Introduction
Cells must ensure that their genetic information is
accurately transmitted to each daughter cell during the cell
division process. To ensure that the DNA has been fully replicated
in an undamaged state, cells progress through a series of cell
cycle checkpoints prior to mitosis. Incomplete DNA replication due
to stalled replication forks, DNA damage caused by ultraviolet
light, chemicals in the environment, gamma radiation or reactive
oxygen species, or treatment with chemotherapeutic agents used in
cancer therapy activate checkpoint signaling pathways that arrest
the cell cycle, providing damaged cells time to complete DNA
synthesis or to repair DNA damage. If the DNA is too severely
damaged, alternate signaling pathways are activated, leading to
permanent growth arrest or programmed cell death (1).
Carcinogenesis is often caused by a functional
mutation or the loss of proteins involved in cell cycle
checkpoints, which provides tumor cells with the selective
advantage of proliferation. Defects in cell cycle checkpoints are
the hallmarks of tumor cells, and also determine the efficacy of
cancer treatment with anti-neoplastic agents. Chemotherapeutic
agents and IR usually damage the DNA of both normal and tumor
cells. Normal cells activate cell cycle checkpoints, leading to
growth arrest and subsequently the DNA repair process. In tumor
cells, however, the loss of appropriate checkpoints due to
mutations in proteins such as p53, Chk1 or Chk2 allows malignant
cells to progress to mitosis with numerous mutations of single- and
double-strand breaks (DSBs) still present in their genomic DNA. As
a result, mutations in the genome accumulate progressively, leading
to apoptosis (2).
Mechanisms of sensing and repairing damaged DNA are
conserved among the species. Ataxia telangiectasia mutated (ATM)
and the catalytic subunit of DNA-dependent protein kinase
(DNA-PKcs) are essential to the DNA damage response when cells are
exposed to DNA insult as described above. Activation of these
kinases has been implicated in the initiation of the DNA repair
process and cell cycle checkpoints. By phosphorylation of their
substrates, these kinases transmit signals to downstream proteins,
including the Mre11/Rad50/Nbs1 (MRN) complex, Fanconi anemia
proteins and the BRCA1 breast cancer tumor-susceptibility protein
(3–6).
We recently identified a novel protein, named BAAT1
(BRCA1-associated protein required for ATM activator-1) (7), which was isolated by yeast two-hybrid
screening using aa1314–1863 of BRCA1 as bait. We found that BAAT1
also binds to ATM, localizes to DSBs and is required for Ser1981
phosphorylation of ATM under conditions of IR treatment.
siRNA-mediated stable or transient depletion of BAAT1 was found to
result in decreased phosphorylation of ATM at Ser1981, resulting in
impaired phosphorylation of NBS1, Chk2 and γH2AX after IR.
Treatment of BAAT1-depleted cells with okadaic acid greatly
restored the phosphorylation of ATM Ser1981, suggesting that BAAT1
is involved in the regulation of ATM phosphatase. PP2A-mediated
dephosphorylation of ATM was partially blocked by purified BAAT1
in vitro. These results suggest that DNA damage-induced ATM
activation requires a coordinated assembly of BRCA1, BAAT1 and
ATM.
In the present study, we detail the functional
interaction of BAAT1 with the proteins involved in the DNA damage
response. The results demonstrate that the DNA-PKcs also requires
BAAT1 for its activation after DNA stress. These results suggest
that BAAT1 is commonly required for the initial stage of the DNA
damage pathway.
Materials and methods
Cell culture
The human embryonic kidney 293T cell line was grown
in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad,
CA) containing 10% fetal bovine serum (FBS; Invitrogen),
penicillin/streptomycin and 10 mM sodium pyruvate (Sigma, St.
Louis, MO). MCF10A cells were cultured on tissue culture flask
(Corning) in a 1:1 mixture of DMEM and F12 medium (DMEM-F12)
supplemented with 5% horse serum, hydrocortisone (0.5 μg/ml),
insulin (10 μg/ml), epidermal growth factor (20 ng/ml) and
penicillinstreptomycin (100 μg/ml each).
Immunoblot analysis and
immunoprecipitation
Cells were treated with the IR mimetic
neocarzinostatin (NCS) (0.5 mg/ml) for the indicated times and then
lysed in ice-cold lysis buffer [(50 mM Tris-HCl (pH 7.6), 150 mM
NaCl, 1 mM EDTA (pH 8.0), 20 mM NaF, 1 mM
Na3VO4, 1% NP-40, 0.5 mM dithiothreitol] in
the presence of protease-inhibitor mix [leupeptin, aprotinin and
phenylmethylsulfonyl fluoride (PMSF) (10 μg/ml), respectively].
After centrifugation (12,000 × g, 10 min), soluble supernatants
were prepared, and the protein concentrations were calculated using
the Bio-Rad protein assay kit. The total cell lysate (20 μg) was
loaded and separated on 6.0% SDS polyacrylamide gels. GSH-beads (AB
Bioscience) were used for GST-pulldown assay using 700 μg of total
cell lysates. Transfer to PVDF membranes (Immobilon-P; Millipore)
was carried out using a semidry transfer method (Trans-Blot;
Bio-Rad) in 25 mM Tris, 192 mM glycine and 10% methanol for 1 h at
20 V. Membranes were blocked in 5% nonfat dried milk in
Tris-buffered saline (TBS)/0.1% Tween-20 and incubated with primary
antibodies and horseradish peroxidase-conjugated secondary
antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) followed by
enhanced chemiluminescence detection. Primary antibodies used in
this study were anti-BAAT1 (Abcam, Cambridge, MA), anti-ATM
(GeneScript, Piscataway, NJ), anti-DNA_PKcs (Calbiochem) anti-SMC1,
anti-NBS1 and anti-GST (Santa Cruz Biotechnology). Additionally
specific anti-phosphorylation antibodies were used against
phospho-ATM (Ser1981; Cell Signaling, Danvers, MA), phospho-SMC1
(Ser966; Bethyl Laboratories, Montgomery, TX), phospho-NBS1
(Ser345) and phospho-DNA-PKcs (Ser2056) (Rockland). The anti-actin
antibody (Santa Cruz Biotechnology) was used to validate the amount
of protein. For immunoprecipitation, FLAG-tagged BAAT1 was
expressed by transient expression. After the cells were lysed,
anti-FLAG M2 beads (Sigma) were added to the lysates. The beads
were extensively washed with lysis buffer and boiled at 95˚C.
Following SDS-PAGE, the samples were transferred to membranes for
immunoblot analysis as described above.
Mouse tissues were obtained from wild-type C57BL/6J
mice (6–7 weeks old; Jackson Laboratory). Tissues were lysed using
a homogenizer (200 series; Pro Scientific Inc., Oxford, CT) in
ice-cold lysis buffer. Denatured proteins from the tissue lysates
were loaded at 35 μg (total protein, Bradford assay) per lane on 8%
SDS-PAGE gel and electro-blotted for 1 h at 25 V onto a PVDF
membrane. Membranes were incubated for 1 h with 5% nonfat dry milk
in TBST contaning 0.1% Tween-20 to block non-specific antibody
binding, and subsequently incubated with the primary antibodies for
BRCA1, SMC1 and BAAT1. The antibody for actin was used to validate
the amount of protein.
Construction of the GST-fusion
protein
GST fragments were generated by PCR from human BAAT1
full-length plasmid containing amino acids 1–123 (#1), 124–301
(#2), 302–400 (#3), 401–540 (#4), 541–700 (#5) and 701–821 (#6),
respectively. Segments of human BAAT1 were PCR amplified using the
following primers: aa1-123, 5′-AA AGG ATC CAA ATG GAC CCAGAA TGC
GCC CAG CTG-3′ and 5′-AA AGC GGC CGC CTA CCA GCC GCT GCG CAC GGT
GGG GAC-3′; aa124-301, 5′-CGC AGC GGC TGG ATC CAG GGC CTG CGC-3′
and 5′-AA AGC GGC CGC CTA CCC CAA AGC CAG GGG TCC CAT GTG-3′;
aa302–400, 5′-CTG GCT TTG GGG ATC CTG AAG CTC GAG-3′ and 5′-AA AGC
GGC CGC CTA GAG CCG CAG GAC AGT CAC TGT AGC-3′; aa401–540, 5′-AA
AGG ATC CAA TGT GAC GGC TCG GCT GCC CCT GCC-3′ and 5′-AA AGC GGC
CGC CTA TGC GCA TCT GAA GTC AGC CTG TCC-3′; aa541–700, 5′-AA AGG
ATC CAA CTC TTG GCT TCA GAG GTG CCT CAG-3′ and 5′-AA AGC GGC CGC
CTA GAG CCC CAC GTG GCA GAG AGC CCT-3′; aa701–821, 5′-AA AGG ATC
CAA TTT GAC TTC GCC TTT TGT GCC TTG-3′ and 5′-AA AGC GGC CGC TCA
GTA GCA GTC GGC CTC GTC CCC-3′. The sequences of the amplified
fragments were confirmed and subcloned into the pEBG vector
(8).
Transfection
Transient transfection with a plasmid expressing
GST-fusion proteins of BAAT1 was performed for 48 h with
Lipofectamine 2000 (Invitrogen) according to the manufacturer's
instructions, followed by NCS treatment. Transfection of BAAT1
siRNA was as described previously (7).
Results and Discussion
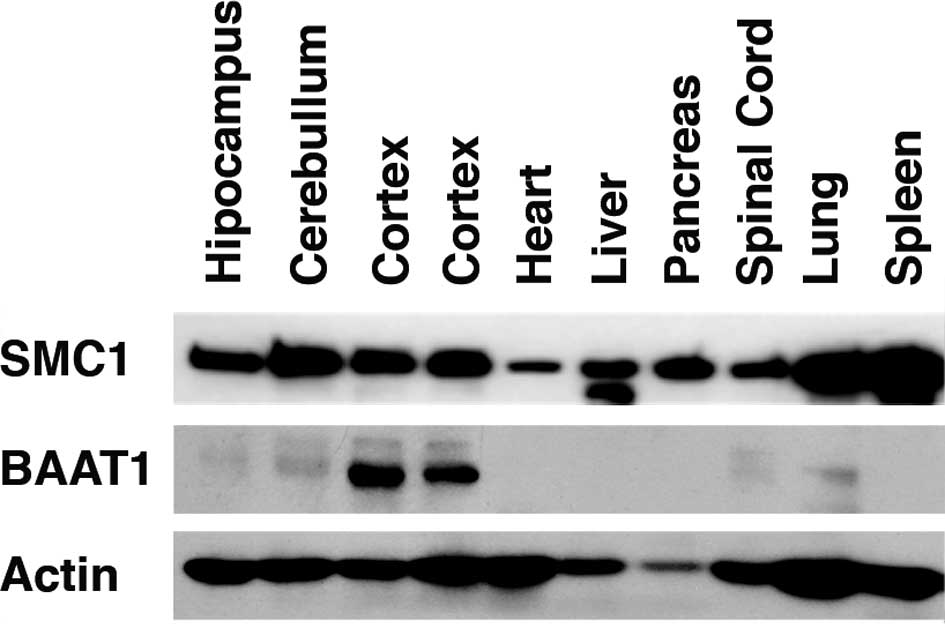
BAAT1 is highly expressed in the
cortex
Recently, using the BRCT domain of BRCA1 as bait,
our group isolated a cDNA encoding a novel protein that we named
BAAT1 (7). Northern blot analysis
indicated that BAAT1 mRNA is ubiquitously expressed, and a much
higher mRNA level is detected in testis (7). In the present study, levels of BAAT1
protein were assessed using the samples obtained from mouse
tissues. As shown in Fig. 1, high
levels of BAAT1 protein were detected in the cortex. Much lower
levels of this protein were detected in the cerebellum, spinal cord
and lung. SMC1 was detected in all of the tissues studied, although
the protein was differentially expressed in these tissues. These
results suggest that BAAT1 is involved in the neuronal system.
BAAT1 binds to ATM, DNA-PKcs and
SMC1
ATM is phosphorylated at Ser1981 when cells are
exposed to cell stress (9). In
BAAT1-depleted cells, this phosphorylation of ATM at Ser1981 was
greatly reduced, indicating that BAAT1 is required for ATM
activation. We further explored whether BAAT1 is involved in other
pathways.
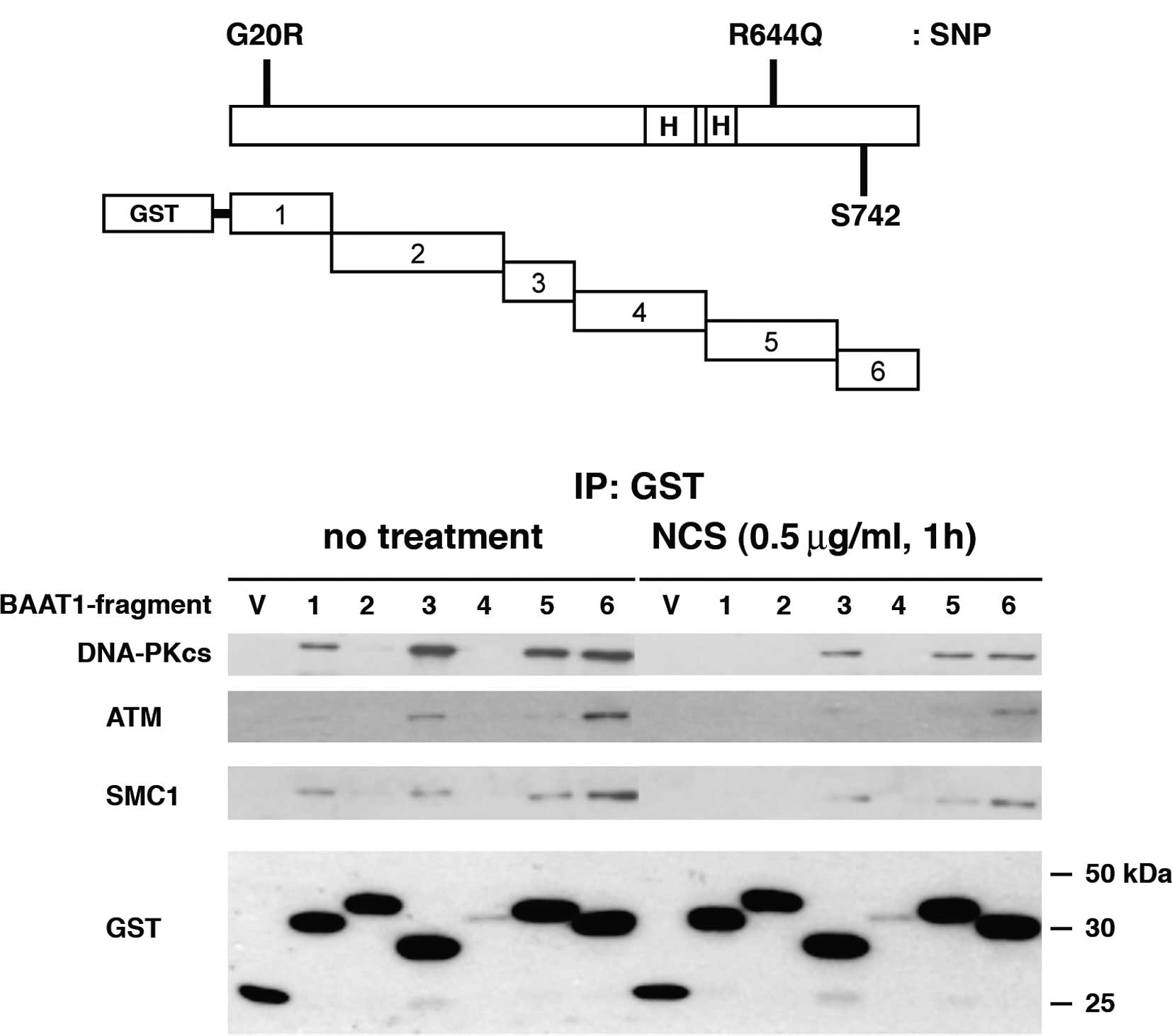
The full-length BAAT1 protein consists of 821 amino
acids of ∼88 kDa, and the human amino acid sequence shares 73.5 and
72.9% identity with mouse and rat sequences, respectively. As shown
in Fig. 2A, the C-terminal half of
the protein contains two HEAT (Huntingtin, elongation factor 3, A
subunit of protein phosphatase 2A and TOR1) repeat domains
(aa495–531, aa544–576) and a putative phosphorylation residue at
amino acid Ser742. From the database, human BAAT1 has two SNP sites
(G20R and R644Q), although it is not known whether these SNPs are
pathogenic. Six fragments of BAAT1 protein were expressed with an
N-terminal GST tag in the 293T cells (Fig. 2A). After 48 h, the cells were
treated with NCS for 1 h to induce DNA DSBs. Each of the GST
fragments were pulled down, and the interaction with cellular
proteins was assessed. Immunoblotting with an anti-GST antibody
revealed the protein levels of the GST-#4 segment of BAAT1 were
lower than the others that were found to be similarly expressed,
suggesting that GST-#4 is unstable.
Immunoblot analysis demonstrated that both the
DNA-PKcs and SMC1 bound to BAAT1 fragments #1, #3, #5 and #6 before
NCS treatment, while interaction with #1 was decreased after NCS
treatment. ATM bound to the #1, #3 and #6 fragments of BAAT1,
although its interaction with BAAT1 #1 was much weaker than with
the others. After NCS treatment, ATM bound primarily to the BAAT1
#6 fragment, although BAAT1 #3 also showed weak interaction.
Interaction between ATM and BAAT1 #1 was not detected after NCS
treatment. These results suggest that protein modification and/or
allosteric change in the conformation that occurs in BAAT1,
DNA-PKcs, ATM and/or SMC1 after NCS treatment determines the
intensity of the interaction among these proteins. We also examined
physical interaction between BAAT1 and Ku70/Ku80, which are
components of the DNA-dependent protein kinase, but
co-immunoprecipitation was not detected (data not shown).
Time-dependent interaction between BAAT1
and ATM/DNA-PKcs
As shown in Fig. 2,
the intensity of the interaction between BAAT1 and other cellular
proteins may be regulated by DNA damage stimuli. We assessed the
interaction of BAAT1 and ATM or the DNA-PKcs with NCS treatment in
a time-dependent manner. Cells (293T) were transfected with
FLA-tagged BAAT1 for 48 h and treated with NCS for 0.5, 1, 1.5, 2
and 3 h. FLAG-BAAT1 was pulled down by anti-FLAG beads, and samples
were immunoblotted with anti-DNA-PKcs or -ATM antibodies (Fig. 3).
Levels of endogenous DNA-PKcs, ATM and exogenous
FLAG-BAAT1 were not altered throughout this time course. After
FLAG-BAAT1 was pulled down, interaction with both the DNA-PKcs and
ATM was detected, but was decreased after 1 h. Interaction was then
recovered 1.5 h after NCS treatment, and decreased significantly
after 3 h. The mechanism involved in this biphasic alteration of
interaction after NCS treatment remains to be elucidated.
BAAT1 knockdown reduces phosphorylation
of the DNA-PKcs at Ser2056 and SMC1 at Ser966 by NCS
Since siRNA-mediated depletion of BAAT1 was
previously found to impair the phosphorylation of ATM at Ser1981
after IR treatment (7), we
investigated whether it also regulates the phosphorylation of the
DNA-PKcs and SMC1. The normal mammary epithelial cell line MCF10A
was transfected with control or BAAT1 siRNA for 48 h, followed by
NCS treatment.
Chen et al found that IR induces the
autophosphorylation of the DNA-PKcs at Ser2056, which is required
for the repair of DSBs by non-homologous end joining (10). As shown in Fig. 4, NCS treatment induced the
autophosphorylation of the DNA-PKcs at Ser2056, however, this
phosphorylation was significantly reduced when BAAT1 was knocked
down by siRNA.
SMC1 proteins plays a pivotal role in sister
chromatid cohesion, chromosome condensation, gender-chromosome
dosage compensation, and DNA recombination and repair (11–14).
Heterodimers of the SMC1 and SMC3 proteins have been implicated
specifically in both sister chromatid cohesion and DNA
recombination. ATM phosphorylates SMC1 protein at Ser957 and Ser966
after IR, and expression of a phospho-deficient form of SMC1
protein mutated at these phosphorylation sites abrogates IR-induced
S phase cell cycle checkpoints (15,16).
In the present study, when cells were treated with NCS, the
phosphorylation of SMC1 at Ser966 was significantly increased,
while this phosphorylation was reduced in BAAT1-knockdown cells. As
shown previously (7),
phosphorylation of NBS1 at Ser343 was also decreased in
BAAT1-knockdown cells.
There are several possible mechanisms involved in
the decreased phosphorylation of DNA-PKcs, SMC1 and NBS1 in
BAAT1-knockdown cells: i) kinase activity responsible for
phosphorylation is inhibited, ii) phosphatase activity is
increased, or iii) localization of DNA-PKcs to the sites of DSBs is
inhibited. Although our previous studies have suggested that BAAT1
may regulate ATM phosphatase (7),
we are also currently investigating other potential mechanisms
using human cell lines in which BAAT1 is stably knocked down or
knocked out (unpublished data).
As described above, BAAT1 has two tandem copies of
the HEAT repeat, which may be important for protein-protein
interaction. Two SNP sites in the coding sequence have been
reported, although it is not clear whether these mutants are
pathogenic. To elucidate the exact function of BAAT1 in the DNA
damage response and cell cycle checkpoints, structural integrity
should also be investigated.
To date, approaches to determining the role of BAAT1
in the DNA damage response have involved the evaluation of the
binding affinity and the modulation of phosphorylation of the
proteins involved in DNA damage signaling using cells in which
BAAT1 is suppressed by knockdown. To elucidate more of the
physiological roles of these proteins, the generation of knockout
mice is essential.
Acknowledgements
The authors would like to acknowledge
Dr Ruth Seal (HUGO Gene Nomenclature Committee) for specific
suggestions and registration of BAAT1/BRCA1-associated
ATM-activator 1 in the Human Genome Organization (HUGO) database.
The authors also thank the members of the Ouchi laboratory for the
helpful discussion. This study was supported by R01CA79892 and
R01CA90631 (both from NIH/NCI to T.O.), the AVON pilot project
grant (to T.O.) and Ms. Susan G. Komen, Investigator Initiated
Research Grant (to T.O.).
References
|
1.
|
Mahaney BL, Meek K and Lees-Miller SP:
Repair of ionizing radiation-induced DNA double-strand breaks by
non-homologous end-joining. Biochem J. 417:639–650. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Pandita TK and Richardson C: Chromatin
remodeling finds its place in the DNA double-strand break response.
Nucleic Acids Res. 37:1363–1377. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lee SH and Kim CH: DNA-dependent protein
kinase complex: a multifunctional protein in DNA repair and damage
checkpoint. Mol Cell. 13:159–166. 2002.PubMed/NCBI
|
|
4.
|
Shiloh Y: ATM and related protein kinases:
safeguarding genome integrity. Nat Rev Cancer. 3:155–168. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Uziel T, Lerenthal Y, Moyal L, Andegeko Y,
Mittelman L and Shiloh Y: Requirement of the MRN complex for ATM
activation by DNA damage. EMBO J. 22:5612–5621. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Tischkowitz MD and Hodgson SV: Fanconi
anaemia. J Med Genet. 40:1–10. 2003. View Article : Google Scholar
|
|
7.
|
Aglipay JA, Martin SA, Tawara H, Lee SW
and Ouchi T: ATM activation by ionizing radiation requires
BRCA1-associated BAAT1. J Biol Chem. 281:9710–9718. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Ouchi T, Lee SW, Ouchi M, Aaronson SA and
Horvath CM: Collaboration of STAT1 and BRCA1 in differential
regulation of IFNg target genes. Proc Natl Acad Sci USA.
97:5208–5213. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Bakkenist CJ and Kastan MB: DNA damage
activates ATM through intermolecular autophosphorylation and dimer
dissociation. Nature. 421:499–506. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Chen BP, Chan DW, Kobayashi J, Burma S,
Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J and Chen DJ: Cell
cycle dependence of DNA-dependent protein kinase phosphorylation in
response to DNA double strand breaks. J Biol Chem. 280:656–662.
2005.PubMed/NCBI
|
|
11.
|
Strunnikov AV and Jessberger R: Structural
maintenance of chromosome (SMC) proteins: conserved molecular
properties for multiple biological functions. Eur J Biochem.
263:6–13. 1999. View Article : Google Scholar
|
|
12.
|
Losada A and Hirano T: Dynamic molecular
linkers of the genome: the first decade of SMC proteins. Genes Dev.
19:1269–1287. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Watrin E and Peters JM: Cohesin and DNA
damage repair. Exp Cell Res. 312:2687–2693. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Wong RW: An update on cohesin function as
a ‘molecular glue’ on chromosomes and spindles. Cell Cycle.
9:1754–1758. 2010.
|
|
15.
|
Kitagawa R, Bakkenist CJ, McKinnon PJ and
Kastan MB: Phosphorylation of SMC1 is a critical downstream event
in the ATM-NBS1-BRCA1 pathway. Genes Dev. 18:1423–1438. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY
and Qin J: SMC1 is a downstream effector in the ATM/NBS1 branch of
the human S-phase checkpoint. Genes Dev. 16:571–582. 2002.
View Article : Google Scholar : PubMed/NCBI
|