Introduction
Colorectal cancer (CRC) is a serious medical
problem. It is one of the most frequently occurring neoplasms and
is responsible for the second highest mortality rate of all cancers
in the more developed regions of the world. Approximately 220,000
new cases of CRC are diagnosed each year in the USA and
Northwestern Europe. The genetic alterations in CRC progression are
determined by one of three pathways: microsatellite instability
(MSI), chromosome instability (CIN) or CpG island methylator
phenotype (CIMP). The progression of CRC is correlated to a number
of characteristic mutations in genes APC, K-RAS,
DCC, P53, transforming growth factor-β/SMAD,
or epigenetic changes. There are a number of other possible
molecular factors involved in CRC progression, one of which appears
to be the gene FJ 194940.1, previously termed
P65.
Previous investigation of the gene encoding
tumor-associated protein (P65) suggested that it is a molecule that
is newly associated with carcinogenesis. Its clinical
characterization as a potential marker of cancer development and
progression has been reported in many tumors (1–3).
Cloning of the complementary DNA (cDNA) encoding this protein
revealed that the sequence corresponds to the 226 24025-226 27543
region of chromosome 1. The gene for this transcript, hereafter
termed FJ 194940.1, consists of 6 exons and probably
undergoes differential splicing in malignant tissues. Moreover,
integration of an additional copy of the rv_001141 provirus in this
region of chromosome 1 was detected. It is possible that this
integration leads to the rearranged transcription of FJ
194940.1 (4).
The aim of this study was to assess the presence of
the expression of each potential exon and exon-exon junction in the
FJ 194940.1 gene transcript. Thereafter, each particular
FJ 194940.1 transcript is compared with certain histological
parameters and grading, as well as clinical staging of the
neoplasms to assess the potential role of the gene transcript as a
prognostic marker for CRC. Finally, a comparison of a particular
FJ 194940.1 transcript with survival time was carried
out.
Materials and methods
Materials
Tissue specimens of colorectal cancer were obtained
from the Oncological Centre of Lodz, Poland. CRC was diagnosed by
histopathological examination using established clinical criteria
(TNM classification by Jass with latest revision Cancer Staging
Manual by AJCC, 1997) at the Department of Pathology, Medical
University of Lodz, Poland. Tissue samples from 102 patients (55
males and 47 females) were frozen immediately following surgery in
liquid nitrogen and stored at −80°C until further examination.
All experiments were carried out with the approval
of the local Ethics Committee (No. RNN/214/00). Informed consent
was obtained from all 102 patients included in the study.
RNA isolation
Total RNA isolation was performed with a Total RNA
Prep Plus Minicolumn Kit (A&A Biotechnology, Poland) as per the
manufacturer’s instructions. This method is based on RNA isolation
methodology developed previously by Chomczynski and Sacchi, 1987
(5). The estimated degree of
contamination of the RNA received was determined using
spectrophotometric analysis. The isolated RNA had an A260:280 ratio
of 1.6–1.8. Purified RNA samples were stored at −80°C until further
use.
Reverse transcriptase polymerase chain
reaction (RT-PCR)
RT-PCR reaction was carried out using Enhanced Avian
HS RT-PCR Kit (Sigma, St. Louis, MO, USA) according to the
manufacturer’s instructions. The cDNA was used immediately or
otherwise stored at −20°C. The presence of cDNA in each sample was
confirmed using PCR with primers complementary to the
β-actin gene. Only samples with a product of this
housekeeping gene were used in subsequent tests.
In the second stage, PCR was performed with
particular primers that are specific for potential exons and exon
junctions of the FJ 194940.1 gene. The primers were designed
using Primer3 software (http://biotools.umassmed.edu/bioapps/primer3_www.cgi).
Amplification conditions were established by gradient PCR.
Amplification of particular exons and exon junctions were carried
out using 0.7 μl 0.5 μM, each forward and reverse primers, and 2.5
μl cDNA as a template. The PCR mixture also contained 0.4 μl 10 mM
mix deoxyribonucleotide triphosphates (dNTPs), 0.2 ml 0.5 units of
Taq polymerase, 2 μl 10X reaction buffer. Parallel negative control
(without cDNA) was amplified. PCR products were detected in 2%
agarose gel electrophoresis.
Primer sequences and the size of the expected PCR
products are shown in Table I.
 | Table I.Primer sequences specific for
potential exons and exon junctions of the FJ 194940.1 gene
and expected product size. |
Table I.
Primer sequences specific for
potential exons and exon junctions of the FJ 194940.1 gene
and expected product size.
| Exon | Primer sequence
| Expected amplicon
length (bp) |
|---|
| Forward primer | Reverse primer |
|---|
| I/II |
TGGTGTCCTATGGAATGCAG |
CAGTTCTTCTGGCCCATCCT | 153 |
| II |
CACTAGAAAACCCTATGACTTTCACA |
GGACAGTTACTTGCAGTTCTTCTG | 103 |
| II/III |
TGTGAAACAAGCAGTGCAAC |
GCTTGTTGGTCAGCCTTCTG | 155 |
| III |
TTTTCCATGTTGATGCTCA |
CGCCTGAGTCTGCAGTAAT | 101 |
| III/IV |
TGCTCATGCATCTCTGCTTT |
TTTGAAATGGGAGCCACTGT | 218 |
| IV |
CAGTGGCTCCCATTTCAAAG |
TCAAACAGGTGATCGTTCCA | 189 |
| IV/V |
CAGCTGGCCTAATCGAAAGA |
TCCAGCATTTCAGCAAGAGA | 155 |
| V |
CTCTCTTGCTGAAATGCTGG |
GGCCCAGGCTTTAAACTATA | 94 |
Statistical analysis
For statistical analysis, the transcript was divided
into two parts: A and B. Part A consisted of exons II and III, as
well as I/II and II/III exon-exon junctions, whereas part B
comprised exons IV and V, as well as III/IV and IV/V exon-exon
junctions. Statistical analyses were performed using STATISTICA 8.0
(StatSoft, Inc., 1984–2008) on the basis of the Chi-square test,
Chi-square test with Yates’ correction and V2 test. P<0.05 was
considered to indicate statistical significance. Time-to-death
distribution for survival in the whole population of the 102
patients was estimated using the Kaplan-Meier method. The log-rank
test was used to test for differences in time-to-death
distribution.
Results
Alternative transcripts
A total of 18 splice variants were identified, which
arose from various combinations of 4 exons (II, III, IV and V) and
exon-exon junctions between exons 1 and 2 (I/II), 2 and 3 (II/III),
3 and 4 (III/IV), as well as 4 and 5 (IV/V) (Table II). In the majority of cases, a
transcript consisting of all the previously mentioned elements was
found. In all 102 samples, PCR products with primers set for exon V
and I/II exon junctions were present. This result indicates that
exon V is common to all mRNA transcripts for the FJ 194940.1
gene. The remaining primer set products yielded inconsistent
results. Of note, in 17 cases, PCR products with primer sets for
exons II and III were found. However, PCR products with a primer
set for the junction of II/III exons were not detected. A similar
situation exists in cases of other primers complementary to the
exon-exon boundaries and separate exons. This suggests the presence
of additional sequences in this area of the FJ 194940.1
transcript.
 | Table II.Combination and frequency (A) of 18
possible splice variants. |
Table II.
Combination and frequency (A) of 18
possible splice variants.
| A | II/III | II | II/III | III | III/IV | IV | IV/V | V |
|---|
| 40 | ------ | ------ | ------ | ------ | ------ | ------ | ------ | ------ |
| 17 | ------ | ------ | | ------ | ------ | ------ | ------ | ------ |
| 8 | ------ | ------ | ------ | | ------ | ------ | ------ | ------ |
| 7 | ------ | | | ------ | ------ | ------ | ------ | ------ |
| 6 | ------ | ------ | ------ | ------ | | | ------ | ------ |
| 4 | ------ | | ------ | ------ | ------ | ------ | ------ | ------ |
| 2 | ------ | ------ | ------ | ------ | | ------ | ------ | ------ |
| 2 | ------ | ------ | ------ | ------ | ------ | ------ | | ------ |
| 2 | ------ | ------ | ------ | ------ | ------ | | | ------ |
| 2 | ------ | | | | ------ | ------ | ------ | ------ |
| 2 | ------ | ------ | | ------ | | | | ------ |
| 2 | ------ | ------ | | ------ | | | ------ | ------ |
| 2 | ------ | | | ------ | | | ------ | ------ |
| 2 | ------ | | | ------ | ------ | | ------ | ------ |
| 1 | ------ | ------ | | | ------ | ------ | ------ | ------ |
| 1 | ------ | ------ | ------ | | | | ------ | ------ |
| 1 | ------ | ------ | | ------ | | ------ | ------ | ------ |
| 1 | ------ | | ------ | ------ | ------ | | ------ | ------ |
FJ 194940.1 gene expression is correlated
with certain clinico-histological features
Statistical analysis was carried out with reference
to the whole FJ 194940.1 transcript and to particular exons
and exon-exon junctions. In addition, the whole transcript was
divided into parts A and B. Part A consisted of exons II and III,
as well as I/II and II/III exon-exon junctions, whereas part B
comprised exons IV and V, as well as III/IV and IV/V exon-exon
junctions.
The patients in this study included 55 females and
47 males. The majority of patients had negative familial history.
No statistically significant correlation was found between gender
and family history and expression of the whole transcript, or parts
A or B.
Tumors in the examined group were localized mainly
in the colon. No statistically significant correlation was noted
between localization of tumor and expression of the whole
transcript, or part A or B.
Examined cases were classified as either tubular or
mucinosum. There was no statistically significant correlation
between histological type and expression of the whole FJ
194940.1 gene transcript, or part A or B. However, there was a
tendency towards a more frequent presence of part A (p=0.0894) in
tubular-type cancer.
Two analyses were conducted between histological
grade and splice variants. In the first approach, cases classified
as G1 were compared with cases classified as G2 or G3. In the
second approach, cases classified as G1 or G2 were compared with
cases classified as G3. Expression of part B of the FJ
194940.1 gene transcript is correlated with well-differentiated
(G1) and moderately differentiated (G2) cases (p=0.0000).
To assess the clinical utility of FJ 194940.1
splice variants, determination of these results was compared with
various clinicopathological parameters, including depth of tumor
invasion (T), lymph node metastases (N) and distant metastases (M).
No statistically significant associations were noted between TNM
classification and the expression of the whole FJ 194940.1
gene transcript, or part A or B.
Lymphocytic tumor infiltration, a favorable
prognostic factor in CRC, was significantly correlated with the
presence of all elements in part A of the FJ 194940.1 gene
transcript (p=0.0477).
Venous invasion was observed in the majority of
patients. There was no statistically significant association
between venous invasion and expression of the whole FJ
194940.1 gene transcript, or part A or B.
The results for the expression of the FJ
194940.1 gene transcript and clinicopathological parameters are
shown in Table III.
 | Table III.Comparison of the expression of a
particular FJ 194940.1 transcript with certain
clinicopathological parameters. |
Table III.
Comparison of the expression of a
particular FJ 194940.1 transcript with certain
clinicopathological parameters.
| Parameters | Expression of the
whole transcript FJ 194940.1 gene
| Expression of part
A transcript FJ 194940.1 gene
| Expression of part
B transcript FJ 194940.1 gene
|
|---|
| − | + | p-value | − | + | p-value | − | + | p-value |
|---|
| Gender | | | | | | | | | |
| Female | 34 | 21 | 0.8170a | 28 | 27 | 0.6797a | 13 | 42 | 0.7762a |
| Male | 28 | 19 | | 22 | 25 | | 10 | 37 | |
| Family history | | | | | | | | | |
| Negative | 55 | 35 | 0.8969b | 45 | 45 | 0.5894c | 19 | 71 | 0.5592b |
| Positive | 7 | 5 | | 5 | 7 | | 4 | 8 | |
| Tumor
localization | | | | | | | | | |
| Rectum | 20 | 15 | 0.5236a | 16 | 19 | 0.5790a | 6 | 29 | 0.3283c |
| Colon | 42 | 24 | | 34 | 32 | | 17 | 49 | |
| Histological
type | | | | | | | | | |
| Tubular | 51 | 36 | | 41 | 46 | 0.3594c | 19 | 68 | 0.9373b |
| Mucinosum | 11 | 4 | 0.2835c | 9 | 6 | | 4 | 11 | |
| Histological
grade | | | | | | | | | |
| G1 or G2 | 41 | 29 | 0.3826a | 38 | 32 | 0.1487a | 8 | 62 | 0.0000c |
| G3 | 21 | 10 | | 12 | 19 | | 15 | 16 | |
| Histological
grade | | | | | | | | | |
| G1 | 9 | 1 | | 8 | 2 | 0.0894b | 2 | 8 | 0.8595b |
| G2 or G3 | 53 | 38 | 0.1061b | 42 | 49 | | 21 | 70 | |
| Invasion of the
intestinal wall | | | | | | | | | |
| pT1 or pT2 | 18 | 11 | 0.8273a | 14 | 15 | 0.9757a | 6 | 23 | 0.7526c |
| pT3 or pT4 | 43 | 29 | | 35 | 37 | | 17 | 55 | |
| Node
involvement | | | | | | | | | |
| pN0 | 36 | 24 | 0.9586a | 30 | 30 | 0.7996a | 12 | 48 | 0.4676c |
| pN1 or pN2 | 23 | 15 | | 18 | 20 | | 10 | 28 | |
| Distant
metastasis | | | | | | | | | |
| pM0 | 53 | 30 | 0.1864c | 44 | 39 | 0.0934c | 19 | 64 | 0.8956b |
| pM1 | 9 | 10 | | 6 | 13 | | 4 | 15 | |
| TNM stage | | | | | | | | | |
| I or II | 35 | 22 | 0.8854a | 29 | 28 | 0.6728a | 12 | 45 | 0.6840a |
| III or IV | 27 | 18 | | 21 | 24 | | 11 | 34 | |
| Lymphocytic
infiltration | | | | | | | | | |
| Absent | 33 | 25 | 0.4036a | 28 | 30 | 0.7742a | 12 | 46 | 0.7585c |
| Present | 28 | 15 | | 22 | 21 | | 10 | 33 | |
| Venous
invasion | | | | | | | | | |
| Absent | 28 | 13 | 0.2029a | 25 | 16 | 0.0477a | 8 | 33 | 0.5494c |
| Present | 34 | 27 | | 25 | 36 | | 15 | 46 | |
FJ19490.1 gene and survival time
Statistical analysis between each separate exon and
exon-exon junction and survival time was carried out. Notably, if
we analyzed the survival time dependent on the presence or absence
of separate exons and boundaries, only the presence of the junction
between exons II/III was likely to be correlated with a longer
survival time. This correlation was statistically significant
(p=0.01972).
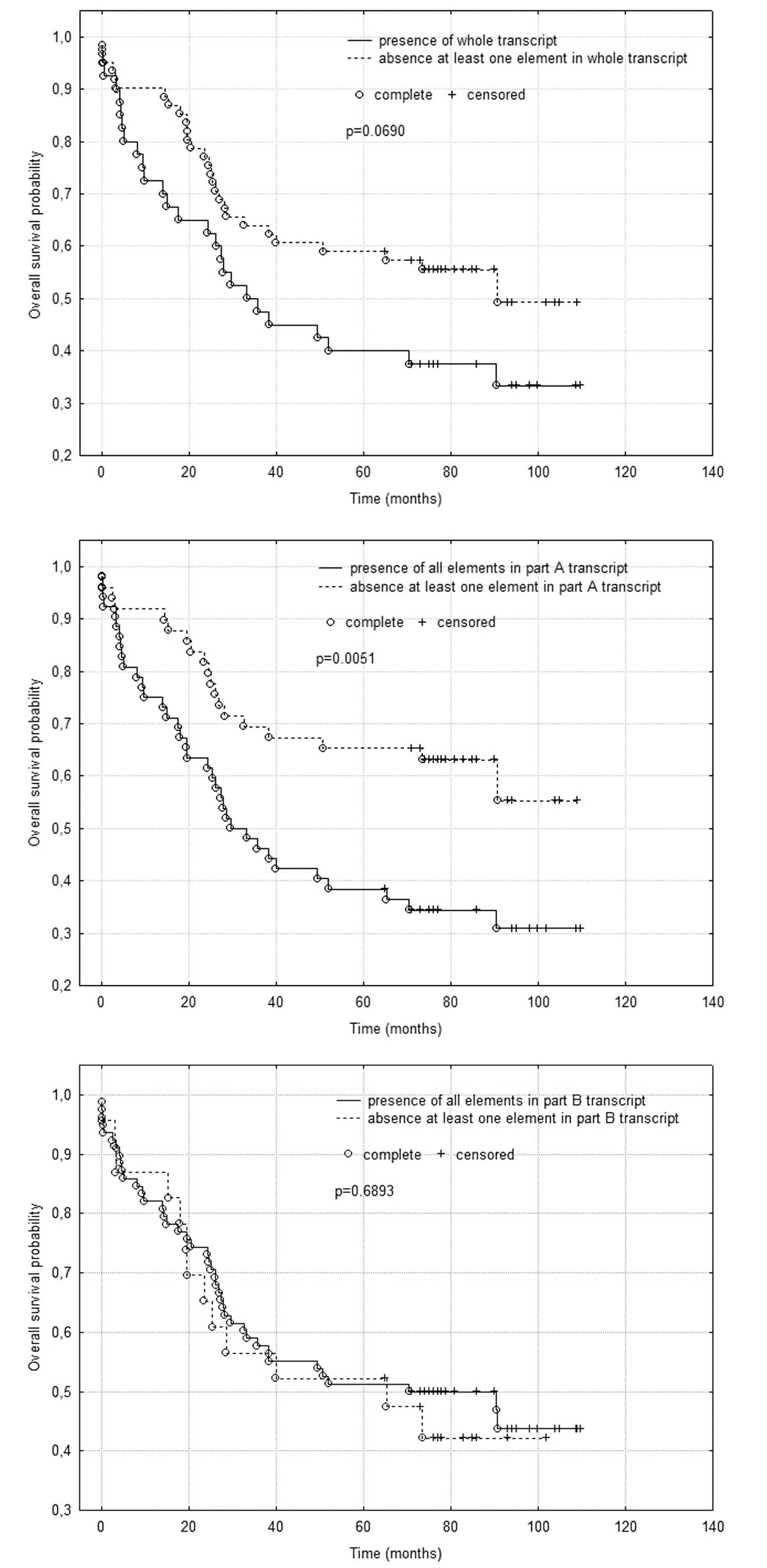
No statistically significant difference was observed
in survival time when comparing patients with the presence and
absence of the whole transcript, despite a marked tendency for
longer survival of patients without at least one element in the
whole transcript (p=0.0690) (Fig.
1A).
A statistically significant difference was found in
survival time when comparing patients with presence or absence of
part A of the FJ 194940.1 gene transcript. Patients who had
all elements in part A of the transcript survived for a shorter
duration (p=0.0051) (Fig. 1B).
There was no statistically significant difference in
survival time when comparing patients with the presence or absence
of part B of the FJ 194940.1 gene transcript (Fig. 1C).
Discussion
Splicing, the production of more than one transcript
from one gene, appears to affect over 50% of human genes. Numerous
cancer-associated genes, including CD44 and WT1, are
alternatively spliced. Variation of the splicing process may occur
during tumor progression and play a significant role in
tumorigenesis (6–12). Furthermore, alternatively spliced
transcripts, which are found mainly in tumor tissues, may be
extremely useful as cancer markers and serve as drug targets. In
addition, clarification and understanding of the changes in
splicing, as well as the characterization of various isoforms, may
improve our knowledge of malignant transformation.
Probably the best characterized genes that exist in
several isoforms and play a role in carcinogenesis are CD44
and the Wilms’ tumor gene, WT1. CD44 standard and
CD44 variants are expressed in a number of cancer cell types
and correlate with the progression and prognosis of certain
malignant tumors. For example, CD44v6 expression was positively
connected with advanced gastric cancer stage, and patients with
this isoform have lower, 3- and 5-year survival times (10). In their study, Aaltomaa et
al demonstrated that the down-regulation of CD44 and its CD44v6
isoform was correlated with tumor malignancy and unfavorable
prognosis in prostate cancer (11). The expression of CD44 isoforms has
also been evaluated in breast cancer. The presence of CD44v3
significantly correlated with the presence of lymph nodes
metastases (13). Thus, CD44
variants may be useful as diagnostic or prognostic markers in
certain human malignant diseases. However, data are conflicting and
further studies are required in order to establish the prognostic
value of CD44 and its variant isoforms. This observation is a good
example of the value of research into the association between
splice variants and cancer.
The WT1 gene has four splice variants but only two
of them are crucial to cancer development. These variants are
termed WT1+KTS and WT1-KTS. The balance between
isoforms with and without the 17-amino acid insertion appears to
affect the regulation of proliferation, differentiation and
apoptosis, and the prevention of Wilms’ tumor formation (14,15).
In light of the most up-to-date FJ 194940.1
investigations, this gene also undergoes alternative splicing, and
its isoforms are present during carcinogenesis.
The FJ 194940.1 gene is localized on
chromosome 1 and is physiologically expressed only in the kidney.
The gene consists of 6 exons and 5 introns (1). The first intron sequence is similar
to a provirus belonging to the HERVL66 family. This provirus is
normally located above the FJ 194940.1 gene. It is likely
that during carcinogenesis, integration of an additional copy of
this virus occurs in the FJ 194940.1 gene region. This
modification alters transcription in the chromosome region and the
gene is then expressed in neoplastic tissue.
Establishing the nucleotide sequence of the FJ
194940.1 gene and its structure has made it possible to design
primers complementary to exons and exon-exon junctions. This
approach enables confirmation of the predicted exon-intron gene
structure and allows us to confirm or contradict alternative
splicing in this gene.
Preliminary results have shown that FJ
194940.1 undergoes alternative splicing in various tumors
(4). In the preliminary
experiment, the RT-PCR product for the junction of exons III/IV,
and IV/V, as well as for exon III, was found in all analyzed
samples (4). Products with other
primers were observed irregularly (4). Findings of those studies are
inconsistent with results presented in this study, in which RT-PCR
products for exon V and the junction of exons I/II were found in
all investigated cases, while other products were irregular. Lack
of a RT-PCR product for exon III and junction of exons III/IV and
IV/V was rare, and appeared in 12, 16 and 6 out of 102 cases,
respectively. This discrepancy between the two experiments may be
explained by the variable number of cases examined by the two
investigators. The preliminary study counted only 30 cases, whereas
in the present study 102 cases were examined.
FJ 194940.1 (previously known as P65)
gene expression assessment in CRC has already been carried out
(1,16). At that time, the primers used were
designed based on amino acid sequences. Presence of the FJ
194940.1 expression was associated with more advanced tumors
with metastases to lymph nodes and distant metastases (16). These findings were confirmed by
means of the real-time PCR technique. Higher levels of FJ
194940.1 expression were observed in more advanced cases,
classified as III and IV according to pTNM classification (1). The results suggest that FJ
194940.1 expression in colon cancer is engaged in the process
of metastasis formation and may be associated with poor prognosis
for the patient.
The presence of part A is correlated with
lymphocytic tumor infiltration, which is a favorable prognostic
factor. However, this factor is associated with shorter survival
time. The presence of part B expression is associated with cases of
low-grade malignancy, which is correlated with better prognosis for
patients.
In conclusion, the investigated FJ 194940.1
gene undergoes alternative splicing. However, the role of its
transcripts and potential proteins remain to be examined in further
detail.
Acknowledgements
This study was supported by grant
502-03/3-015-02/502-34-014 from the Medical University of Lodz,
Poland.
References
|
1.
|
E BalcerczakM BalcerczakM
MirowskiQuantitative analysis of the p65 gene expression in
patients with colorectal cancerInt J Biomed
Sci3287291200723675055
|
|
2.
|
E BalcerczakJ BartkowiakJZ BłońskiT RobakM
MirowskiExpression of gene encoding p65 oncofetal protein in acute
and chronic leukemiasNeoplasma49295299200212458326
|
|
3.
|
W CzyżE BalcerczakM RudowiczH
NiewiadomskaZ PasiekaK KuzdakM MirowskiExpression of C-ERBB2 and
P65 genes and their protein products in follicular neoplasms of
thyroid glandFol Histochem Cytobiol419195200312722795
|
|
4.
|
E BalcerczakT MalewskiM BartczakM
MirowskiAlternation of FJ 194940.1 transcript expression after
provirus integration in human neoplasmInt J Integrat
Biol1158632011
|
|
5.
|
B Kwabi-AddoF RopiquetD GiriM
IttmannAlternative splicing of fibroblast growth factor receptors
in human prostate
cancerProstate46163172200110.1002/1097-0045(20010201)46:2%3C163::AID-PROS1020%3E3.0.CO;2-T11170144
|
|
6.
|
P ChomczynskiN SacchiSingle-step method of
RNA isolation by acid guanidinium thiocyanate-phenol-chloroform
extractionAnal
Biochem1621569198710.1016/0003-2697(87)90021-22440339
|
|
7.
|
TI OrbanE OlahExpression profiles of BRCA1
splice variants in asynchronous and in G1/S synchronized tumor cell
linesBiochem Biophys Res
Commun2803238200110.1006/bbrc.2000.406811162473
|
|
8.
|
D BaudryM HamelinMO CabanisJC FournetMF
TournadeS SarnackiC JunienC JeanpierreWT1 splicing alterations in
Wilms’ tumorsClin Cancer Res6395739652000
|
|
9.
|
M MixonF KittrellD MedinaSplice variant
expression of CD44 in patients with breast and ovarian cancerOncol
Rep8145151200111115587
|
|
10.
|
M HoriJ ShimazakiS InagawaM ItabashiM
HoriAlternatively spliced MDM2 transcripts in human breast cancer
in relation to tumor necrosis and lymph node involvementPathol
Int50786792200010.1046/j.1440-1827.2000.01119.x11107050
|
|
11.
|
Y XinA GraceMM GallagherBT CurranMB
LeaderEW KayCD44v6 in gastric carcinoma: a marker of tumor
progressionAppl Immunohistochem Mol
Morphol9138142200110.1097/00129039-200106000-0000611396631
|
|
12.
|
S AaltomaaP LipponenM Ala-OpasVM
KosmaExpression and prognostic value of CD44 standard and variant
v3 and v6 isoforms in prostate cancerEur
Urol39138144200110.1159/00005242811223672
|
|
13.
|
J RysA KruczakB LackowskaA
Jaszcz-GruchałaA BrandysA StelmachM ReinfussThe role of CD44v3
expression in female breast carcinomasPol J
Pathol54243247200314998292
|
|
14.
|
SM HewittGF SaundersDifferentially spliced
exon 5 of the Wilms’ tumor gene WT1 modifies gene
functionAnticancer Res1662162619968687106
|
|
15.
|
C EnglertX HouS MaheswaranWT1 suppresses
synthesis of the epidermal growth factor receptor and induces
apoptosisEMBO J144662467519957588596
|
|
16.
|
M BalcerczakE BalcerczakG Pasz-WalczakR
KordekM MirowskiExpression of the p65 gene in patients with
colorectal cancer: comparison with some histological typing,
grading and clinical stagingEur J Surg
Oncol30266270200410.1016/j.ejso.2003.11.00615028307
|