Introduction
Breast cancer is a complex, multifactorial disease
with a strong interplay between genetic and environmental factors
(1). Worldwide, it is the most
common malignancy in women accounting for 23% of all cancers and
the primary cause of death. It is the second most common cancer
when both genders are considered together (2). At present, in Sri Lanka, breast
cancer in women diagnosed at a median age of 50 years contributes
to approximately 25% of all cancers (3). The age-standardized incidence rate
(ASR) of breast cancer per 100,000 women significantly differs
between developed and developing countries, and ASR for female
breast cancer is 18.4 in Sri Lanka (3–5).
Although several risk factors including family
history predispose to breast cancer, the most potent appear to be
mutations in the breast cancer susceptibility genes BRCA1
(MIM 113705) and BRCA2 (MIM 600185) (6). Even though familial breast cancer
accounts for only 5–10% of all breast cancers, individuals carrying
mutations in one of these genes have a 40–80% chance of developing
breast cancer (7). The lifetime
risk of developing breast cancer by the age of 70 years is 46–87%
for BRCA1 and 26–84% for BRCA2 mutation carriers
(8–10). The BRCA1 gene located on the
long arm of chromosome 17 (17q21) consists of 24 exons coding for
1863 amino acids (11). The
BRCA2 gene located on chromosome 13 (13q12.3) consists of 27
exons coding for 3418 amino acids (12). Both genes apparently function as
tumour-suppressor genes and play a pivotal role in the control of
homologous recombination and double-strand break repair in response
to DNA damage (13–15).
We recently reported BRCA1 mutations in a
cohort of Sri Lankan breast cancer patients (16) but BRCA2 germline mutations
have not been previously characterized. Mutations in both genes
together account for the majority of families with hereditary
susceptibility to breast and ovarian cancer (17). Furthermore, mutations in the
central portion of the BRCA2 gene [ovarian cancer cluster
region (OCCR)] are reported to be associated with a significantly
higher ratio of cases of ovarian:breast cancer in female carriers
than the mutations in the 5′ or 3′ region (18). Mutations in the BRCA2 gene
in men lead to an increased risk of cancers in the breast, prostate
and pancreas (19).
BRCA1- and BRCA2-associated risks of
developing cancer differ in geographical and historically defined
groups. Ethnic group-specific mutations showing a high frequency of
occurrence are referred to as founder mutations. Identification of
a founder mutation is a vital step towards the improvement of
genetic screening and counselling. It also facilitates more
specific approaches to molecular testing (7,20).
In the present study, 109 breast cancer patients, 20 at-risk
individuals and 20 healthy controls were screened for BRCA2
mutations. Here, we report novel and previously reported pathogenic
and possibly pathogenic mutations, intronic variants and common
polymorphisms observed.
Materials and methods
Study participants
A total of 109 patients (n=55 with a family history
of breast cancer and n=54 sporadic breast cancer), 20 at-risk
individuals and 20 healthy controls without a personal or family
history of any cancer were studied. They were study participants in
whom we previously investigated BRCA1 mutations, and their
characteristics have been previously described (16). The number of patients studied in
the present study was slightly lower than that in the BRCA1
study. At-risk individuals were limited to members of a family with
a history of breast and other cancers where one breast cancer
patient and two unaffected family members carry a deleterious
BRCA1 mutation but included an additional study participant
than in the BRCA1 study. Characteristics of the study
participants are summarised in Table
I. Mean age at diagnosis was 47.76±9.55 and 47.60±10.49 years
for familial and sporadic breast cancer patients, respectively.
Fourteen familial and 10 sporadic breast cancer patients were
diagnosed below 40 years of age. Among the familial cases 34, 17
and 3 patients had one, two and three affected family members,
respectively. One patient had 4 affected family members. The
majority of the patients and controls and all at-risk individuals
were ethnically Sinhalese. There were no descendents of Europeans.
Ethical approval from the Institution Review Board and written
informed consent from the study participants were obtained prior to
the study. Socio-demographic and clinical data were obtained from
the study participants, and cancer diagnoses were confirmed by
reviewing medical reports and pathology reports.
 | Table ICharacteristics of the patients
selected for the study. |
Table I
Characteristics of the patients
selected for the study.
| Familial breast
cancer (n=55) | Sporadic breast
cancer (n=54) |
|---|
| Age (years) at
diagnosis | | |
| Mean ± SD | 47.76±9.55 | 47.60±10.49 |
| <40 | 15 | 11 |
| ≥40 | 39 | 41 |
| N/A | 1 | 2 |
| No. of family
members with breast cancer | | |
| 0 | 0 | 54 |
| 1 | 34 (1st, 13; 2nd,
21) | 0 |
| 2 | 17 (1st, 4; 2nd, 9;
1st and 2nd both, 4) | 0 |
| 3 | 3 (1st, 1; 2nd, 0;
1st and 2nd both, 2) | 0 |
| 4 | 1 (1st and 2nd
) | 0 |
| No. of family
members with other cancersa | | |
| 1 | 16 | 0 |
| 2 | 5 | 0 |
| 3 | 1 | 0 |
| 4 | 1 | 0 |
Samples
Genomic DNA was extracted using the protocol
described by Miller et al (21) from aliquots of peripheral blood
samples that had been stored at −20°C.
Primer design and PCR conditions
Nine sets of overlapping primers were designed for
the exon 11 coding region and adjacent intronic region of
BRCA2 using the website ‘Primer3: WWW primer tool’ (22). Specific primers for other exons of
the BRCA2 gene for PCR amplification were selected from the
BIC primer database.
Polymerase chain reaction (PCR) amplification was
performed using each of the 40 primer pairs in a 25-μl volume
containing 100 ng genomic DNA, 1.0–3.5 mmol/l MgCl2, 1X
PCR buffer [10 mM Tris-HCl (pH 8.3) and 50 mM KCl], 2.5 mmol/l
dNTPs (Promega, Madison, WI, USA), 5 pmols of each primer and 0.5
units of Taq polymerases (Promega). The PCR reaction was carried
out for 33 cycles, and the thermal regime consisted of an initial
denaturation at 94°C for 7 min, subsequent denaturation at 94°C for
1 min, annealing at the respective optimal temperature for 1 min
and extension at 72°C for 2 min and final extension at 72°C for 10
min. The optimal annealing temperature was obtained by optimizing
the PCR condition for each primer set for the exons of
BRCA2, and this varied from 51 to 66°C.
Mutation detection
Single-strand conformation
polymorphism
Single-strand conformation polymorphism (SSCP)
analysis was performed on all the exons except exon 11 in the
familial and sporadic breast cancer patients, at-risk individuals
from family F-01 and healthy controls as previously described
(16). Equal volumes of the PCR
product of the sample and denaturing loading buffer (95% formamide,
0.05% xylene cyanol, 0.05% bromophenol blue) were heated at 95°C
for 5 min. Denatured DNA samples were separated by 10%
polyacrylamide gel electrophoresis in non-denaturing gel and run at
100 V for 8–10 h in 0.5X TBE at 10–15°C. Separated DNA fragments
were visualized by silver staining (23). Samples which showed abnormal
migration patterns on SSCP were reconfirmed by performing a second
PCR reaction followed by a second SSCP run.
DNA sequencing
Samples to be sequenced were PCR amplified and the
products were purified using GFX™ PCR DNA and Gel Band Purification
(GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). Purified
PCR products were directly sequenced using the commercially
available DYEnamic™ ET Dye Terminator kit (GE Healthcare) and
MegaBACE 1000 automated DNA sequencer. Mutations and sequence
variants detected were reconfirmed by performing a second PCR and
direct sequencing. Exon 11 was analyzed by direct sequencing. This
analysis was limited to 55 patients with a family history, 20
at-risk individuals and 20 healthy controls. Direct sequencing was
also carried out for the samples showing abnormal banding patterns
during SSCP screening and for some representative samples not
showing any abnormal banding patterns to confirm the mutations and
sequence variants.
Results
This is the first report on BRCA2 mutations
and polymorphisms in Sri Lankan breast cancer patients in whom
BRCA1 mutations and sequence variants have been
characterized recently (16). A
total of 149 including 109 patients were studied, and twenty-three
sequence variants were identified.
Clearly and possibly pathogenic mutations are shown
in Table II. Two novel frame-shift
mutations, c.2403 insA and c.2667 insT in exon 11, and two novel
possibly pathogenic mutations, c.1191 A>C in exon 10 and c.5695
A>C in exon 11, and previously reported possibly pathogenic
intronic variant, IVS8-1 G>C in intron 8, were detected.
Sequence variants with unknown significance and polymorphisms are
shown in Table III. Twelve
previously reported polymorphisms in the exon 2 untranslated region
(5′UTR) and in exon 10, 11 and 14 were identified (Table III). Six novel polymorphisms in
exons 9, 10, 11, 14 and IVS15-21 insTT in intron 15 were
observed.
 | Table IIClearly pathogenic and possibly
pathogenic BRCA2 mutations identified. |
Table II
Clearly pathogenic and possibly
pathogenic BRCA2 mutations identified.
| E/I | NT | Base change | Codon | AA change | Designation | Variation type | BIC entry | Cases (%) | Pathogenic |
|---|
| I-8 | 910-1 | G>C | Non-coding | - | IVS8-1 G>C | IVS | Yes | 1 (0.78) | Clearly |
| 10 | 1191 | A>C | 321 | Lys>Asn | 1191 A>C | M-UV | No | 4 (3.1) | Possibly |
| 11a | 2403 | ins A | 726 | Val>Ser | 2403 insA | F | No | 4 (4.2) | Clearly |
| 11a | 2667 | ins T | 813 | Pro>Ser | 2667 insT | F | No | 1 (1.05) | Clearly |
| 11a | 5695 | A>C | 1823 | Lys>Gln | 5695 A>C | M-UV | No | 6 (6.3) | Possibly |
 | Table IIISequence variants with unknown
significance and known polymorphisms of the BRCA2 gene
identified. |
Table III
Sequence variants with unknown
significance and known polymorphisms of the BRCA2 gene
identified.
| E/I | NT | Base | Codon change | AA change | Designation | Variation type | BIC Entry | No. of cases
carrying the sequence variant
|
|---|
| F (n=55) | S (n=54) | R (n=20) | C (n=20) |
|---|
| 2 | 203 | G>A | - | - | 203 G>A | 5′UTR | Yes | 36 | 35 | 12 | 2 |
| 9 | 969 | C>T | 247 | Iso>Iso | 969 C>T | Silent-P | No | 1 | 0 | 0 | 0 |
| 9 | 971 | C>G | 247 | Ala>Gly | 971 C>G | M-P | No | 1 | 0 | 0 | 0 |
| 10 | 1093 | A>C | 289 | Asn>His | 1093 A>C | M-P | Yes | 6 | 3 | 3 | 0 |
| 10 | 1342 | A>C | 372 | Asn>His | 1342 A>C | M-P | Yes | 30 | 13 | 16 | 5 |
| 10 | 1352 | C>T | 375 | Pro>Leu | 1352 C>T | M-P | Yes | 4 | 0 | 1 | 0 |
| 10 | 1353 | C>T | 375 | Pro>Pro | 1353 C>T | Silent-P | No | 1 | 0 | 2 | 0 |
| 10 | 1593 | A>G | 455 | Ser>Ser | 1593 A>G | Silent-P | Yes | 1 | 0 | 4 | 0 |
| 11a | 2457 | T>C | 743 | His>His | 2457 T>C | Silent-P | Yes | 0 | - | 1 | 0 |
| 11a | 2766 | A>C | 846 | Ser>Ser | 2766 A>C | Silent-P | No | 0 | - | 1 | 0 |
| 11a | 3538 | A>C | 1103 | Thr>Pro | 3538 A>C | M-P | Yes | 0 | - | 1 | 0 |
| 11a | 3624 | A>G | 1132 | Lys>Lys | 3624 A>G | Silent-P | Yes | 0 | - | 6 | 1 |
| 11a | 4035 | T>C | 1269 | Val>Val | 4035 T>C | Silent-P | Yes | 0 | - | 1 | 0 |
| 11a | 4791 | A>G | 1521 | Leu>Leu | 4791 A>G | Silent-P | Yes | 50 | - | 20 | 17 |
| 11a | 6741 | G>C | 2171 | Val>Val | 6741 G>C | Silent-P | Yes | 47 | - | 20 | 18 |
| 14 | 7452 | A>G | 2408 | Pro>Pro | 7452 A>G | Silent-P | No | 15 | 13 | 20 | 0 |
| 14 | 7470 | A>G | 2414 | Ser>Ser | 7470 A>G | Silent-P | Yes | 0 | 0 | 1 | 0 |
| I-15 | 7845-21 | insTT | N-C | - | IVS15-21insTT | UV | No | 1 | 0 | 0 | 0 |
c.2403 insA in exon 11 (codon 726), a novel frame
shift insertion, created a stop codon at codon 750 of the BRCA2
protein in four unrelated patients with a family history of breast
cancer. All four of them were Sinhalese. Three of them, diagnosed
with breast cancer at an early age (35, 38, 47 years) had one
affected second degree relative each. The fourth patient diagnosed
at the age of 51 years is unmarried and has a sister affected with
breast cancer. Recurrence of the disease after 10 years of initial
diagnosis was reported in one. Mutation c.2667 insT in exon 11
which is also novel created a stop codon at codon 825 in the BRCA2
protein. It was found in one patient diagnosed with breast cancer
at 53 years of age having two affected first degree relatives.
Possibly pathogenic novel missense alteration c.5695
A>C (exon 11) was found in six study participants from family
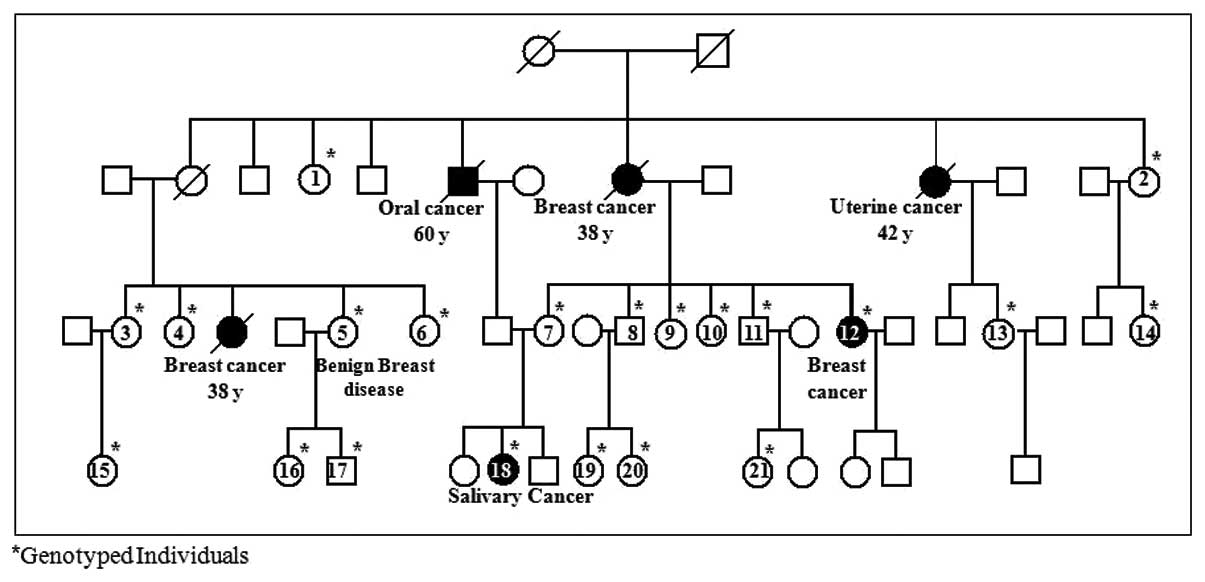
F-01. The pedigree of this family is shown in Fig. 1. Pedigree numbers and sequence
variants found in this family are shown in Table IV. This family consists of 8
siblings in the 2nd generation of which 4 are deceased, one each
from breast (II.8 at the age of 38 years), uterine (II.10 at the
age of 42 years) and oral (II.6 at the age of 60 years) cancers.
The possibly pathogenic mutation (c.5695 A>C in exon 11) was
found in 3 members of the 3rd generation (III.3, III.6, III.19) and
3 members of the 4th generation (IV.1, IV.2, IV.5). One member
(III.19) carrying this mutation also has a deleterious mutation in
exon 21 of BRCA1 (c.5405 delG) (16). Another (IV.5) was diagnosed with
salivary gland cancer at 13 years of age and her paternal
grandfather (II.6) and maternal grandmother (II.8) had died of oral
and breast cancer, respectively. Her mother (III.9) carries a novel
possibly pathogenic missense mutation, c.1191 A>C in exon 10.
The same mutation was also found in 2 other at-risk individuals
from the F-01 family (III.02, III.13) and in another familial
breast cancer patient from a different family diagnosed at the age
of 49 years and having one first and one second degree relative
affected.
 | Table IVMutations and polymorphisms in the
BRCA2gene among the F-01 family members. |
Table IV
Mutations and polymorphisms in the
BRCA2gene among the F-01 family members.
Individuals
|
BRCA2mutations and
polymorphismsa
|
|---|
| No. | Pedigree no. | 1191
A>C
10 | 5695
A>C
11 | 203
G>A
2 | 1093
A>C
10 | 1342
A>C
10 | 1352
C>T
10 | 1353
C>T
10 | 1593
A>G
10 | 2457 T/C
11 | 2766
A>C
11 | 3538
A>C
11 | 3624
A>G
11 | 4035
T>C
11 | 7470
A>G
11 |
|---|
| 1 | II.4 | | | | | √ | | | | | | | √ | | |
| 2 | II.13 | | | | | √ | | | | | | | | | |
| 3 | III.02 | √ | | √ | √ | √ | √ | √ | √ | | | | | | |
| 4 | III.03 | | √ | | | √ | | | | | | | √ | | |
| 5 | III.06 | | √ | √ | | √ | | | | | | | | | |
| 6 | III.07 | | | | | √ | | | | | | | √ | | √ |
| 7 | III.09 | √ | | √ | | √ | | | | | | | | | |
| 8 | III.11 | | | | | | | | √ | | | | √ | | |
| 9b | III.12 | | | √ | | √ | | | | | | | √ | | |
| 10 | III.13 | √ | | √ | | √ | | | | | | | | | |
| 11 | III.14 | | | | | √ | | | | | | | | | |
| 12b | III.16 | | | √ | | √ | | | | | | √ | √ | | |
| 13b | III.19 | | | √ | | √ | | | | | √ | | | | |
| 14 | III.22 | | | | | √ | | | | | | | | | |
| 15 | IV.01 | | | √ | √ | | | | √ | √ | | | | | |
| 16 | IV.02 | | | √ | | √ | | | | | | | | | |
| 17 | IV.03 | | | √ | | √ | | | | | | | | √ | |
| 18 | IV.05 | | | √ | | √ | | | | | | | | | |
| 19 | IV.07 | | | √ | | | | | | | | | | | |
| 20 | IV.08 | | | | √ | | | | √ | | | | | | |
| 21 | IV.09 | | | √ | | √ | | √ | | | | | | | |
A previously reported possibly pathogenic intronic
variant, IVS8-1 G>C in intron 8, was detected in one patient
with breast cancer and ovarian cancer both occurring at the age of
46 years. She also has co-existing novel silent (c.969 C>T) and
missense (c.971 C>G) mutations in exon 9. This patient has two
second degree relatives affected with breast cancer.
Novel unclassified variant IVS15-21 insTT in intron
15 was found in one patient diagnosed at 43 years of age whose
mother also had breast cancer. Analysis of the IVS15-21 insTT
intronic variant using Human Splicing Finder version 2.4 showed
that the acceptor motif of the exon 16 (at −12 region) is located
several bases away (downstream) from the detected variant (24).
We found four previously reported (c.1093 A>C,
c.1342 A>C (p.N372H), c.1352 C>T, c.1593 A>G) and one
novel (c.1353 C>T) polymorphism in exon 10. Missense mutation
c.1093 A>C was identified in six familial and three sporadic
breast cancer patients, 3 at-risk individuals but in none of the
healthy controls. c.1342 A>C (p.N372H) was the most prevalent
polymorphism, and the prevalence was significantly higher among
familial breast cancer patients and at-risk individuals (80%) than
among sporadic breast cancer patients (24%) and healthy controls
(25%). Missense mutation c.1352 C>T was detected in four
familial breast cancer patients and 1 at-risk individual, silent
mutation c.1593 A>G in one familial breast cancer patient and 4
at-risk individuals and novel silent mutation c.1353 C>T in one
familial breast cancer patient and 2 at-risk individuals. These
three mutations were not found in sporadic breast cancer patients
or healthy controls. All five polymorphisms in exon 10 co-existed
in one at-risk individual from the F-01 family (III.02).
In addition to the clearly and possibly pathogenic
mutations, we detected seven other sequence variants in exon 11.
These were three reported silent mutations (c.2457 T>C in one
at-risk individual, c.3624 A>C in 6 at-risk individuals and one
healthy control and c.4035 T>C in 1 at-risk individual), one
novel silent mutation (c.2766 A>C in 1 at-risk individual), a
reported missense mutation (c.3538 A>C in one familial breast
cancer patient) and two reported common polymorphisms (c.4791
A>G and c.6741 G>C). Prevalence of the two common
polymorphisms did not significantly differ between familial breast
cancer patients, at-risk individuals and healthy controls and these
were not studied in sporadic breast cancer patients.
We found the previously reported polymorphism c.203
G>A in exon 2 in >60% of the familial and sporadic breast
cancer patients and at-risk individuals but at a lower prevalence
(16.5%) among the healthy controls. A novel common polymorphism
c.7452 A>G in exon 14 was also found in the familial and
sporadic breast cancer patients and at-risk individuals but not in
any healthy controls. One at-risk individual also carried the
reported silent mutation c.7470 A>G in exon 14 in addition to
this polymorphism. The most common four polymorphisms, c.203 G>A
in exon 2, c.1342 A>C in exon 10, and c.4791 A>G and c.6741
G>C in exon 11, identified in the present study co-existed in
several familial patients, at-risk individuals and healthy
controls.
Discussion
After screening a predominantly Sinhalese cohort of
familial and sporadic breast cancer patients, at-risk individuals
from a family carrying a deleterious BRCA1 mutation and
healthy controls, we identified 23 sequence variants in the
BRCA2 gene. Two novel pathogenic frame-shift mutations
resulting in a premature stop codon in exon 11 and two novel
possibly pathogenic mutations one each in exon 10 and 11 and a
previously reported pathogenic variant in intron 8 were detected.
One novel intronic variant in intron 15 and five novel
polymorphisms were observed, two in exon 9 and one each in exons
10, 11 and 14. Twelve previously reported polymorphisms were
identified, one in the exon 2 untranslated region (5′UTR), four in
exon 10, six in exon 11 and one in exon 14.
The two clearly pathogenic mutations identified in
the present study (c.2403 insA and c.2667 insT) are frame-shift
insertions in exon 11 leading to a premature stop codon and thus
resulting in a truncated protein. Of these, c.2403 insA occurred in
four unrelated familial breast cancer patients of Sinhalese
ethnicity indicating that it is likely be a common mutation among
familial breast cancer patients in this ethnic group. The number of
study participants from other ethnic groups was small, thus one
cannot exclude the presence of this mutation in other ethnic groups
in Sri Lanka. The other deleterious mutation was found only in one
patient.
Mutation c.5695 A>C found in several members of
the F-01 family is located in the sixth BRC repeat (BRC6 repeat)
out of the eight BRC repeats encoded by exon 11. The BRC repeats
mediate the binding of DNA repair protein Rad51 (25). DNA recombinase RAD51 and BRCA2
protein complex is crucial for the repair of double-strand DNA
breaks (26). As this mutation
alters lysine (basic polar amino acid) at codon 1823 to glutamine
(neutral polar amino acid) it may change conformation of the
protein rendering pathogenicity. One at-risk individual found to
have this mutation was affected with salivary cancer, diagnosed at
the age of 13 years and another was diagnosed with a benign breast
tumour at the age of 32 years. The remaining four members carrying
this mutation are, at present, free from benign or malignant breast
disease or any other cancer despite one of them also carrying a
deleterious BRCA1 mutation.
Novel missense mutation c.1191 A>C in exon 10
found in one familial breast cancer patient and 3 at-risk
individuals in the present study, alters amino acid lysine (basic
polar amino acid) to asparagine (neutral polar amino acid) at codon
321. This too can be functionally significant, as it may affect the
conformation of the protein.
Nearly 23 and 6.5% of unclassified variants in
BRCA1 and BRCA2, respectively, are located in
intronic sequences but only few studies have been carried out to
identify whether these variants affecting RNA splicing are involved
in the pathogenicity of breast cancer (27). According to the BIC database, the
intronic variant IVS8-1 G>C located at the splice-site of the
intron is clinically important (28). To date, none of the Asian countries
have reported this mutation although this has been reported in
Western Europe. In exon 9, novel missense mutation c.971 C>G
alters amino acid alanine (nonpolar aliphatic) to glycine (nonpolar
aliphatic) at codon 247. This co-existed with the novel silent
mutation c.969 C>T in the same patient, and both these sequence
variants are unlikely to be of clinical importance. However, the
patient who carries these mutations has a previously reported
possibly pathogenic intronic variant, IVS8-1 G>C in intron
8.
Only ten intronic variations have been identified in
intron 15 and none of the Asian countries have reported variants in
this intron (28). In the present
study, we found a novel unclassified variant, IVS15-2 1insTT in
intron 15, in one familial breast cancer patient. However, results
from the bioinformatic analysis using Human Splicing Finder version
2.4 did not indicate this variant to be pathogenic (24).
One novel and four previously reported polymorphisms
were found in exon 10. Missense polymorphism c.1342 A>C
(BRCA2 p.N372H) occurred more frequently in the familial
breast cancer patients and at-risk individuals from family F-01
when compared to the sporadic breast cancer patients and healthy
controls suggesting a predisposition to familial breast cancer in
its presence. This polymorphism results in non-conservative amino
acid substitution [asparagine (N) to histidine (H)], hence may
affect the structure and function of the BRCA2 protein. N372H
locates within a region of BRCA2 (residues 290–453) that has been
shown to interact with a transcriptional co-activator protein,
P/CAF, which possesses histone acetyltransferase activity (29). The HH genotype of the N372H
polymorphism in the BRCA2 gene was reported to be associated
with a 1.3-fold increased risk of breast cancer in a combined
analysis of British, German and Finnish women (30) and in Australian women (31) but others have failed to find a
significant effect in older Caucasian women (32). The N372H polymorphism had an allele
frequency of 0.45 in the present study, which was higher than the
average frequency of 0.26 reported in British, German and Finnish
populations (30). This
polymorphism was also reported in a Malaysian population among
Malay, Chinese and Indian ethnic groups (33). A more recent meta-analysis failed
to show an effect of this polymorphism on breast cancer even when a
subgroup analysis by ethnicity was carried out but showed an effect
in population-based studies (34).
These authors have suggested the BRCA2 p.N372H allele as a
low-penetrant risk factor for developing breast cancer.
Missense mutation c.1093 A>C found in six
familial breast cancer patients with a strong family history of
breast cancer and 3 at-risk individuals has also been reported in
studies in Denmark and two Asian countries, Japan (35) and Malaysia (33). Two patients had mothers, three
patients had maternal aunts and two patients had maternal
grandmothers affected with breast cancer. Apart from that, there
were two paternal relations of these patients affected with breast
cancer. The c.1593 A>G polymorphism detected in one familial
breast cancer patient and 4 at-risk individuals has been reported
from Western countries (28) and
from Asia (33,35,36).
Co-existence of all five polymorphisms identified in exon 10
observed in one at-risk individual does not appear to have been
previously reported.
In exon 11, we identified two pathogenic mutations
and one possibly pathogenic mutation described above and seven
other sequence variants. Of the latter, silent mutations, c.2457
T>C and c.4035 T>C, have been reported in Europe and
Malaysia. The missense mutation c.3538 A>C detected in a
familial breast cancer patient carrying a deleterious BRCA1
mutation, alters threonine (hydrophilic polar uncharged) to proline
(aliphatic nonpolar) which may be significant for protein folding
or binding. However, according to the BIC database this transition
effect is recorded as ‘unknown’. This variant was reported in
Western European communities (28). The allele frequency of c.3624
A>G in exon 11 was much less than what was reported in a
European population (allele frequency of 0.08 vs. 0.31). c.4791
A>G and c.6741 G>C in exon 11 showed a very high allele
frequency of 0.86 in the Sri Lankan population whereas according to
the BIC database these polymorphisms were recorded only in
Africa.
Reported silent mutation c.7470 A>G in exon 14
found in one at-risk individual has been reported in several
non-Asian (28) and Asian
countries (33,35,36).
The novel common polymorphism, c.7452 A>G in exon 14, had an
allele frequency of 0.2 but allele frequencies did not
significantly differ between study groups.
The frequency of the common polymorphisms differed
from those reported for other populations (28). Common polymorphism c.203G>A in
exon 2 had an allele frequency of 0.39 in the present study whereas
a frequency of 0.25 was reported in predominantly European
populations. The four most common polymorphisms co-existed in
several familial breast cancer patients and at-risk individuals.
Co-existence of these four polymorphisms has not been reported
previously, but the presence of c.203 G>A in exon 2, c.1342
A>C in exon 10 and c.3624 A>G in exon 11 has been reported in
the Spanish population (28).
The prevalence of clearly pathogenic BRCA2
mutations was 11% (6/55) and, clearly and possibly pathogenic
BRCA2 mutations was 12.73% (7/55) among the familial breast
cancer patients. Thus when compared to our previous observation of
a prevalence of 6.25% for BRCA1 mutations (16), BRCA2 mutations appear to be
more frequent in Sri Lankan familial breast cancer patients.
In Sri Lanka, approximately 19% of female breast
cancers are diagnosed in women below 50 years of age suggesting a
significant genetic susceptibility. However, genetic screening
programmes are not in place due to the lack of data on mutations
associated with breast cancer for the Sri Lankan population and
high cost. We identified two pathogenic and three possibly
pathogenic mutations in the BRCA2 gene in familial breast
cancer. Of these, c.2403 insA in exon 11, the pathogenic mutation
which occurred in 4 out of 51 unrelated Sinhalese patients, is
likely to be useful in screening programmes.
Acknowledgements
We thank the director and staff of the
National Cancer Institute, Maharagama, Sri Lanka for their
cooperation, Dr Nalinda Silva for assistance with sample
collection, Dr Mohamed H. Ziard for assistance with sample
collection from the F-01 family. This study was supported by the
Sida/Secretariat for Research Cooperation Grant for Molecular
Biology and Biotechnology awarded to E.H.K. and K.H.T. and
constituted partly to the PhD programme of S.deS.
References
|
1.
|
Martin AM, Blackwood MA, Antin-Ozerkis D,
et al: Germline mutations in BRCA1 and BRCA2 in
breast-ovarian families from a breast cancer risk evaluation
clinic. J Clin Oncol. 19:2247–2253. 2001.
|
|
2.
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
3.
|
Cancer Incidence Data: Sri Lanka Year
2001–2005. Cancer Registry, National Cancer Control Programme;
Colombo 5, Sri Lanka: 2009
|
|
4.
|
Parkin DM, Whelan SL, Ferlay J, Teppo L
and Thomas DB: Cancer Incidence in Five Continents. VIII. IARC
Scientific Publications no. 143. IARC; Lyon: 1997
|
|
5.
|
Liede A and Narod SA: Hereditary breast
and ovarian cancer in Asia: genetic epidemiology of BRCA1
and BRCA2. Hum Mutat. 20:413–424. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Newman B, Austin MA, Lee M and King MC:
Inheritance of human breast cancer: evidence for autosomal dominant
transmission in high risk families. Proc Natl Acad Sci USA.
85:3044–3048. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Fackenthal JD and Olopade OI: Breast
cancer risk associated with BRCA1 and BRCA2 in
diverse populations. Nat Rev Cancer. 7:937–948. 2007. View Article : Google Scholar
|
|
8.
|
Satagopan JM, Offit K, Foulkes W, et al:
The lifetime risks of breast cancer in Ashkenazi Jewish carriers of
BRCA1 and BRCA2 mutations. Cancer Epidemiol
Biomarkers Prev. 10:467–473. 2001.PubMed/NCBI
|
|
9.
|
Antoniou A, Pharoah PDP, Narod S, et al:
Average risks of breast and ovarian cancer associated with
BRCA1 or BRCA2 mutations detected in case series
unselected for family history: a combined analysis of 22 studies.
Am J Hum Genet. 72:1117–1130. 2003.PubMed/NCBI
|
|
10.
|
Lahad EL and Friedman E: Cancer risks
among BRCA1 and BRCA2 mutation carriers. Br J Cancer.
96:11–15. 2007. View Article : Google Scholar
|
|
11.
|
Hall JM, Lee MK, Newman B, Morrow JE,
Anderson LA, Huey B and King MC: Linkage of early-onset familial
breast cancer to chromosome 17q21. Science. 250:1684–1689. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wooster R, Neuhausen SL, Mangion J, et al:
Localization of a breast cancer susceptibility gene, BRCA2,
to chromosome 13q12–13. Science. 265:2088–2090. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Narod SA and Foulkes WD: BRCA1 and
BRCA2: 1994 and beyond. Nat Rev Cancer. 4:665–676. 2004.
View Article : Google Scholar
|
|
14.
|
Scully R and Livingston DM: In search of
the tumour-suppressor function of BRCA1 and BRCA2.
Nature. 408:429–432. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Venkitaraman AR: Cancer susceptibility and
the functions of BRCA1 and BRCA2. Cell. 108:171–182.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
De Silva W, Karunanayake EH, Tennekoon KH,
Allen M, Amarasinghe I, Angunawala P and Ziard MH: Novel sequence
variants and a high frequency of recurrent polymorphisms in
BRCA1 gene in Sri Lankan breast cancer patients and at risk
individuals. BMC Cancer. 8:2142008.PubMed/NCBI
|
|
17.
|
Ford D, Easton DF, Stratton M, et al:
Genetic heterogeneity and penetrance analysis of the BRCA1
and BRCA2 genes in breast cancer families. The Breast Cancer
Linkage Consortium. Am J Hum Genet. 62:676–689. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Thompson D and Easton D: Variation in
cancer risks, by mutation position, in BRCA2 mutation
carriers. Am J Hum Genet. 68:410–419. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Liede A, Karlan BY and Narod SA: Cancer
risks for male carriers of germline mutations in BRCA1 and
BRCA2: a review of the literature. J Clin Oncol. 22:735–742.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Ferla R, Calo V, Cascio S, et al: Founder
mutations in BRCA1 and BRCA2 genes. Ann Oncol.
18(Supp 6): S93–S98. 2007. View Article : Google Scholar
|
|
21.
|
Miller SA, Dykes DD and Poleskey HF: A
simple salting out procedure for extracting DNA from human
nucleated cells. Nucleic Acids Res. 16:12151988. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Rozen S and Skaletsky HJ: Primer3, 1996.
Code available at http://www-genome.wi.mit.edu/genome_software/other/primer3.htmluri.
|
|
23.
|
Wallace AJ: SSCP/Heteroduplex analysis.
PCR Mutation Detection Protocols. 187. Theophilus BDM and Rapley R:
Humana Press; Totowa, New Jersey: pp. 151–163. 2002, View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Desmet FO, Hamroun D, Lalande M,
Collod-Béroud G, Claustres M and Béroud C: Human Splicing Finder:
an online bioinformatics tool to predict splicing signals. Nucleic
Acid Res. 37:e672009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Bork P, Blomberg N and Nilges M: Internal
repeats in BRCA2 protein sequence. Nat Genet. 13:22–23. 1996.
View Article : Google Scholar
|
|
26.
|
Lord CJ and Ashworth A: RAD51,
BRCA2 and DNA repair: a partial resolution. Nat Struct Mol
Biol. 14:461–462. 2007.
|
|
27.
|
Vreeswijk MPG, Kraan JN, van der Klift HM,
et al: Intronic variants in BRCA1 and BRCA2 that
affect RNA splicing can be reliably selected by splice-site
prediction programs. Hum Mutat. 30:107–114. 2009.
|
|
28.
|
Breast Cancer Information Core (BIC)
Database: Available at: http://research.nhgri.nih.gov/bic/uri.
Last modified: Wednesday, 29 September 2010.
|
|
29.
|
Fuks F, Milner J and Kouzarides T:
BRCA2 associates with acetyltransferase activity when bound
to P/CAF. Oncogene. 17:2351–2354. 1998. View Article : Google Scholar
|
|
30.
|
Healey CS, Dunning AM, Teare MD, et al: A
common variant in BRCA2 is associated with both breast
cancer risk and prenatal viability. Nat Genet. 26:362–364. 2000.
View Article : Google Scholar
|
|
31.
|
Spurdle AB, Hopperm JL, Chen X, et al: The
BRCA2 372 HH genotype is associated with risk of breast
cancer in Australian women under age 60 years. Cancer Epidemiol
Biomarkers Prev. 11:413–416. 2002.PubMed/NCBI
|
|
32.
|
Cox DG, Hankinson SE and Hunter DJ: No
association between BRCA2 N372H and breast cancer risk.
Cancer Epidemiol Biomarkers Prev. 14:1353–1354. 2005.
|
|
33.
|
Toh GT, Kang P, Lee SSW, et al:
BRCA1 and BRCA2 germline mutations in Malaysian women
with early onset breast cancer without a family history. PLoS One.
3:e20242008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Qiu LX, Yao L, Xue K, et al: BRCA2
N372H polymorphism and breast cancer susceptibility: a
meta-analysis involving 44,903 study participants. Breast Cancer
Res Treat. 123:487–490. 2010. View Article : Google Scholar
|
|
35.
|
Ikeda N, Miyoshi Y, Yoned K, Shiba E,
Sekihara Y, Kinoshita M and Noguchi S: Frequency of BRCA1
and BRCA2 germline mutations in Japanese breast cancer
families. Int J Cancer. 91:83–88. 2001.
|
|
36.
|
Saxena S, Chakraborthy A, Kaushal M, et
al: Contribution of germline BRCA1 and BRCA2 sequence
alterations to breast cancer in Northern India. BMC Med Genet.
7:752006. View Article : Google Scholar
|