Introduction
Esophageal cancer is the sixth most common cancer
worldwide. The prognosis of patients with esophageal cancer remains
poor, prompting the search for new treatment strategies. The
overall 5-year survival rate is less than 50%, despite the use of
multi-modality therapy. Many patients in the early stage of the
disease develop local tumor recurrence or distant metastasis within
a short period after surgery. Therefore, more effective therapies
for esophageal cancer must be developed.
The control of cellular proliferation is extremely
important to an individual, therefore the cell cycle is strictly
controlled during individual development and cell differentiation.
Malignant tumors result from the loss of normal cell-cycle control,
and tumor cells multiply in a disordered manner. Several genes and
molecules are involved in the origin and/ or progression of
esophageal cancer, including TP53 (1,2),
deleted in esophageal cancer 1 (DEC1) (3–5),
deleted in colorectal cancer (DCC) (6–8),
deleted in lung cancer 1 (DLC1) (9), cyclin D1 (10–12),
transforming growth factor-β receptor type II (TGFBRII) (13,14),
adenomatous polyposis coli (APC) (15), survivin (16,17)
and murine double minute 2 (MDM2) (18). However, the precise mechanisms that
underlie the development and progression of esophageal squamous
cell carcinoma (ESCC) remain unclear.
The ubiquitin-proteasome system regulates important
cellular processes including development and differentiation,
apoptosis, protein transportation, immunologic and inflammatory
responses, cell-cycle progression and cellular division (19). The cell cycle and cellular division
are primarily coordinated by two ubiquitin ligases, SCF ubiquitin
ligase and anaphase-promoting complex (19,20).
The SCF ubiquitin ligases are comprised of the F-box protein
family, Cul1, Rbx1 and Skp1. Approximately 70 F-box proteins are
present in humans and provide substrate specificity (19,20).
FBXW7, one of the F-box proteins, induces the
degradation of positive cell-cycle regulators such as c-Myc, cyclin
E, c-Jun and Notch (19,20). These regulators are known as
oncoproteins, and the genes encoding these proteins are oncogenes
whose mutation and overexpression have been reported in humans.
FBXW7 is therefore focused on as a tumor suppressor gene
(21–23), and has been reported to be a
clinicopathologic factor in glioma (24), prostate cancer (25), colorectal cancer (26), gastric cancer (27) and T-cell acute lymphocytic leukemia
(28,29). However, the role of FBXW7 in
esophageal cancers is uncertain. In the present study, we examined
the relationship between the expression of FBXW7 and the
clinicopathological factors and prognosis of patients with
ESCC.
Materials and methods
Cell lines and cell culture
Esophageal cancer cell lines (TE1-15 and KYSE30-520)
were purchased from the Japanese Collection of Research
Bioresources (JCRB). The normal human esophageal mucosa Het-1A cell
line was purchased from the American Type Culture Collection
(ATCC). Esophageal cancer cell lines were grown in RPMI-1640 medium
(Sigma) supplemented with 10% fetal bovine serum (FBS) (Gibco) in
tissue culture dishes at 37°C in a humidified 5% CO2
incubator. Het-1A cells were grown in LHC-9 serum-free medium
(Biofluids, Rockville, MD, USA) at 37°C in a humidified 5%
CO2 incubator.
Tissue samples
Esophageal cancer (primary ESCC) samples and paired
non-cancerous samples were obtained from 43 patients who had
undergone a radical esophagectomy at Nagoya City University
Hospital between 1996 and 2005 without preoperative chemotherapy or
radiation. The study design was approved by the Institutional
Review Board of Nagoya City University Hospital, and written
consent was obtained from all patients. Samples were snap frozen in
liquid nitrogen and stored at −80°C until RNA and DNA extraction.
Patient characteristics are presented in Table I.
 | Table I.Correlation of FBXW7 mRNA
expression in esophageal cancer with clinicopathological
factors. |
Table I.
Correlation of FBXW7 mRNA
expression in esophageal cancer with clinicopathological
factors.
|
Characteristics | No. of patients
(n=43) | FBXW7
expression | P-value |
|---|
| Tissue
(samples) | | | |
| Normal | 43 |
16.293±64.896a | |
| Tumor | 43 | 2.793±2.648a | 0.9003 |
| Age at surgery | | | |
| ≤65 | 22 | 1.405±1.317b | |
| <65 | 21 | 1.442±1.303b | 0.8841 |
| Gender | | | |
| Male | 37 | 1.345±1.148b | |
| Female | 6 | 1.903±2.067b | 0.8334 |
| pStage | | | |
| 0-I | 8 | 2.154±1.204b | |
| II–IV | 35 | 1.256±1.272b | 0.0289 |
| Primary tumor | | | |
| T1 | 14 | 1.848±1.124b | |
| T2–T4 | 29 | 1.218±1.339b | 0.0315 |
| Lymph node
metastasis | | | |
| Negative | 13 | 1.421±0.977b | |
| Positive | 30 | 1.424±1.425b | 0.6154 |
| Lymphatic
invasion | | | |
| − | 10 | 1.957±1.163b | |
| + | 31 | 1.252±1.308b | 0.0336 |
| Unknown | 2 | | |
| Vessel
invasion | | | |
| − | 16 | 1.633±1.179b | |
| + | 25 | 1.276±1.379b | 0.1608 |
| Unknown | 2 | | |
| Ki-67 | | | |
| − | 16 | 1.659±1.124b | |
| + | 25 | 1.171±1.146b | 0.0733 |
| Unknown | 2 | | |
RNA extraction and real-time reverse
transcription polymerase chain reaction analysis
Total RNA was extracted from the esophageal cancer
tissue and the corresponding normal esophageal mucosa using the
real-time reverse transcription polymerase chain reaction (RT-PCR)
Absolutely RNA™ Miniprep kit (Stratagene, La Jolla, CA, USA)
according to the manufacturer’s instructions. The concentration of
total RNA was adjusted to 200 ng/ml using a spectrophotometer.
Reverse transcription reactions were performed at 42°C for 90 min
and at 95°C for 5 min followed by incubation at 72°C for 15 min
using 1 μg of total RNA, 0.5 μg oligo (dT) primer and Superscript
II enzyme (Gibco BRL, Gaithersburg, MD, USA).
Real-time quantitative reverse
transcription polymerase chain reaction with TaqMan probes
Real-time quantitative RT-PCR amplification of the
cDNA template corresponding to 20 ng of total RNA was performed
using TaqMan® Fast Universal PCR Master Mix (Applied
Biosystems, Foster City, CA, USA) in an ABI 7500 Fast Real-Time PCR
System (Applied Biosystems). PCR was conducted at 95°C for 10 min,
followed by 40 cycles at 95°C for 3 sec and 60°C for 30 sec.
FBXW7-specific TaqMan probes were designed to amplify a
70-bp PCR product encoding the common region among three
FBXW7 isoforms (FBXW7α, FBXW7β,
FBXW7γ) (TaqMan® Gene Expression Assays,
assay ID: Hs01015623_m1, Applied Biosystems). The relative
expression levels of FBXW7 mRNA were obtained by normalizing
the amount of FBXW7 mRNA to that of GAPDH mRNA (TaqMan® Gene
Expression Assays, assay ID: Hs99999905_m1, Applied Biosystems) as
an endogenous control in each sample.
Immunohistochemistry
Immunohistochemical staining was performed on
formalin-fixed paraffin-embedded primary human ESCC tissues using a
1:150 dilution of monoclonal anti-Ki-67 antibodies (Dako, Denmark
A/S). Paraffin-embedded sections of tumor were deparaffinized and
rehydrated. The sections were then heat-treated by microwaving in
10 mM citrate buffer for 10 min to facilitate antigen retrieval,
then cooled to room temperature. The sections were treated with
0.3% H2O2 in methanol for 30 min to
neutralize endogenous peroxidases, blocked with non-specific goat
serum for 10 min and incubated with anti-Ki-67 antibody overnight
at room temperature in a humidified chamber. Immunoreactive protein
was detected with a Dako Envision™+ System, HRP (DAB). The sections
were then counterstained with hematoxylin. The expression of Ki-67
was scored using light microscopy according to the proportion of
positive staining throughout the entire slide: (−), negative or
<5%; (+), <33%; and (++), >33%. Ki-67 immunohistochemical
staining was classified as negative for scores of (−) and positive
for scores of (+) or (++).
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD). Statistical analysis was performed using the Stat-View
software package (Abacus Concepts, Berkeley, CA, USA). The
Mann-Whitney U test was used to evaluate the significance of
differences in the expression levels of FBXW7/ GAPDH mRNA.
The survival of patients with ESCC was examined by the Kaplan-Meier
method, and the survival time was compared using the log-rank test.
Survival was measured from the day of surgery. Multivariate
analysis was performed using Cox’s regression model and the
logistic multivariate regression model. Differences were considered
statistically significant at P<0.05, and a tendency was
determined at P<0.1.
Results
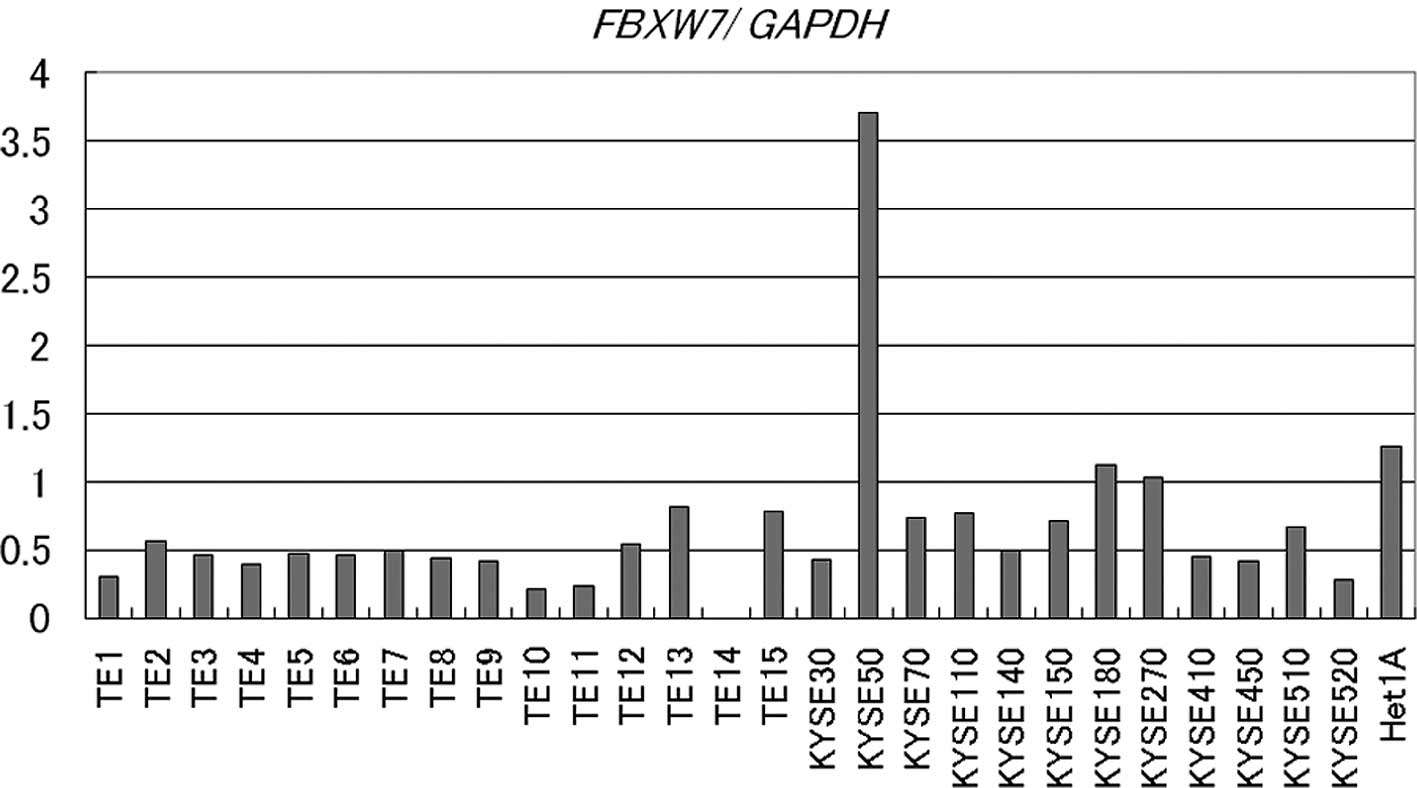
Quantitative RT-PCR was used to examine the relative
expression level of FBXW7 mRNA by normalization to
GAPDH in two esophageal cancer cell lines (TE1-15,
KYSE30-520) and one normal human esophageal mucosa cell line
(Het-1A). FBXW7 mRNA was detectable in all cell lines except
TE14. KYSE50 had the highest expression level of FBXW7 mRNA.
Only this cell line exhibited increased expression of FBXW7
mRNA compared to Het1A; the other cell lines had lower expression
(Fig. 1). The expression of
FBXW7 mRNA was examined in the 43 ESCC tissue specimens and
the paired normal esophageal mucosal tissue of patients who had not
received preoperative therapy. The mean expression level of
FBXW7 mRNA in the ESCC tissue was lower than that of the
corresponding normal tissue, although the difference was not
statistically significant.
Next, the relationship between the ratio of
FBXW7 mRNA expression in the tumor to that in the normal
esophageal mucosa (T/N ratio) and the clinicopathological factors
of the 43 patients were examined. There were no significant
differences in FBXW7 mRNA expression levels with respect to
age, gender, lymph node status or blood vessel invasion. However,
the expression levels were significantly correlated with the
progression of the cancer and its local invasiveness (stage,
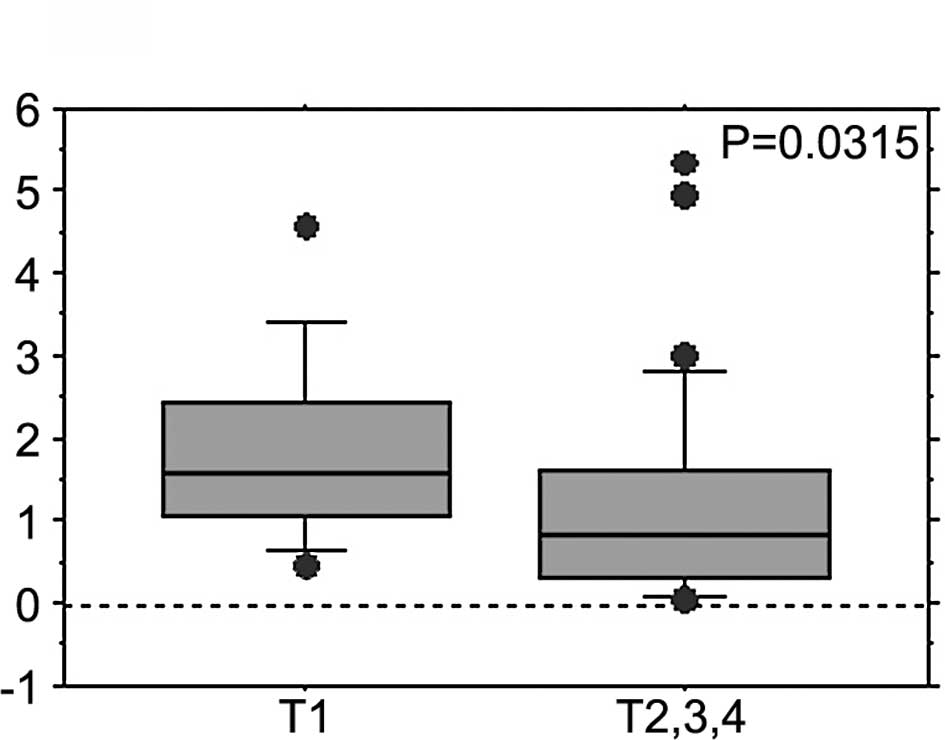
t-factor and lymphatic invasion) (Table 1, Fig.
2). The FBXW7 mRNA expression levels in patients with
muscle-invasive tumors (T2–4) were significantly lower than those
in patients with less invasive T1 tumors (1.218±1.339 vs.
1.848±1.124; P=0.0315, Mann-Whitney U test) (Table I, Fig.
2A). The FBXW7 mRNA expression levels were significantly
lower in stage 0-I ESCC than in stage II–IV ESCC (1.256±1.272 vs.
2.154±1.204; P=0.0289, Mann-Whitney U test) (Table I, Fig.
2B). Moreover, FBXW7 mRNA expression levels were
significantly lower in patients with lymphatic invasion than in
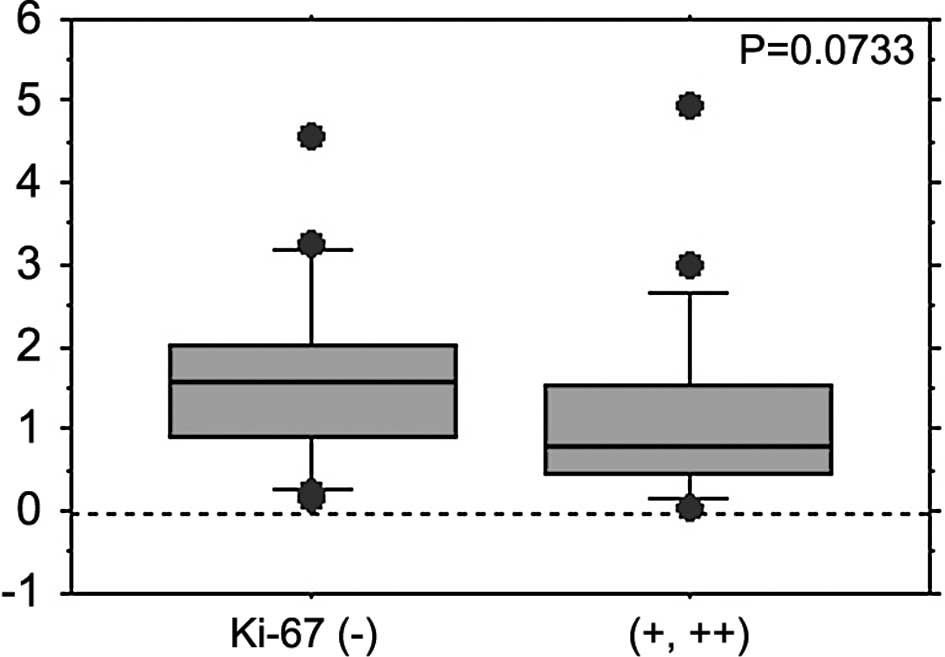
those without (1.252±1.308 vs. 1.957±1.163; P=0.0336) (Table I). Ki-67 as a proliferation marker
was also detected by immunostaining, and its correlation with the
expression levels of FBXW7 was evaluated in the 41 cases
investigated in this study. The Ki-67-positive cases [scored as (+)
or (++)] tended to express decreased levels of FBXW7 mRNA as
compared to the negative cases (1.171±1.146 vs. 1.659±1.124;
P=0.0733) (Table I, Fig. 3).
The correlation between the expression levels of
FBXW7 and the postoperative survival period of patients with
ESCC was investigated (median follow-up, 30.0 months). The 43 cases
were divided into the high FBXW7 mRNA expression group [the
ratio of FBXW7 mRNA expression in the tumor to that in
normal esophageal mucosa (T:N ratio) >1.0] and the low
FBXW7 mRNA expression group (T:N ratio <1.0). The
patients with low levels of FBXW7 mRNA expression had a
significantly shorter postoperative survival time than the patients
with high levels of FBXW7 mRNA expression [24.9±19.5 (n=21)
vs. 34.3±19.5 months (n=22); P=0.0255, log-rank test) (Fig. 4)].
Univariate analysis (Table II) revealed the following
prognostic factors to be statistically significant: the extent of
the primary tumor (risk ratio 9.524, P<0.0001), lymph node
metastasis (risk ratio 7.752, P<0.0001), lymphatic invasion
(risk ratio 6.061, P=0.0006), vessel invasion (risk ratio 3.175,
P=0.0006) and FBXW7 mRNA expression (risk ratio 2.688,
P=0.0332). However, accoding to the multivariate analysis,
FBXW7 mRNA expression was not an independent prognostic
factor (data not shown).
 | Table II.Univariate analysis. |
Table II.
Univariate analysis.
| Parameter | Risk ratio | 95% CI | P-value |
|---|
| Age at surgery | | | |
| ≤65 | 1 | | |
| <65 | 0.933 | 0.547–1.591 | 0.7992 |
| Gender | | | |
| Male | 1 | | |
| Female | 1.331 | 0.713–2.485 | 0.3688 |
| Primary tumor | | | |
| T1 | 1 | | |
| T2–T4 | 9.524 | 3.401–27.027 | <0.0001 |
| Lymph node
metastasis | | | |
| Negative | 1 | | |
| Positive | 7.752 | 3.067–19.608 | <0.0001 |
| Lymphatic
invasion | | | |
| − | 1 | | |
| + | 6.061 | 2.169–16.949 | 0.0006 |
| Vessel
invasion | | | |
| − | 1 | | |
| + | 3.175 | 1.645–6.135 | 0.0006 |
| FBXW7
expression (T/N) | | | |
| High | 1 | | |
| Low | 2.688 | 1.082–6.667 | 0.0332 |
| Ki-67 | | | |
| − | 1 | | |
| + | 1.447 | 0.762–2.747 | 0.2587 |
Discussion
FBXW7 is an F-box protein that contains
components of the SCF ubiquitin ligase complexes, which are
involved in the ubiquitin-proteasome pathway. FBXW7 is
localized to chromosome region 4q32 and has three isoforms
(FBXW7α, FBXW7β, FBXW7γ).
FBXW7 is important for the degradation of positive
cell-cycle regulators such as c-Myc, cyclin E, c-Jun and Notch
(20,21,30–32).
c-Myc is an oncoprotein that is an important substrate of
FBXW7. c-Myc helps to control the G1-to-S phase transition.
It promotes the entry of cells into the G1 phase, and its
expression is maintained throughout the cell cycle. Degradation of
c-Myc by FBXW7 leads to cell-cycle exit (G0 phase) and
maintains cell-cycle arrest. Conversely, the loss of FBXW7
promotes cell-cycle progression and cell proliferation, and is
therefore considered one of the major causes of carcinogenesis or
carcinoma development (33–35).
Onoyama et al reported that mice carrying an FBXW7
T-cell conditional knockout developed thymic hyperplasia and thymic
lymphomas (35). Furthermore,
decreased expression of FBXW7 is correlated with poor
prognosis in gastric cancer (27)
and colorectal cancer (26).
In the present study, we investigated the
relationship between the expression of FBXW7 and the
clinicopathological factors and prognosis of patients with ESCC who
did not receive preoperative therapy. FBXW7 expression
levels were significantly correlated with the progression of the
cancer and local invasiveness. In cases with muscle-invasive
(T2–4), lymphatic invasive and stage II–IV tumors, FBXW7
expression was significantly lower than in other cases (1.218±1.339
vs. 1.848±1.124, P=0.0315; 1.252±1.308 vs. 1.957±1.163, P=0.0336;
1.256±1.272 vs. 2.154±1.204, respectively; P=0.0289, Mann-Whitney U
test) (Table I).
These results indicate that decreased expression of
FBXW7 may contribute to tumor growth and invasion in ESCC.
Tumors with decreased FBXW7 expression may undergo active
cellular division due to the unregulated cell cycle and may become
more invasive. This concept is supported by our finding that
FBXW7 expression levels tended to be lower in Ki-67-positive
cases. We also found that the low FBXW7 expression group had
a significantly poorer prognosis than the high FBXW7
expression group [24.9±19.5 (n=21) vs. 34.3±19.5 (n=22) months,
P=0.0255, log-rank test; Fig. 4].
This was due to the increased proportion of advanced cases in the
low FBXW7 expression group as compared to the high
FBXW7 expression group.
Esophageal cancer has a very poor prognosis, and the
molecular mechanism of carcinogenesis in this cancer is unclear. In
the present study, we identified a relationship between
FBXW7 expression and tumor progression. It is therefore
possible that FBXW7 is a molecular prognostic marker that
can be used to elucidate the mechanism of carcinogenesis.
References
|
1.
|
Hu N, Huang J, Emmert-Buck MR, et al:
Frequent inactivation of the tp53 gene in esophageal squamous cell
carcinoma from a high-risk population in China. Clin Cancer Res.
7:883–891. 2001.PubMed/NCBI
|
|
2.
|
Audrezet MP, Robaszkiewicz M, Mercier B,
et al: Tp53 gene mutation profile in esophageal squamous cell
carcinomas. Cancer Res. 53:5745–5749. 1993.PubMed/NCBI
|
|
3.
|
Leung AC, Wong VC, Yang LC, et al:
Frequent decreased expression of candidate tumor suppressor gene,
DEC1, and its anchorage-independent growth properties and impact on
global gene expression in esophageal carcinoma. Int J Cancer.
122:587–594. 2008. View Article : Google Scholar
|
|
4.
|
Yang L, Leung AC, Ko JM, et al: Tumor
suppressive role of a 2.4 mb 9q33–q34 critical region and DEC1 in
esophageal squamous cell carcinoma. Oncogene. 24:697–705.
2005.PubMed/NCBI
|
|
5.
|
Nishiwaki T, Daigo Y, Kawasoe T and
Nakamura Y: Isolation and mutational analysis of a novel human
cDNA, DEC1 (deleted in esophageal cancer 1), derived from the tumor
suppressor locus in 9q32. Genes Chromosomes Cancer. 27:169–176.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Park HL, Kim MS, Yamashita K, et al: DCC
promoter hypermethylation in esophageal squamous cell carcinoma.
Int J Cancer. 122:2498–2502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Wang M, Lu R and Fang D: The possible role
of loss of heterozygosity at APC, MCC and DCC genetic loci in
esophageal carcinoma. Zhonghua Zhong Liu Za Zhi. 21:16–18.
1999.PubMed/NCBI
|
|
8.
|
Miyake S, Nagai K, Yoshino K, Oto M, Endo
M and Yuasa Y: Point mutations and allelic deletion of tumor
suppressor gene DCC in human esophageal squamous cell carcinomas
and their relation to metastasis. Cancer Res. 54:3007–3010.
1994.PubMed/NCBI
|
|
9.
|
Seng TJ, Low JS, Li H, et al: The major
8p22 tumor suppressor DLC1 is frequently silenced by methylation in
both endemic and sporadic nasopharyngeal, esophageal, and cervical
carcinomas, and inhibits tumor cell colony formation. Oncogene.
26:934–944. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Shiozaki H, Doki Y, Yamana H and Isono K:
A multi-institutional study of immunohistochemical investigation
for the roles of cyclin d1 and e-cadherin in superficial squamous
cell carcinoma of the esophagus. J Surg Oncol. 79:166–173. 2002.
View Article : Google Scholar
|
|
11.
|
Nagasawa S, Onda M, Sasajima K, et al:
Cyclin d1 overexpression as a prognostic factor in patients with
esophageal carcinoma. J Surg Oncol. 78:208–214. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Sarbia M, Stahl M, Fink U, et al:
Prognostic significance of cyclin d1 in esophageal squamous cell
carcinoma patients treated with surgery alone or combined therapy
modalities. Int J Cancer. 84:86–91. 1999. View Article : Google Scholar
|
|
13.
|
Souza RF, Garrigue-Antar L, Lei J, et al:
Alterations of transforming growth factor-beta 1 receptor type II
occur in ulcerative colitis-associated carcinomas, sporadic
colorectal neoplasms, and esophageal carcinomas, but not in gastric
neoplasms. Hum Cell. 9:229–236. 1996.
|
|
14.
|
Garrigue-Antar L, Souza RF, Vellucci VF,
Meltzer SJ and Reiss M: Loss of transforming growth factor-beta
type II receptor gene expression in primary human esophageal
cancer. Lab Invest. 75:263–272. 1996.PubMed/NCBI
|
|
15.
|
Kawakami K, Brabender J, Lord RV, et al:
Hypermethylated APC DNA in plasma and prognosis of patients with
esophageal adenocarcinoma. J Natl Cancer Inst. 92:1805–1811. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Rosato A, Pivetta M, Parenti A, et al:
Survivin in esophageal cancer: an accurate prognostic marker for
squamous cell carcinoma but not adenocarcinoma. Int J Cancer.
119:1717–1722. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kato J, Kuwabara Y, Mitani M, et al:
Expression of survivin in esophageal cancer: correlation with the
prognosis and response to chemotherapy. Int J Cancer. 95:92–95.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Shimada Y, Imamura M, Shibagaki I, et al:
Genetic alterations in patients with esophageal cancer with short-
and long-term survival rates after curative esophagectomy. Ann
Surg. 226:162–168. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Reed SI: The ubiquitin-proteasome pathway
in cell cycle control. Results Probl Cell Differ. 42:147–181. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Nakayama KI and Nakayama K: Ubiquitin
ligases: cell-cycle control and cancer. Nat Rev Cancer. 6:369–381.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Akhoondi S, Sun D, von der Lehr N, et al:
FBXW7/hCDC4 is a general tumor suppressor in human cancer.
Cancer Res. 67:9006–9012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Fujii Y, Yada M, Nishiyama M, et al:
FBXW7 contributes to tumor suppression by targeting multiple
proteins for ubiquitin-dependent degradation. Cancer Sci.
97:729–736. 2006. View Article : Google Scholar
|
|
23.
|
Welcker M, Orian A, Jin J, et al: The FBW7
tumor suppressor regulates glycogen synthase kinase 3
phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad
Sci USA. 101:9085–9090. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Hagedorn M, Delugin M, Abraldes I, et al:
FBXW7/hCDC4 controls glioma cell proliferation in vitro and
is a prognostic marker for survival in glioblastoma patients. Cell
Div. 2:92007. View Article : Google Scholar
|
|
25.
|
Koh MS, Ittmann M, Kadmon D, Thompson TC
and Leach FS: CDC4 gene expression as potential biomarker for
targeted therapy in prostate cancer. Cancer Biol Ther. 5:78–83.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Iwatsuki M, Mimori K, Ishii H, et al: Loss
of FBXW7, a cell cycle regulating gene, in colorectal
cancer: clinical significance. Int J Cancer. 126:1828–1837.
2009.
|
|
27.
|
Yokobori T, Mimori K, Iwatsuki M, et al:
P53-altered FBXW7 expression determines poor prognosis in
gastric cancer cases. Cancer Res. 69:3788–3794. 2009.
|
|
28.
|
Mansour MR, Sulis ML, Duke V, et al:
Prognostic implications of NOTCH1 and FBXW7 mutations in
adults with t-cell acute lymphoblastic leukemia treated on the MRC
UKALLXII/ECOG E2993 protocol. J Clin Oncol. 27:4352–4356.
2009.PubMed/NCBI
|
|
29.
|
Baldus CD, Thibaut J, Goekbuget N, et al:
Prognostic implications of NOTCH1 and FBXW7 mutations in
adult acute t-lymphoblastic leukemia. Haematologica. 94:1383–1390.
2009.PubMed/NCBI
|
|
30.
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: a tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Welcker M, Orian A, Grim JE, Eisenman RN
and Clurman BE: A nucleolar isoform of the FBW7 ubiquitin ligase
regulates c-Myc and cell size. Curr Biol. 14:1852–1857. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Mao JH, Perez-Losada J, Wu D, et al:
FBXW7/CDC4 is a p53-dependent, haploinsufficient tumour
suppressor gene. Nature. 432:775–779. 2004. View Article : Google Scholar
|
|
33.
|
Tsunematsu R: Cell cycle regulation by
F-BOX protein FBXW7. Fukuoka Igaku Zasshi. 100:87–94.
2009.PubMed/NCBI
|
|
34.
|
Onoyama I and Nakayama K: Regulation of
cell-cycle exit and g0-phase maintenance during differentiation by
FBXW7. Tanpakushitsu Kakusan Koso. 53:1217–1224.
2008.PubMed/NCBI
|
|
35.
|
Onoyama I, Tsunematsu R, Matsumoto A, et
al: Conditional inactivation of FBXW7 impairs cell-cycle
exit during t cell differentiation and results in
lymphomatogenesis. J Exp Med. 204:2875–2888. 2007.
|