Introduction
The epidermal growth factor receptor (EGFR)
signaling pathway plays a crucial role in many carcinogenic
processes, such as proliferation, angiogenesis, invasion and
metastasis, and resistance to apoptosis (1,2).
Since deregulation of EGFR pathway genes has been observed
frequently in various types of tumors, including non-small cell
lung cancer (NSCLC), the development of targeted agents for lung
cancer therapy has focused mainly on EGFR and its downstream
networks (3), such as RAS/RAF/MAP
kinase and PI3K/AKT, being the two major pathways (3,4).
The pathway linking receptor tyrosine kinases to the
Ras family to the Raf serine-threonine kinase to the MAP kinase
cascade is critical for cell proliferation and is frequently
activated in human cancers (5).
MAP kinase, also known as extracellular signal-regulated protein
kinase (ERK), is crucial for the transduction of growth signals
from several key growth factors, such as EGF. ERK1 and ERK2,
downstream effectors of the RAS-RAF-MEK-ERK-MAP kinase pathway, are
constitutively active in many NSCLCs. Mutations of Braf were
first reported in melanomas (over 60%) and colorectal cancers. The
V600E (previously reported as V599E) mutant form of Braf
activates the RAF/MEK/ERK pathway in human melanoma cells in
vitro, and transformation of a melanocyte cell line with mutant
Braf was found to activate the MAP kinase pathway (6). Braf mutations have since been
reported in 3% (7) and 1.6%
(8) of NSCLCs. Recently,
additional studies have shown that somatic mutations of the
Braf gene are found in approximately 2% of Caucasians
(9,10) and in 1.2% of East Asians (11).
The erbB family comprises four structurally related
receptors: erbB1 (EGFR) to erbB4. On ligand stimulation, the
receptor forms either homodimers or heterodimers, which activate
their cytoplasmic domain. Several reports have shown that somatic
mutations of the EGFR gene are found in 25–40% of Japanese
NSCLC patients (12,13), but only in 10% of NSCLC patients in
the US (14,15). EGFR mutations were
predominantly found in female, non-smokers with adenocarcinomas
(12–16). Actually, EGFR mutations in
NSCLC have been correlated with clinical response to gefitinib
therapy (17–19). In addition, it has been reported
that erbB2 mutations at the kinase domain are present in 4%
of European-derived NSCLC patients (20). Somatic erbB2 mutations are
more frequent in never-smoker and adenocarcinoma histology
(21). Although the erbB2
mutation was also investigated in Japanese NSCLC (13,22),
the Braf mutation frequency in Japanese NSCLC is not well
known. We previously described a single Braf mutation case
(23), however, in this study, we
investigated the Braf mutation status in 305 surgically
treated NSCLC cases.
Patients and methods
Patients
This is a retrospective study. The study group
included 305 lung cancer patients who had undergone surgery at the
Department of Surgery II, Nagoya City University Medical School
between 1997 and 2009. We also investigated the EGFR,
erbB2 (n=249) and Kras mutation status for the same
patient group. All tumor samples were immediately frozen and stored
at −80°C until assayed. The clinical and pathological
characteristics of the 305 lung cancer patients are as follows: 180
cases at stage I, 51 at stage II and 74 at stage III–IV. The mean
age was 66.1 years (range 39–88). Among the 305 lung cancer
patients, 261 (85.6%) were diagnosed as adenocarcinoma and 38
(12.5%) were squamous cell carcinoma. The study was approved by the
institutional ethics board and written
PCR assays for Braf and erbB2
Total RNA was extracted from lung cancer tissues and
adjacent non-malignant lung tissues using the Isogen kit (Nippon
Gene, Tokyo, Japan) according to the manufacturer’s instructions.
RNA concentration was determined by spectrophotometer and adjusted
to a concentration of 200 ng/ml. Approximately 20 cases were
excluded since tumor cells were too few to sufficiently extract
tumor RNA. RNA (1 μg) was reverse-transcribed by Superscript
II enzyme (Gibco BRL, Gaithersburg, MD, USA) with 0.5 μg
oligo(dT)12–16 (Amersham Pharmacia Biotech Inc., Piscataway, NJ,
USA). The reaction mixture was incubated at 42°C for 50 min and
then at 72°C for 15 min. We then used 1 μl of each DNA for
PCR analyses. The PCR reactions were performed using the LA-Taq kit
(Takara Bio Inc., Shiga, Japan) in a 25-μl reaction volume.
The primer sequences for the Braf gene for the kinase domain
(exon 11–15) were as follows: the forward primer,
5-GACGGGACTCGAGTGATGAT-3 and the reverse primer,
5-CCACAAATGGATCCAGACA-3 (532 bp). The cycling conditions were as
follows: initial denaturation at 94°C for 5 min, followed by 35
cycles at 94°C for 40 sec, 60°C for 40 sec and 72°C for 45 sec. The
primer sequences for erbB2 gene for kinase domain (exon
19–22) were as follows: the forward primer,
5-CGCTTTTGGCACAGTCTACA-3 and the reverse primer,
5-GGGATCCCATCGTAAGGTTT-3 (594 bp). The cycling conditions were as
follows: initial denaturation at 94°C for 5 min, followed by 35
cycles at 94°C for 40 sec, 60°C for 40 sec and 72°C for 45 sec. The
products were purified by the Qiagen PCR purification kit (Qiagen,
Valencia, CA, USA). Amplified cDNAs were separated on 1% agarose
gels, and the bands were visualized by ethidium bromide and
photographed under ultraviolet transillumination. These samples
were sequenced using the ABI Prism 3100 analyzer (Applied
Biosystems Japan Ltd., Tokyo, Japan) and analyzed by BLAST and
chromatograms by manual review from forward and reverse, both side.
EGFR and Kras sequencing methods were previously
submitted elsewhere (12,13,16,24).
Results
Braf gene mutation status in Japanese
lung cancer patients
Of the 305 patients, 93 had EGFR mutations
within the kinase domain, including 45 exon 19 deletions, 41 L858R,
5 exon 20 insertion, 3 G719S and 2 L861Q. Twenty-two had
Kras mutations at codon 12 or 13. Braf mutation was
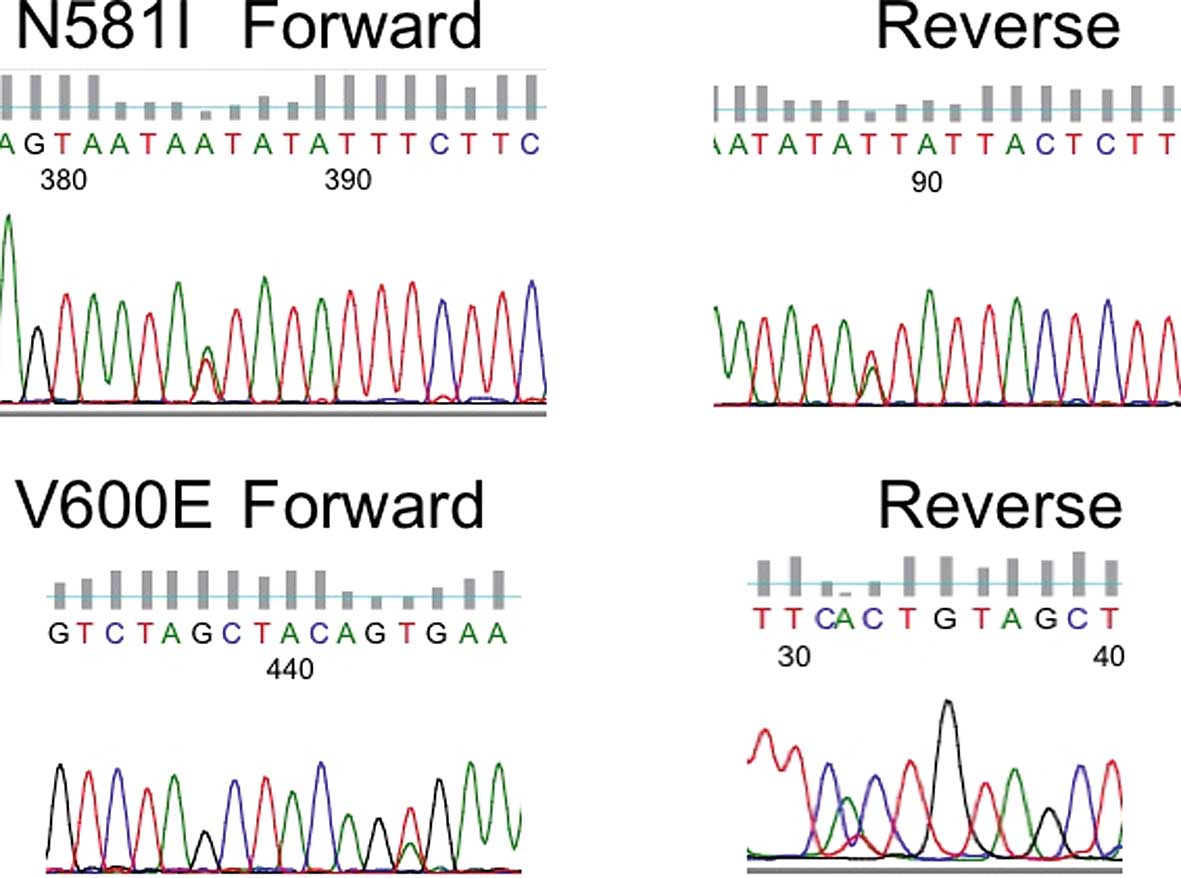
found in 6 (1.96%) of the 305 NSCLC patients. Reverse sequence
reading was also confirmed (Fig.
1). Five consisted of V600E (1799 thymine to adenine; valine to
glutamic acid) and one was N581I (1742 adenine to thymine;
asparagine to isoleucine). Four were male and 2 were female. Five
were smokers and 1 was a never smoker. All were adenocarcinoma
cases. Within the total cohort, there was no significant difference
in the Braf mutation rate according to gender (p= 0.8234),
age (p=0.5050), pathological stage (p=0.7004) or smoking status
(never smoker vs. smoker; p=0.6195). However, in the adenocarcinoma
cases, heavy-smoker group (Brinkman index >400) had a
significantly higher Braf mutation rate than in the
light-smoker or never-smoker group (p<0.05). There was no
significant difference in the Braf mutation rate according
to gender (p=0.6449), age (p=0.4665) and pathological stage
(p=0.7704) among the adenocarcinoma cases (Table I).
 | Table I.Clinicopathological data of the 261
lung adenocarcinoma patients. |
Table I.
Clinicopathological data of the 261
lung adenocarcinoma patients.
| Factors | Braf gene
status
| p-value |
|---|
| Mutant patients
(n=6) | Wild-type patients
(n=255) |
|---|
| Mean age (years;
65.8±9.0) | 69.0±8.7 | 65.7±9.0 | 0.4665 |
| Stage | | | 0.7704 |
| I | 4 (2.5%) | 155 (97.5%) | |
| II–IV | 2 (2.0%) | 100 (98.0%) | |
| Lymph node
metastasis | | | 0.8182 |
| N0 | 4 (2.2%) | 181 (97.8%) | |
| N+ | 2 (2.6%) | 74 (97.4%) | |
| Smoking status | | | 0.0476 |
| BI ≤400 | 1 (0.7%) | 146 (99.3%) | |
| BI >400 | 5 (4.4%) | 109 (95.3%) | |
| EGFR
mutation | | | 0.0630 |
| Mutation | 0 (0%) | 94 (100%) | |
| Wild-type | 6 (3.6%) | 161 (96.7%) | |
| Kras
mutation | | | 0.4521 |
| Mutation | 0 (0%) | 22 (100%) | |
| Wild-type | 6 (2.5%) | 233 (97.5%) | |
| Age (years) | | | 0.5054 |
| ≤65 | 2 (1.6%) | 120 (98.4%) | |
| >65 | 4 (2.9%) | 135 (97.1%) | |
| Gender | | | 0.6449 |
| Male | 4 (2.7%) | 109 (97.3%) | |
| Female | 2 (1.8%) | 146 (98.2%) | |
erbB2 gene mutation status in Japanese
lung cancer patients
We also sequenced the kinase domain of erbB2
for 248 NSCLC patients. Among the 248 patients, 5 (2%) had
erbB2 mutations. All mutations were at exon 20. Three were
female. All were non-smokers. Four had a 12 amino acid insertion
mutation (2324–2325 ins ATACGTGATGGC), located in exon 20 at the
kinase domain (775–776 ins YVMA). One had an amino acid insertion
mutation (2326 G to TTGT) located in the exon 20 at kinase domain
(776 glycine to leucine plus cysteine). Reverse sequence for the
erbB2 gene was also confirmed. Never smokers had a
significantly higher erbB2 mutation rate than the smokers
(p=0.0052). There was no significant difference in the erbB2
mutation rate according to gender (p=0.2471), age (p=0.5142) and
pathological stage (p=0.3863) (Table
II).
 | Table II.Clinicopathological data of 248 NSCLC
patients. |
Table II.
Clinicopathological data of 248 NSCLC
patients.
| Factors | erbB2 gene
status
| p-value |
|---|
| Mutant patients
(n=5) | Wild-type patients
(n=243) |
|---|
| Mean age (years;
65.7±8.7) | 68.0±5.0 | 65.7±8.8 | 0.5142 |
| Stage | | | 0.3863 |
| I | 2 (1.4%) | 144 (98.6%) | |
| II–IV | 3 (2.9%) | 99 (97.1%) | |
| Lymph node
metastasis | | | 0.7083 |
| N0 | 3 (1.8%) | 165 (98.2%) | |
| N+ | 2 (2.5%) | 78 (97.5%) | |
| Smoking status | | | 0.0052 |
| Never smoker | 5 (5.1%) | 93 (94.9%) | |
| Smoker | 0 (0%) | 150 (100%) | |
| EGFR
mutation | | | 0.1828 |
| Mutation | 0 (0%) | 64 (100%) | |
| Wild-type | 5 (2.7%) | 179 (97.3%) | |
| Pathology | | | 0.3846 |
|
Adenocarcinoma | 5 (2.3%) | 211 (97.7%) | |
|
Non-adenocarcinoma | 0 (0%) | 32 (100%) | |
| Age (years) | | | 0.7046 |
| ≤65 | 2 (1.7%) | 118 (98.3%) | |
| >65 | 3 (2.3%) | 125 (97.7%) | |
| Gender | | | 0.2471 |
| Male | 2 (1.3%) | 158 (98.7%) | |
| Female | 3 (3.4%) | 85 (96.6%) | |
Within these NSCLCs, all four gene (EGFR,
erbB2, Kras and Braf) mutations existed
exclusively. All patients with Braf or erbB2
mutations were alive at this point and we did not perform survival
analysis (data not shown).
Discussion
In the present study, we found six Braf
mutations in 305 Japanese NSCLC cases. The Braf mutation was
exclusively found without EGFR or erbB2 mutations.
Braf mutations were predominantly found in heavy smokers
among the adenocarcinoma cases. This population was also thought to
have a higher incidence of Kras gene mutations (10,24).
On the other hand, in our analysis, erbB2 gene mutations
were predominantly found in non-smokers with adenocarcinomas.
The v-raf murine sarcoma viral oncogene homolog B1
(Braf) encodes a serine/threonine kinase that acts in the
MAP kinase pathway, through both receptor tyrosine kinases and
G-protein coupled receptors (5).
Mutations in Braf were first reported in melanomas and
colorectal cancers, but have since been reported in a variety of
solid tumors (6,7), including stage I lung adenocarcinomas
(7). Activating Braf
mutations, especially common mutant V600E, induce constitutive
activation of the signal transduction pathway, providing a potent
promitogenic force that drives malignant transformation (6). The Braf V600E mutant showed
greatly increased activity in the Raf/MEK/Erk pathway both in
vitro and in vivo (6,25).
An inducible transgenic mouse model of Braf V600E developed
by Ji et al (26)
demonstrated that mutant Braf was sufficient for the
development of lung adenocarcinomas. Since the incidence of
Braf mutations is highest in melanomas, the bulk of the
clinical trials to date have focused on this disease, targeting
either BRAF itself or MEK 1/2, the latter of which is associated
with growth-dependency in Braf mutant cell lines (27,28).
The most promising of specific agents has been PLX4032, which was
associated with an 80% response rate in the extension phase of a
recent multicenter phase I study that included 32 patients with
advanced stage melanoma with Braf mutations (29).
Mutations in the erbB2 gene were found in
approximately 2% of primary NSCLCs, predominantly in never
smoker-like EGFR mutations (12–16).
It has been shown that the common erbB2 mutation, A775 ins
YVMA, led to oncogenic transformation in a cellular assay (30). The mutations target residues that
are highly conserved in the erbB family, and are homologous to the
exon 20 insertion mutations of EGFR. Murine cells
transformed with the mutant were relatively resistant to the
reversible EGFR inhibitor, resembling the resistant phenotype found
in cells carrying the homologous mutations in exon 20 of
EGFR (31,32). However, the mutant cells exhibited
high sensitivity to the irreversible dual-specificity EGFR/ERBB2
kinase inhibitor HKI-272 (30).
In our analysis, Braf or erbB2
mutations were only found in the adenocarcinomas, but not in the
squamous cell carcinoma cases. It was also suggested that
Braf mutations in Japanese NSCLC patients are not common and
have an even lower frequency than that in US patients (6,7), or
in in vitro analysis from lung cancer cell lines (11%)
(5). Our data showed that
mutations of the Braf or erbB2 gene as a mechanism of
tumorigenesis are unlikely to be associated with many cases of
Japanese NSCLCs. Despite the promise of anti-BRAF therapy or
irreversible erbB inhibitors, our findings indicate that a small
percentage of Japanese NSCLC patients actually harbor the
Braf or erbB2 mutation and, in turn, a few patients
with these tumors may likely benefit from these anticancer agents.
However, completely exclusive EGFR, erbB2 and
Braf mutation status would help us to choose individualized
molecular targeting therapy for NSCLC. Further studies are required
to confirm the mechanisms of Braf and erbB2 mutations
involved in the sensitivity or resistance of targeted therapy for
lung cancer.
Acknowledgements
This study was supported by
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (JSPS) (nos. 23659674, 21390394, 21591820) and
a grant for cancer research from the Program for Developing the
Supporting System for Upgrading Education and Research (2009) from
the Ministry of Education, Culture, Sports, Science and Technology
of Japan. The authors would like to thank Mrs. Akiha Kuramoto and
Miki Mochizuki for their excellent technical assistance.
References
|
1.
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signaling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Rowinsky EK: The erbB family: targets for
therapeutic development against cancer and therapeutic strategies
using monoclonal antibodies and tyrosine kinase inhibitors. Annu
Rev Med. 55:433–457. 2004. View Article : Google Scholar
|
|
3.
|
Wieduwilt MJ and Moasser MM: The epidermal
growth factor receptor family: biology driving targeted
therapeutics. Cell Mol Life Sci. 65:1566–1584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Laurent-Puig P, Lievre A and Blons H:
Mutations and response to epidermal growth factor receptor
inhibitors. Clin Cancer Res. 15:1133–1139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Peyssonnaux C and Eychene A: The
Raf/MEK/ERK pathway: new concepts of activation. Biol Cell.
93:53–62. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Davies H, Bignell GR, Cox C, et al:
Mutations of the BRAF gene in human cancer. Nature. 417:949–954.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Brose MS, Voipe P, Feldman M, et al: BRAF
and RAS mutations in human lung cancer and melanoma. Cancer Res.
62:6997–7000. 2002.PubMed/NCBI
|
|
8.
|
Naoki K, Chen T-H, Richards WG, Sugarbaker
DJ and Meyerson M: Missense mutations of the BRAF gene in human
lung adenocarcinoma. Cancer Res. 62:7001–7003. 2002.PubMed/NCBI
|
|
9.
|
Hoque MO, Brait M, Rosenbaum E, et al:
Genetic and epigenetic analysis of erbB signaling pathway genes in
lung cancer. J Thorac Oncol. 5:1887–1893. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Paik PK, Arcila ME, Fara M, et al:
Clinical characteristics of patients with lung adenocarcinomas
harboring Braf mutations. J Clin Oncol. 29:2046–2051. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Lee YS, Kim MJ, Jin G, et al: Somatic
mutations in epidermal growth factor receptor signaling pathway
genes in non-small cell lung cancers. J Thorac Oncol. 5:1734–1740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Sasaki H, Shimizu S, Endo K, et al: EGFR
and erbB2 mutation status in Japanese lung cancer patients. Int J
Cancer. 118:180–184. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. New Eng J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Pao W, Miller V, Zakowski M, et al: EGF
receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and elrotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
|
|
16.
|
Sasaki H, Endo H, Konishi A, et al: EGFR
mutation status in Japanese lung cancer patients: genotyping
analysis using LightCycler. Clin Cancer Res. 11:2924–2929. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Mok TS, Wu Y-L, Thongprasert S, et al:
Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma.
New Eng J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer habouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomized phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
19.
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. New Eng J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Stephens P, Hunter C, Bignell G, et al:
Intragenic erbB2 kinase mutations in tumors. Nature. 431:525–526.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Shigematsu H, Takahashi T, Nomura M, et
al: Somatic mutations of the HER2 kinase domain in lung
adenocarcinomas. Cancer Res. 65:1642–1646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tomizawa K, Suda K, Onozato R, et al:
Prognostic and predictive implications of HER2/ERBB2/neu gene
mutations in lung cancers. Lung Cancer. 74:139–144. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Sasaki H, Kawano O, Endo K, et al:
Uncommon V599E Braf mutations in Japanese patients with lung
cancer. J Surg Res. 133:203–206. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sasaki H, Hikosaka Y, Kawano O, Moriyama
S, Yano M and Fujii Y: Evaluation of Kras mutation and copy number
gain in non-small cell lung cancer. J Thorac Oncol. 6:15–20. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ikenoue T, Hikiba Y, Kanai F, et al:
Functional analysis of mutations within the kinase activation
segment of B-Raf in human colorectal tumors. Cancer Res.
63:8132–8137. 2003.PubMed/NCBI
|
|
26.
|
Ji H, Wang Z, Perera SA, et al: Mutations
in Braf and Kras converse on activation of the mitogen-activated
protein kinase pathway in lung cancer mouse models. Cancer Res.
67:4933–4939. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Welbrock C and Huristone A: BRAF as
therapeutic target in melanoma. Biochem Pharmacol. 80:561–567.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Solit DR, Garraway LA, Pratilas CA, et al:
BRAF mutation predicts sensitivity to MEK inhibition. Nature.
439:358–362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Flaherty KT, Puzanov I, Kim KB, et al:
Inhibition of mutated activated Braf in metastatic melanoma. N Eng
J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Minami Y, Shimamura T, Shah K, et al: The
major lung cancer-derived mutants of ERBB2 are oncogenic and are
associated with sensitivity to the irreversible EGFR/ERBB2
inhibitor HKI-272. Oncogene. 26:5023–5027. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Greulich H, Chen TH, Feng W, et al:
Oncogenic transformation by inhibitor-sensitive and -resistant EGFR
mutants. PLoS Med. 2:e2132005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Sasaki H, Endo K, Takada M, et al: EGFR
exon20 insertion mutation in Japanese lung cancer. Lung Cancer.
58:324–328. 2007. View Article : Google Scholar : PubMed/NCBI
|