenotype of newborn granule neurons can be reversed in
GSK-3β-overexpressing mice. Mol Psychiatry. 18:395–95.
2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Llorens-Martín M, Fuster-Matanzo A,
Teixeira CM, Jurado-Arjona J, Ulloa F, Defelipe J, Rábano A,
Hernández F, Soriano E and Avila J: GSK-3β overexpression causes
reversible alterations on postsynaptic densities and dendritic
morphology of hippocampal granule neurons in vivo. Mol Psychiatry.
18:451–460. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lim NK, Hung LW, Pang TY, Mclean CA,
Liddell JR, Hilton JB, Li QX, White AR, Hannan AJ and Crouch PJ:
Localized changes to glycogen synthase kinase-3 and collapsin
response mediator protein-2 in the Huntington's disease affected
brain. Hum Mol Genet. 23:4051–4063. 2014.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gray JE, Infante JR, Brail LH, Simon GR,
Cooksey JF, Jones SF, Farrington DL, Yeo A, Jackson KA, Chow KH, et
al: A first-in-human phase I dose-escalation, pharmacokinetic, and
pharmacodynamic evaluation of intravenous LY2090314, a glycogen
synthase kinase 3 inhibitor, administered in combination with
pemetrexed and carboplatin. Invest New Drugs. 33:1187–1196.
2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Rizzieri DA, Cooley S, Odenike O, Moonan
L, Chow KH, Jackson K, Wang X, Brail L and Borthakur G: An
open-label phase 2 study of glycogen synthase kinase-3 inhibitor
LY2090314 in patients with acute leukemia. Leuk Lymphoma.
57:1800–1806. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang Z, Smith KS, Murphy M, Piloto O,
Somervaille TC and Cleary ML: Glycogen synthase kinase 3 in MLL
leukaemia maintenance and targeted therapy. Nature. 455:1205–1209.
2008.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Suzuki E, Kajita S, Takahashi H, Matsumoto
T, Tsuruta T and Saegusa M: Transcriptional upregulation of HNF-1β
by NF-κB in ovarian clear cell carcinoma modulates susceptibility
to apoptosis through alteration in bcl-2 expression. Lab Invest.
95:962–972. 2015.PubMed/NCBI View Article : Google Scholar
|