Introduction
Ovarian cancer is a fatal gynecological malignant
tumor. Although ovarian cancer has a lower incidence rate than
endometrial cancer, with 313,959 new cases in 2020, it is
associated with a high mortality rate of 207,252 cases, ranking it
third among all gynecological malignancies (1). Based on histological differences, as
demonstrated by the World Health Organization, ovarian cancer is
divided into four categories: i) Epithelial; ii)
gonadal-mesenchymal; iii) germ cell; and iv) metastatic. Epithelial
ovarian cancer (EOC) is the most common type of ovarian cancer. Of
note, >70% of patients with EOC are diagnosed at an advanced
stage, and the 5-year survival is ~48% (2). With extensive research performed on
EOC, several treatment modalities are available, including surgical
treatment, chemotherapy, immunotherapy, targeted therapies and
others; however, primary debulking surgery and combination
chemotherapeutic regimens, including paclitaxel (Taxol), remain the
standard of care for patients with advanced-stage EOC (3–5).
Although patients with EOC initially respond to treatment, the
majority of them experience relapse within a few years due to
chemotherapeutic resistance, which is one of the reasons for the
low survival rate of patients with EOC (6,7).
Paclitaxel is one of the first-line drugs approved
for the treatment of EOC, which has a unique mechanism of action,
and is considered to be one of the most successful natural
anticancer drugs (8,9). The mechanism of Taxol involves binding
with the 31-amino acid N-terminal from the β-microtubule protein
subunit to induce microtubule stability and prevent its
depolymerization, generating G2/M phase accumulation in tumor
cells, thus inhibiting mitosis and cell proliferation, and
promoting cell apoptosis (10–12).
However, the development of resistance to Taxol severely limits the
clinical chemotherapeutic efficacy in patients with EOC (13). It has been reported that the
upregulation of cytoplasmic polyadenylation element binding protein
4 promotes Taxol resistance in ovarian cancer via the translational
regulation of CSAG family member 2 in vitro (14). Feng et al (15) demonstrated that glucose-6-phosphate
dehydrogenase promoted Taxol resistance in EOC cells by regulating
the expression of glutathione S-transferase P1. However, Taxol
resistance is a complex process. It is important to identify novel
promising gene targets associated with Taxol resistance for EOC
drug therapy, targeted elimination of drug resistance, and
improvement of treatment efficacy and patient prognosis.
In the present study, transcriptome sequencing
technology and the Gene Expression Omnibus (GEO; an international
public repository of microarray chips, second-generation sequencing
and other forms of high-throughput genomic data uploaded by
researchers worldwide) dataset were used to explore the genes
related to paclitaxel resistance in an EOC cell line. Through
bioinformatics analysis and verification in vitro, the
present study aimed to find new targets with potential as a
molecular marker of EOC resistance, and also to provide a new basis
for the clinical prediction of the key molecular mechanisms of
Taxol resistance.
Materials and methods
Cell culture
The human EOC A2780 cell line and the
A2780-Taxol-resistant cell line were purchased from ImmoCell
Biotechnology Co., Ltd. (provided by American Tissue Culture
Collection). A2780 and A2780/Taxol cells were cultured in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Zhejiang Tianhang Biotechnology Co., Ltd.) and 1%
penicillin-streptomycin solution (Biosharp Life Sciences), at 37°C
in a humidified incubator with 5% CO2. The medium for
the A2780 cell line was additionally supplemented with 1%
L-glutamine (Procell Life Science & Technology Co., Ltd.). The
medium for the A2780/Taxol cell line was also supplemented with 60
ng/ml Taxol (cat. no. H20203702; Sichuan Huiyu Pharmaceutical Co.,
Ltd.).
RNA-seq analysis
Total RNA was extracted from ~1×106 cells
using Trizol (Beyotime Institute of Biotechnology), followed by
RNA-seq analysis performed by BGI [Platform: DNBSEQ (Homo
sapiens)]. The RNA-seq data were filtered with SOAPnuke (version,
1.5.2). Reads were mapped to the reference genome (version,
GCF_000001405.39_GRCh38.p13) and aligned using HISAT2 (version,
2.0.4) and Bowtie2 (version, 2.2.5) for comparison with the human
reference genome.
DEG analysis
Gene expression levels were calculated for both the
A2780 and the A2780/Taxol cell lines in the present study using
RSEM (version 1.2.8) to screen DEGs1 with |log2(Fold Change)|≥1 and
FDR≤0.001. The gene expression level file GSE159791 (https://www.ncbi.nlm.nih.gov/geo/) of A2780/Taxol
cells was downloaded from the GEO database (16). Combination with DEGs1, common genes
were selected and analyzed using the R software package DESeq2
(version 1.42.1) with the A2780 cell as the control. DEGs were
screened from common genes with |log2(Fold Change)|≥1 and
Padj<0.01. Results were visualized using the R
packages pheatmap (version 1.0.12) and ggplot2 (version 3.4.3).
Gene ontology (GO) and Kyoto
encyclopedia of genes and genomes (KEGG) enrichment analysis
GO terms and pathways were obtained from the GO
(ftp://ftp.ncbi.nih.gov/gene/DATA/gene2go.gz) and KEGG databases
(version 101.0). The P-value was calculated using hypergeometric
test in the function phyper of R, and the package qvalue (version
2.4.2) was used to perform a multiple positive test on the P-value.
Q-value (corrected P-value) of <0.05 as the threshold to select
the significantly enriched GO term and pathway, visualized with R
language package GOplot (version 1.0.2) and ggplot2. The files of
protein-protein interaction (PPI) analysis for DEGs in the pathway
were downloaded from the STRING database (https://cn.string-db.org/), and visualized with
Cytoscape (version 3.8.0).
MTT cell viability assay
A2780 and A2780/Taxol cells (6×103
cells/well) were cultured into 96-well plates, treated with varying
concentrations of Taxol solution (0, 30, 60, 120, 240, or 480
ng/ml), with five replication wells per group. After 48 h, 10 µl
MTT (Biofroxx; NeoFroxx) was added to each well and incubated at
37°C for 4 h. The supernatant was discarded, and 150 µl DMSO
solution was added to measure absorbance at 490 nm and calculate
the IC50.
Western blotting
The A2780 and A2780/Taxol cells were lysed to
extract total protein with RIPA and PMSF solution (100:1 ratio;
Beyotime Institute of Biotechnology). Total protein concentration
was quantified with the BCA Protein Concentration Assay Kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. The protein samples (20 µg per lane)
were separated on 10% gels using SDS-PAGE. PVDF membranes (Beyotime
Institute of Biotechnology) were blocked with non-fat powder milk
for 2 h at room temperature and incubated overnight at 4°C with
rabbit anti-MMP1 (cat. no. AF0231; Beyotime Institute of
Biotechnology), rabbit anti-ZYX (cat. no. 38377; SAB
Biotherapeutics, Inc.) and rabbit anti-UNC5C (cat. no. 44671; SAB
Biotherapeutics, Inc.) diluted at a ratio of 1:1,000. Membranes
were washed with TBST (contain 0.1% Tween) and incubated with
HRP-conjugated affinipure goat anti-rabbit IgG (H+L) (1:1,000
dilution; cat. no. A0208; Beyotime Institute of Biotechnology) at
room temperature for 2 h. Bands were visualized using a
chemiluminescence kit (Beyotime Institute of Biotechnology).
Protein hybridization results were observed using an automated
chemiluminescence instrument (Tanon-5200; Ewell Biotechnology), and
abundance was processed using Fiji (−win64; ImageJ, National
Institutes of Health).
Immunohistochemistry
A total of 4×104 cells/ml were cultured
in 12-well plates overnight. Cells were fixed with 4%
paraformaldehyde (Wuhan Servicebio Technology Co., Ltd.) for 1 h at
room temperature. Cells were then washed with PBS, and 50–100 µl
film breaking solution was added to the plate for incubation at
room temperature for 20 min. A total of 3% BSA was added for
blocking for 30 min at room temperature, followed by incubation in
a wet box at 4°C overnight with the primary antibody (anti-MMP1:
1:100 dilution; cat. no. AF0231; Beyotime Institute of
Biotechnology; anti-ZYX: 1:100 dilution; cat. no. 38377; SAB
Biotherapeutics, Inc; and anti-UNC5C: 1:100 dilution; cat. no.
bs-11493R; Beijing Bioss Biotechnology Co., Ltd.). The plate was
incubated for 50 min with HRP-labeled secondary antibody (1:100
dilution; cat. no. GB23303; Wuhan Servicebio Technology Co., Ltd.),
and freshly prepared DAB color developing solution was added
controlling the color developing time. Hematoxylin was used as a
counterstain at room temperature for 3 min, and the hematoxylin
fractionation solution fractionated for a few sec. Hematoxylin
re-blueing solution (Wuhan Servicebio Technology Co., Ltd.) was
used for staining and rinsed. The slivers were dehydrated and
sealed with neutral gum. After light microscopic examination,
images were collected for analysis.
Cellular immunofluorescence (IF)
assay
For the IF assay, the medium was aspirated, and the
cell crawls were washed three times with cold PBS. After being
fixed with 4% paraformaldehyde for 30 min, the cells were
penetrated with membrane breaking working solution for 20 min.
Subsequently, the cells were blocked with 3% BSA for 30 min (Wuhan
Servicebio Technology Co., Ltd.), washed three times with PBS and
incubated with the primary antibody (same as aforementioned
immunohistochemistry antibodies) overnight. After washing three
times with PBS, the cells were incubated with the secondary
antibody (1:100 dilution; cat. no. E032420; EarthOx Life Sciences)
coupled with DyLight 594-TFP ester for 2 h. After washing three
times with PBS, the cells were stained with DAPI dye solution, and
incubated for 10 min in the dark. Finally, the cell crawls were
washed three times with PBS, and blocked with an anti-fluorescence
quencher. Images were captured using a fluorescence microscope
(BX51-32FL; Olympus Corporation). The average fluorescence density
was calculated using Fiji.
Analysis on clinical information and
RNA-seq data from the cancer genome atlas (TCGA)
Clinical information of patients with EOC and other
cancers was obtained from TCGA database using the R package
TCGAbiolinks (version 3.14). RNA-seq data and survival-related
files of patients from TCGA database were downloaded using the UCSC
Xena online tool (https://xenabrowser.net/datapages/) for subsequent
analysis.
Statistical analysis
Kaplan-Meier survival curves were created and
visualized using the packages survminer (version 0.4.9), survival
(version 3.5–7) and TSHRC (version 0.1–6; http://cran.r-project.org/web/packages/TSHRC/TSHRC.pdf)
in R (version 4.3.1). Statistical analysis was carried out using
the log-rank test for Kaplan-Meier survival curves. One-way ANOVA
and least significant difference tests were used for the
statistical analysis and were performed using SPSS (version 26.0;
IBM Corp.); visualization was carried out using GraphPad Prism
(version 5; Dotmatics). P<0.05 was considered to indicate a
statistically significant difference. A total of three biologically
independent repeats were carried out, and the data are presented as
mean ± SD.
Results
DEGs in A2780/Taxol cells
The drug resistance index (RI) of the A2780/Taxol
cell line in the present study was 33.62, exhibiting highly
resistant characteristics (Fig.
S1B). The common gene expression profile was similar to that of
other Taxol-resistant cell lines in the GEO database, but different
from that of the A2780 cell line [Figs.
1A and S1B; other RI values
from the data in the study by Szenajch et al (16)]. Through differential expression
analysis, 6,226 DEGs (DEGs1) were identified in the transcriptome
sequencing data (Fig. 1B). The
A2780 cells were used in the present study as control to reprocess
the drug resistance data of the GEO database; 498 DEGs (DEGs2) were
finally obtained based on DEGs1 (Fig.
1C; screening method, Fig. S1C and
D; gene names, Table SI).
Discovering DEGs significantly
associated with the overall survival (OS) of patients with EOC and
Taxol resistance
As chemoresistance is associated with a decreased
survival rate of patients, genes in DEGs2 that were both up or
downregulated in the A2780/Taxol and drug resistance datasets were
selected for survival curve analysis. A total of 27 genes were
found to be significantly associated with the OS of patients with
EOC (Figs. 1D, S2 and S3). Considering that the log-rank test
might lose power when the survival curves crossed at a later stage
(17), a two-stage test (TS)
weighted analysis was carried out for POLR3GL, ZNF239, FLRT3 and
ZFHX4. The TS P-values of POLR3GL, ZNF239, FLRT3 and ZFHX4 were
0.66, 0.35, 0.35 and 0.28, respectively (Figs. S2 and S3). Since these values are >0.05, they
were excluded from subsequent analysis. Of these survival-related
genes, increased levels of GDF15 were found to be associated with
enzalutamide and EPI-001 resistance in prostate cancer cells
(18). NR1D2 was shown to be
associated with enzalutamide resistance in neuroendocrine prostate
cancer, and PHLDA1 was found to be associated with Lewis(y) highly
expressing chemoresistant ovarian cancer cell (19,20).
Imiquimod facilitates chemoresistance via the upregulation of MMP1,
and RNA interference targeting ZYX reduces tumor cell HN12
resistance to cisplatin (DDP) (21,22).
The downregulation of NCAM2 was shown to be associated with
resistance to the monoclonal antibody drug trastuzumab in
HER2+ breast cancer, and FBXL7 knockdown affects DDP
resistance in nasopharyngeal carcinoma (23,24).
In summary, these 23 genes were intimately associated with
resistance to chemotherapy in various types of cancer,
demonstrating the feasibility and accuracy of the screening
performed in the present study for these Taxol
resistance-associated genes that were closely associated with the
OS of patients with EOC.
KEGG and GO enrichment analysis of the
DEGs, MMP1, ZYX and UNC5C
To investigate the role of DEGs on the basis of cell
integral changes, the DEGs1 genes were subjected to GO and KEGG
enrichment analysis. A Q value of <0.05 was considered to
indicate significant enrichment. As shown by the KEGG enrichment
analysis, the most significant pathway was ‘focal adhesion’, while
in cellular component and biological process of GO, ‘cell junction’
and ‘cell adhesion’ were the significant pathways, indicating that
they were associated with Taxol resistance (Figs. 2A and S4A-C). In addition, the results of the
enrichment analysis indicated the involvement of the cell migration
process (25–27), suggesting that cell migration
affects the occurrence of Taxol resistance in A2780 cells. The 23
survival-related genes were mapped into the significant enrichment
pathways, and only MMP1, ZYX and UNC5C were enriched in the ‘PPAR
signaling’, ‘focal adhesion’ and ‘axon guidance’ pathways,
respectively (Fig. 2B). Therefore,
these three genes were used as gene targets for follow-up
experiments.
Role of MMP1, ZYX and UNC5C in KEGG
pathways through PPI network analysis
Through KEGG pathway analysis, the enrichment
pathways of MMP1, ZYX and UNC5C was identified. However, the
mechanisms through which these genes play a role in the pathway
remain unknown. Therefore, PPI network analysis was performed on
the DEGs enriched in ‘PPAR signaling’, ‘focal adhesion’ and ‘axon
guidance’ pathways. MMP1 interacted with angiopoietin-like 4
(ANGPTL4; Fig. 2C). It was
hypothesized that MMP1 may affect cell migration through ANGPTL4,
leading to the generation of drug resistance. The results from the
study by Liao et al (28)
confirmed that the downregulation of MMP1 hindered the migration
and invasion of head and neck squamous cell carcinoma cells
enhanced by EGF and recombinant ANGPTL4. ZYX interacted with
paxillin (PXN) and vinculin (VCL) (Fig.
2D). It was hypothesized that ZYX may affect cell adhesion
through PXN and VCL, thus mediating the generation of drug
resistance. The study by Legerstee et al (29) on protein pairs related to the
function of ‘focal adhesion’ demonstrated that the binding of ZYX
and PXN, and that of VCL and vasodilator stimulated phosphoprotein
affected cell adhesion and migration. In the PPI network of the
‘axon guidance pathway’ shown in Fig.
2E, FYN, the largest node, was the core of the network and
interacted with UNC5C. According to literature, the knockdown of
UNC5C enhances the phosphorylation of FAK and SRC (30), and FYN is a specific member of the
SRC kinase family (31). Therefore,
it was hypothesized that UNC5C mediated the generation of drug
resistance by affecting Src kinase activity through FYN.
Furthermore, these results confirmed the reliability of PPI network
analysis to examine the interactions between proteins, and provided
molecular information that the three targets may participate in the
drug resistance mechanisms of EOC.
At the same time, in order to investigate the
association of Taxol resistance of EOC with disease stage, the
associations between MMP1, ZYX and UNC5C, and the disease stage of
EOC (stage II, III and IV) were examined. The results indicated
that MMP1, ZYX and UNC5C were not associated with the disease stage
of patients with EOC (P>0.05, not statistically significant;
Fig. S4D-F).
MMP1, ZYX and UNC5C were significantly
associated with carboplatin (CBP), DDP and doxorubicin (DOX)
resistance in patients with EOC, respectively
Chemotherapeutic drugs for EOC, in addition to
Taxol, include CBP, DDP and DOX. CBP, DDP and DOX limit DNA
replication and transcription via various mechanisms. Although DNA
replication affected by CBP, DDP and DOX occurs during the S phase,
and Taxol induces G2/M phase accumulation, these drugs ultimately
lead to apoptosis, and they may share common signaling pathways and
networks in the final stage (32–34).
Therefore, it was hypothesized that Taxol resistance may cause
resistance to other chemotherapeutic drugs as well. In the present
study, CBP, DDP and DOX, as well as Taxol, were selected to analyze
the association between MMP1, ZYX and UNC5C and the OS of patients
with EOC in TCGA. The results revealed that MMP1, ZYX and UNC5C
were significantly associated with the survival of patients treated
with CBP, DDP and DOX, respectively, and this trend was consistent
with Taxol, indicating that Taxol may share key resistance-related
targets with CBP, DDP and DOX (Fig.
3). This finding preliminarily confirmed one of the ways that
Taxol resistance causes resistance to other chemotherapeutic
drugs.
Analysis of the association between
MMP1, ZYX and UNC5C and the OS of Taxol-treated patients with head
and neck squamous cell carcinoma (HNSC), lung cancer (LUNG) and
uterine corpus endometrial cancer (UCEC)
Taxol is also used clinically in the treatment of
patients with LUNG, HSSC and UCEC, as well as other types of cancer
(35–37). Regarding HNSC, LUNG and UCEC,
patients with a history of treatment with Taxol in TCGA, excluding
patients with free tumor tissue, were selected for Kaplan-Meier
survival curve analysis to investigate the association between
MMP1, ZYX and UNC5C, and the OS of other patients with cancer
treated with Taxol. The results revealed that only UNC5C was
significantly associated with the survival of patients with HNSC
treated with Taxol. MMP1, ZYX and UNC5C were not significantly
associated with the survival of patients with LUNG and UCEC treated
with Taxol (Fig. 4). The results
indicated that MMP1 and ZYX may be specific in Taxol resistance in
EOC, while the common resistance occurrence and mechanisms of UNC5C
in EOC and HNSC remain to be explored.
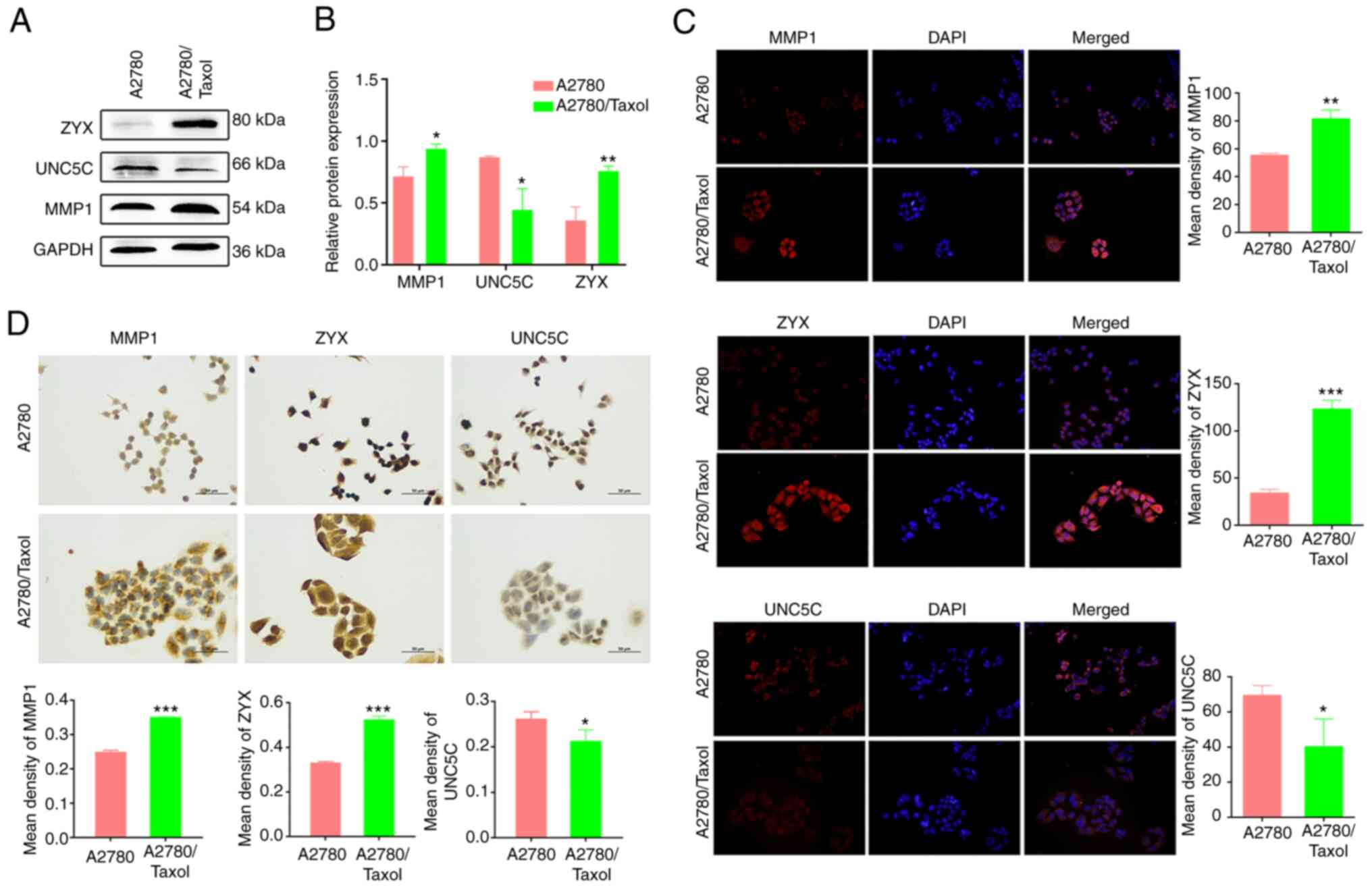
Expression of MMP1 and ZYX was
increased in A2780/Taxol cells, while UNC5C expression was
decreased
The protein expression of MMP1 and ZYX was
significantly increased in A2780/Taxol cells (P<0.05 and
P<0.01, respectively), while the expression of UNC5C was
significantly decreased (P<0.05; Fig. 5A). The same results were obtained by
immunofluorescence and immunohistochemistry (Fig. 5B and C), and were consistent with
the RNA-seq data. This demonstrated the reliability of the
selection of Taxol resistance-associated gene targets through
RNA-seq, and the stability and accuracy of the multiple validation
work. These in vitro cellular results confirmed that the
expression of MMP1 and ZYX was significantly upregulated, and the
expression of UNC5C was significantly downregulated in A2780/Taxol
cells, demonstrating that the three gene targets are potential and
promising molecular markers of Taxol resistance in EOC.
Discussion
The standard treatment for EOC is surgery and
chemotherapy. However, surgery for advanced-stage ovarian cancer
often leads to severe post-operative complications, including
patient mortality or the impossibility of the administration of
subsequent oncological treatments, which can directly affect the
survival rate (38). By contrast,
treatment with chemotherapeutic drugs is safer for patients.
However, Taxol is clinically ineffective as it often induces drug
resistance, leading to multidrug resistance. Therefore, in the
present study, Taxol resistance-related gene targets in patients
with EOC were selected to break through the reversal of drug
resistance, promote the clinical efficacy of Taxol and ultimately
improve the survival rate of patients with EOC.
In the present study, it was found that DEGs were
most significantly enriched in the focal adhesion pathway. Focal
adhesions are subcellular structures that provide strong adhesion
to the extracellular matrix (ECM) and serve as scaffolds for a
number of signaling pathways involving integrins or mechanical
forces applied to cells. Currently, focal adhesions have been
revealed to be a key determinant of cell migration and play a
critical role in promoting tumor cell invasion (39). The most significant enrichment
results in the GO analysis of DEGs were also associated with cell
connection or migration, indicating that DEGs may contribute to
Taxol resistance in EOC through cell migration.
Through Kaplan-Meier survival analysis and KEGG
pathway enrichment analyses of the DEGs, three Taxol
resistance-related gene targets were finally obtained: MMP1, ZYX
and UNC5C. MMP1 can affect ECM and basement membrane degradation or
increase AKT phosphorylation to activate the AKT pathway, leading
to cell migration (40,41). ZYX is an adhesion protein that
affects cell adhesion and cytoskeletal rearrangement, leading to
cell proliferation and migration (42,43).
UNC5C affects FAK and FYN activity, and mediates cell migration by
promoting integrin-dependent cell adhesion and increasing skeletal
rearrangements. The downregulation of UNC5C may also activate the
PI3K/AKT pathway and MMP9 expression, leading to cell migration and
proliferation (30,44). Moreover, FYN interacts with PXN,
both of which affect cell migration (45). Combined with the results of PPI
network analysis in the present study, the potential mechanism by
which MMP1, ZYX and UNC5C induce the development of EOC Taxol
resistance by promoting cell migration was obtained (Fig. 6). In addition, MMP1 was upregulated
in A2780/Taxol cells; however, MMP1 overexpression increased the OS
of Taxol-treated patients with EOC. It was hypothesized that MMP1
upregulation activated the AKT pathway to induce mitophagy,
promoting cell apoptosis, thus producing a potent self-protective
effect on the organism (46). This
eventually revealed the development of Taxol resistance in patients
with EOC, but an increase in survival. The downregulation of UNC5C
resulted in Taxol resistance; however, the increase in the OS of
patients with EOC may be due to the activation of AKT, similar to
MMP1. The present study analyzed the association between the
upregulation of ZYX expression and the OS of patients with EOC
treated with Taxol; the results revealed a significant reduction in
OS which was anticipated considering that ZYX upregulation could
initiate the cell migration pathway that induced Taxol resistance
and thus hindered the chemotherapeutic effect. These results
indicated that ZYX may be used as a potential marker of Taxol
resistance.
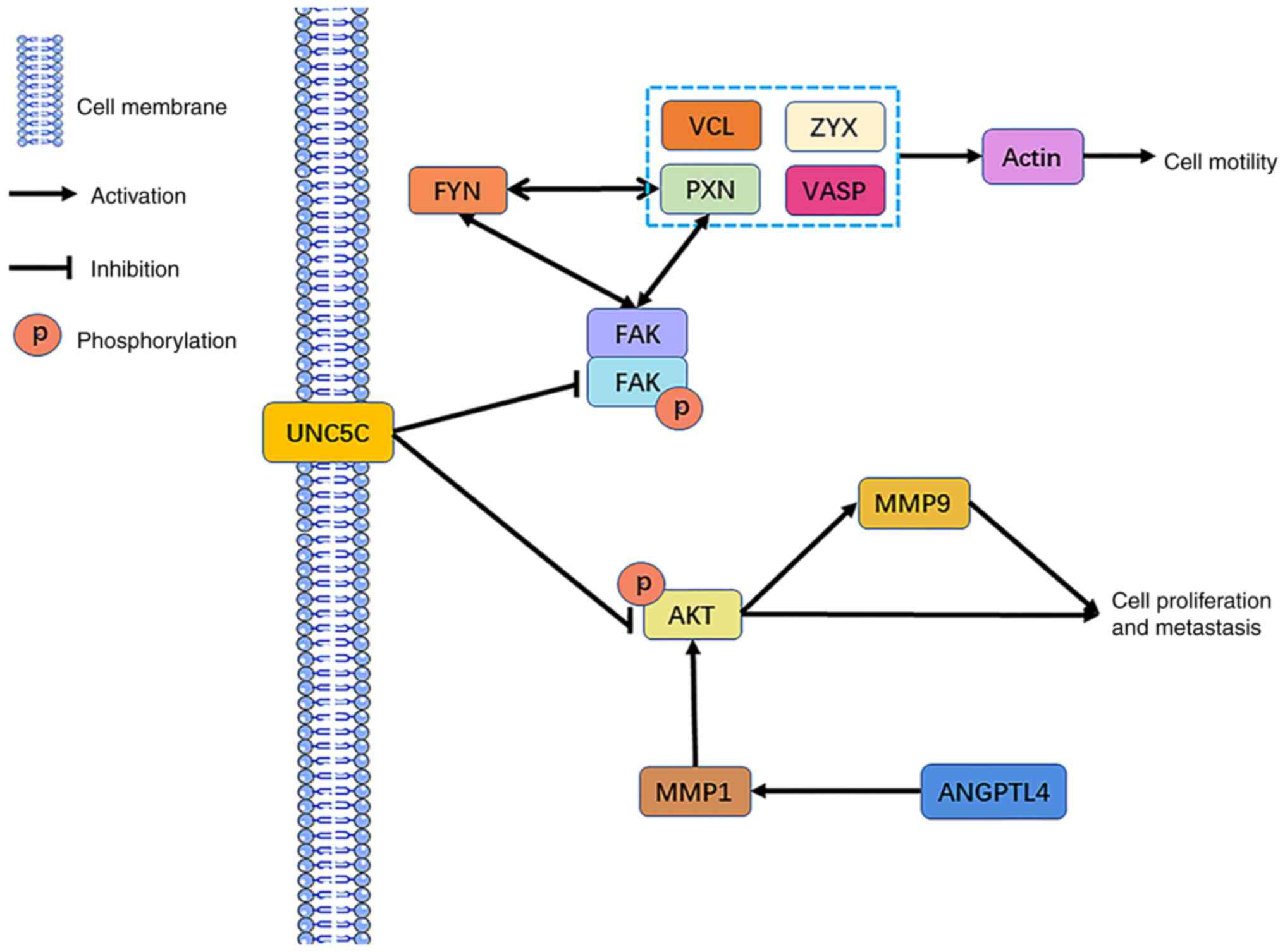 | Figure 6.Potential mechanisms of MMP1, ZYX and
UNC5C in Taxol resistance. ZYX, zyxin; MMP, matrix metalloprotease;
UNC5C, Unc-5 netrin receptor C; p, phosphorylated; VCL, vinculin;
PXN, paxillin; VASP, vasodilator stimulated phosphoprotein; FAK,
protein tyrosine kinase 2; ANGPTL4, angiopoietin like 4. |
In conclusion, the present study demonstrated that
Taxol resistance in A2780/Taxol cells originated from the
activation of molecular mechanisms related to cell migration.
Epithelial-mesenchymal transformation (EMT) is considered a
promoter of metastasis, during which cancer cells acquire mobility
and the ability to migrate from the primary site (47). Simultaneously, EMT mediates the
generation of chemical resistance in cancer (48). In the process of enhanced cell
migration, there is an inevitable generation of an EOC cell
population in the EMT transition state (49), which leads to the development of
Taxol resistance. On the other hand, it has been shown that
physical confinement during cancer cell migration triggers
therapeutic resistance (50). These
factors confirm the credibility of the results of the present
study.
In the present study, the three Taxol
resistance-related gene targets MMP1, ZYX and UNC5C in EOC
A2780/Taxol cells were selected using RNA-seq and bioinformatics
analysis, and validated in vitro in cellular experiments.
The present study provides novel drug resistance molecular targets
and insights for their clinical application in patients with EOC
with Taxol-induced multidrug resistance. These targets may enhance
the efficacy of treatment and improve the prognosis of
patients.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Medical and Health
Technology Project of Shenzhen Longgang District (grant no.
LGKCYLWS2021000023), the National Natural Science Foundation of
China (grant no. 81102753) and the ‘ovarian cancer chromosome
instability region molecular marker target and clinical application
research’ Enterprise Horizontal Project.
Availability of data and materials
The RNA-seq data are available in the GEO database
(accession no. GSE230667; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE230667).
Authors' contributions
RY, HZ, ZC, TZ, PW, HL, YH, CZ, XW and YZ
contributed to the study conception and design. Experimental
design, manuscript writing and experimental cost management were
performed by YZ, XW, RY and HZ. RY, ZC, TZ and PW conducted the
experiments and collected the data. Statistical analysis was
performed by RY, HL, YH and CZ. RY and YZ confirm the authenticity
of all the raw data. All authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EOC
|
epithelial ovarian cancer
|
|
DEGs
|
differentially expressed genes
|
|
RNA-seq
|
RNA-sequencing
|
|
OS
|
overall survival
|
|
paclitaxel
|
Taxol
|
|
MMP1
|
matrix metalloproteinase 1
|
|
ZYX
|
zyxin
|
|
UNC5C
|
Unc-5 netrin receptor C
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuroki L and Guntupalli SR: Treatment of
epithelial ovarian cancer. BMJ. 371:m37732020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lheureux S, Gourley C, Vergote I and Oza
AM: Epithelial ovarian cancer. Lancet. 393:1240–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elyashiv O, Wong YNS and Ledermann JA:
Frontline maintenance treatment for ovarian cancer. Curr Oncol Rep.
23:972021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haunschild CE and Tewari KS: The current
landscape of molecular profiling in the treatment of epithelial
ovarian cancer. Gynecol Oncol. 160:333–345. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang C, Xia BR, Zhang ZC, Zhang YJ, Lou G
and Jin WL: Immunotherapy for ovarian cancer: Adjuvant,
combination, and neoadjuvant. Front Immunol. 11:5778692020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gupta S, Nag S, Aggarwal S, Rauthan A and
Warrier N: Maintenance therapy for recurrent epithelial ovarian
cancer: Current therapies and future perspectives-a review. J
Ovarian Res. 12:1032019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Khayrani AC, Mahmud H, Oo AKK, Zahra MH,
Oze M, Du J, Alam MJ, Afify SM, Quora HAA, Shigehiro T, et al:
Targeting ovarian cancer cells overexpressing CD44 with
immunoliposomes encapsulating glycosylated taxol. Int J Mol Sci.
20:10422019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L and Chen L: Progress in research on
Taxol and tumor immunotherapy. Cell Mol Biol Lett. 24:402019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashrafizadeh M, Mirzaei S, Hashemi F,
Zarrabi A, Zabolian A, Saleki H, Sharifzadeh SO, Soleymani L,
Daneshi S, Hushmandi K, et al: New insight towards development of
paclitaxel and docetaxel resistance in cancer cells: EMT as a novel
molecular mechanism and therapeutic possibilities. Biomed
Pharmacother. 141:1118242021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu W, Wei T, Li Z and Zhu J: p53-dependent
apoptosis is essential for the antitumor effect of paclitaxel
response to DNA damage in papillary thyroid carcinoma. Int J Med
Sci. 18:3197–3205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao S, Tang Y, Wang R and Najafi M:
Mechanisms of cancer cell death induction by paclitaxel: An updated
review. Apoptosis. 27:647–667. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nan G, Zhao SH, Wang T, Chao D, Tian RF,
Wang WJ, Fu X, Lin P, Guo T, Wang B, et al: CD147 supports
paclitaxel resistance via interacting with RanBP1. Oncogene.
41:983–996. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Gan H, Zhao F, Ma X, Xie X, Huang
R and Zhao J: CPEB4-promoted paclitaxel resistance in ovarian
cancer in vitro relies on translational regulation of CSAG2. Front
Pharmacol. 11:6009942021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng Q, Li X, Sun W, Sun M, Li Z, Sheng H,
Xie F, Zhang S and Shan C: Targeting G6PD reverses paclitaxel
resistance in ovarian cancer by suppressing GSTP1. Biochem
Pharmacol. 178:1140922020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szenajch J, Szabelska-Beręsewicz A,
Świercz A, Zyprych-Walczak J, Siatkowski I, Góralski M, Synowiec A
and Handschuh L: Transcriptome remodeling in gradual development of
inverse resistance between paclitaxel and cisplatin in ovarian
cancer cells. Int J Mol Sci. 21:92182020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H, Han D, Hou Y, Chen H and Chen Z:
Statistical inference methods for two crossing survival curves: A
comparison of methods. PLoS One. 10:e01167742015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang R, Wen P, Yang G, Feng Y, Mi Y, Wang
X, Zhu S and Chen YQ: N-glycosylation of GDF15 abolishes its
inhibitory effect on EGFR in AR inhibitor-resistant prostate cancer
cells. Cell Death Dis. 13:6262022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He MX, Cuoco MS, Crowdis J, Bosma-Moody A,
Zhang Z, Bi K, Kanodia A, Su MJ, Ku SY, Garcia MM, et al:
Transcriptional mediators of treatment resistance in lethal
prostate cancer. Nat Med. 27:426–433. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu J, Zheng M, Qi Y, Wang H, Liu M, Liu Q
and Lin B: Lewis(y) antigen-mediated positive feedback loop induces
and promotes chemotherapeutic resistance in ovarian cancer. Int J
Oncol. 53:1774–1786. 2018.PubMed/NCBI
|
|
21
|
Zhu S, Yang N, Niu C, Wang W, Wang X, Bai
J, Qiao Y, Deng S, Guan Y and Chen J: The miR-145-MMP1 axis is a
critical regulator for imiquimod-induced cancer stemness and
chemoresistance. Pharmacol Res. 179:1061962022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sansing HA, Sarkeshik A, Yates JR, Patel
V, Gutkind JS, Yamada KM and Berrier AL: Integrin αβ1, αvβ, α6β
effectors p130Cas, Src and talin regulate carcinoma invasion and
chemoresistance. Biochem Biophys Res Commun. 406:171–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Von Der Heyde S, Wagner S, Czerny A,
Nietert M, Ludewig F, Salinas-Riester G, Arlt D and Beißbarth T:
mRNA profiling reveals determinants of trastuzumab efficiency in
HER2-positive breast cancer. PLoS One. 10:e01178182015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dong X, Liu W, Li X, Gan Y, Zhou L, Li W
and Xie L: Butein promotes ubiquitination-mediated survivin
degradation inhibits tumor growth and overcomes chemoresistance.
Sci Rep. 12:206442022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tribollet V, Cerutti C, Géloën A, Berger
E, De Mets R, Balland M, Courchet J, Vanacker JM and Forcet C: ERRα
coordinates actin and focal adhesion dynamics. Cancer Gene Ther.
29:1429–1438. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Janiszewska M, Primi MC and Izard T: Cell
adhesion in cancer: Beyond the migration of single cells. J Biol
Chem. 295:2495–2505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu QR, Du XH, Huang TT, Zheng YC, Li YL,
Huang DY, Dai HQ, Li EM and Fang WK: Role of cell-cell junctions in
oesophageal squamous cell carcinoma. Biomolecules. 12:13782022.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liao YH, Chiang KH, Shieh JM, Huang CR,
Shen CJ, Huang WC and Chen BK: Epidermal growth factor-induced
ANGPTL4 enhances anoikis resistance and tumour metastasis in head
and neck squamous cell carcinoma. Oncogene. 36:2228–2242. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Legerstee K, Geverts B, Slotman JA and
Houtsmuller AB: Dynamics and distribution of paxillin, vinculin,
zyxin and VASP depend on focal adhesion location and orientation.
Sci Rep. 9:104602019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yuan M, Xie F, Xia X, Zhong K, Lian L,
Zhang S, Yuan L and Ye J: UNC5C-knockdown enhances the growth and
metastasis of breast cancer cells by potentiating the integrin
α6/β4 signaling pathway. Int J Oncol. 56:139–150. 2020.PubMed/NCBI
|
|
31
|
Du G, Wang J, Zhang T, Ding Q, Jia X, Zhao
X, Dong J, Yang X, Lu S, Zhang C, et al: Targeting Src family
kinase member Fyn by Saracatinib attenuated liver fibrosis in vitro
and in vivo. Cell Death Dis. 11:1182020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ashrafizaveh S, Ashrafizadeh M, Zarrabi A,
Husmandi K, Zabolian A, Shahinozzaman M, Aref AR, Hamblin MR,
Nabavi N, Crea F, et al: Long non-coding RNAs in the doxorubicin
resistance of cancer cells. Cancer Lett. 508:104–114. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lohan-Codeço M, Barambo-Wagner ML,
Nasciutti LE, Ribeiro Pinto LF, Meireles Da Costa N and Palumbo A
Jr: Molecular mechanisms associated with chemoresistance in
esophageal cancer. Cell Mol Life Sci. 79:1162022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Rojas P, Mao J, Mustè Sadurnì M,
Garnier O, Xiao S, Higgs MR, Garcia P and Saponaro M: Persistence
of RNA transcription during DNA replication delays duplication of
transcription start sites until G2/M. Cell Rep. 34:1087592021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang YH, Mao JW and Tan XL: Research
progress on the source, production, and anti-cancer mechanisms of
paclitaxel. Chin J Nat Med. 18:890–897. 2020.PubMed/NCBI
|
|
36
|
Kitamura N, Sento S, Yoshizawa Y, Sasabe
E, Kudo Y and Yamamoto T: Current trends and future prospects of
molecular targeted therapy in head and neck squamous cell
carcinoma. Int J Mol Sci. 22:2402020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miller DS, Filiaci VL, Mannel RS, Cohn DE,
Matsumoto T, Tewari KS, DiSilvestro P, Pearl ML, Argenta PA, Powell
MA, et al: Carboplatin and paclitaxel for advanced endometrial
cancer: Final overall survival and adverse event analysis of a
phase III trial (NRG oncology/GOG0209). J Clin Oncol. 38:3841–3850.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Llueca A, Serra A, Climent MT, Segarra B,
Maazouzi Y, Soriano M and Escrig J; on behalf MUAPOS Working Group,
: Outcome quality standards in advanced ovarian cancer surgery.
World J Surg Oncol. 18:3092020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shen J, Cao B, Wang Y, Ma C, Zeng Z, Liu
L, Li X, Tao D, Gong J and Xie D: Hippo component YAP promotes
focal adhesion and tumour aggressiveness via transcriptionally
activating THBS1/FAK signalling in breast cancer. J Exp Clin Cancer
Res. 37:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhu Y, Tao Z, Chen Y, Lin S, Zhu M, Ji W,
Liu X, Li T and Hu X: Exosomal MMP-1 transfers metastasis potential
in triple-negative breast cancer through PAR1-mediated EMT. Breast
Cancer Res Treat. 193:65–81. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang G, Li T, Tan G, Song Y, Liu Q, Wang
K, Ai J, Zhou Z and Li W: Identity of MMP1 and its effects on tumor
progression in head and neck squamous cell carcinoma. Cancer Med.
11:2516–2530. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yan R, Ge X, Pang N, Ye H, Yuan L, Cheng
B, Zhou K, Yang M, Sun Y, Zhang S, et al: Essential role of zyxin
in platelet biogenesis and glycoprotein Ib-IX surface expression.
Cell Death Dis. 12:9552021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Partynska A, Gomulkiewicz A, Dziegiel P
and Podhorska-Okolow M: The role of zyxin in carcinogenesis.
Anticancer Res. 40:5981–5988. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cooper J and Giancotti FG: Integrin
signaling in cancer: Mechanotransduction, stemness, epithelial
plasticity, and therapeutic resistance. Cancer Cell. 35:347–367.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lu Q, Lai Y, Zhang H, Ren K, Liu W, An Y,
Yao J and Fan H: Hesperetin inhibits TGF-β1-induced migration and
invasion of triple negative breast cancer MDA-MB-231 cells via
suppressing Fyn/Paxillin/RhoA pathway. Integr Cancer Ther.
21:153473542210869002022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Katreddy RR, Bollu LR, Su F, Xian N,
Srivastava S, Thomas R, Dai Y, Wu B, Xu Y, Rea MA, et al: Targeted
reduction of the EGFR protein, but not inhibition of its kinase
activity, induces mitophagy and death of cancer cells through
activation of mTORC2 and Akt. Oncogenesis. 7:52018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song H, Liu D, Dong S, Zeng L, Wu Z, Zhao
P, Zhang L, Chen ZS and Zou C: Epitranscriptomics and epiproteomics
in cancer drug resistance: Therapeutic implications. Signal
Transduct Target Ther. 5:1932020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ashrafizadeh M, Zarrabi A, Hushmandi K,
Kalantari M, Mohammadinejad R, Javaheri T and Sethi G: Association
of the epithelial-mesenchymal transition (EMT) with cisplatin
resistance. Int J Mol Sci. 21:40022020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tulchinsky E, Demidov O, Kriajevska M,
Barlev NA and Imyanitov E: EMT: A mechanism for escape from
EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev
Cancer. 1871:29–39. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shen Q, Hill T, Cai X, Bui L, Barakat R,
Hills E, Almugaiteeb T, Babu A, Mckernan PH, Zalles M, et al:
Physical confinement during cancer cell migration triggers
therapeutic resistance and cancer stem cell-like behavior. Cancer
Lett. 506:142–151. 2021. View Article : Google Scholar : PubMed/NCBI
|