Introduction
Lung cancer is one of the most common malignant
tumors and the leading cause of cancer-related mortality worldwide
(1). Small-cell lung carcinoma
(SCLC) is the most aggressive subtype of all lung tumors and is
associated with poor patient survival (2). Although the investigation of known
genes and proteins has yielded much new information, previously
unknown biomarkers such as non-coding RNA gene products may also
lend insight into the biology of lung cancer. Further investigation
of the use of differential microRNA (miR) expression in the
diagnosis, prognosis and treatment of lung cancer is warranted.
miRs are a class of short, single-stranded,
endogenous and highly conserved non-coding RNAs that are involved
in numerous developmental processes and the modulation of gene
expression. Recent studies have demonstrated that various miRs,
such as let-7, miR-15 and miR-16, are downregulated in human cancer
cells and may function as tumor suppressors (3–6).
Conversely, various miRs, including miR-21, miR-31 and miR-155, are
upregulated in human cancer cells and may function as oncomirs
(oncogene miRs) (7–9). Members of the let-7 family of miRs
are expressed at an extremely low level in lung cancer (10) and act as tumor suppressors by
repressing cell proliferation and regulating multiple oncogenes,
such as RAS and C-MYC (11–14).
In addition, the C-MYC oncogene was found to regulate the
expression of the pluripotency factor LIN-28, which modulates the
process of mature let-7 (15).
Therefore, the upregulation of let-7 expression may inhibit cell
proliferation by downregulating the C-MYC and LIN-28
expression signaling pathway in lung cancer cells.
Tea, one of the most popular beverages consumed
worldwide, has received much attention due to its disease
prevention effects (16,17). Studies have shown that these
effects are attributed to the polyphenolic constituents which are
present in high amounts in green tea (18). The inhibitory effects of tea
catechins against experimental carcinogenesis have also been
demonstrated in many animal models (19–21).
In recent years, it has been reported that tea catechins induce the
modification of the expression profile of miRs and mediate the
apoptotic effect in human cancer cells (22,23).
Furthermore, let-7 is known as a tumor suppressor in lung cancer
cells or animal models by repressing cell proliferation (10,24–26)
and its expression profile may be altered by various agents
(23,27,28).
However, the effect and mechanism of tea catechins on let-7
expression in human lung cancer remain unknown. Here, we identified
let-7a-1, let-7g, C-MYC and LIN-28 expression levels
in human lung cancer cells after tea catechin treatment and found
that tea catechins repressed cell proliferation of lung cancer
cells through upregulation of let-7 and downregulation of the
C-MYC, LIN-28 signaling pathway.
Materials and methods
Cell lines and tea catechins
NCI-H446 and MSTO-211H lung cancer cells were
purchased from the Cell Resource Centre of the Shanghai Institutes
for Biological Sciences of the Chinese Academy of Sciences
(Shanghai, China) and cultured in RPMI-1640 medium (Invitrogen Life
Technologies, Carlsbad, CA, USA) containing 10% fetal bovine serum
(Hyclone, USA). These two cell lines overexpress the C-MYC
protooncogene. The tea catechins were extracted from green tea by
the All China Federation of Supply and Marketing Cooperatives
(ACFSMC) (Hangzhou, China) and the purity was 85%. The catechins
were dissolved in double-distilled H2O and stored at
−20°C for research use.
Tea catechin stimulation
Prior to tea-catechin treatment, the cells were
maintained in a humidified incubator at 37°C in 5% CO2
and the media were replaced two times/week. Logarithmically growing
lung cancer cells were harvested and seeded in 6-well plates
(1×105 cells/well) or 96-well plates (1×104
cells/well). After overnight proliferation, the adherent cells were
incubated with tea catechins at final concentrations of 0, 50, 100
and 200 μg/ml for 24–96 h. At the end of each treatment, the cells
were used for cell growth assay.
Quantitative real-time RT-PCR for miRs
and relative gene expression
Forty-eight hours post-treatment, total-RNA was
isolated from the cells using RNAiso reagent (Takara Bio, Inc.,
Shiga, Japan) according to the manufacturer’s instructions. One
microgram of total-RNA was subjected to reverse transcription
reaction. Quantitative real-time RT-PCR of miRs (let-7a-1/7g) was
performed using the Hairpin-it™ miR qPCR quantitation kit (Shanghai
GenePharma, Co., Ltd., Shanghai, China) and the SYBR-Green PCR
master mix (Takara Bio, Inc.) in the Mx3005P real-time PCR system
(Stratagene, USA) for quantitative miR detection. The reactions
were performed in 8-strip tubes (Axygen, USA) at 95°C for 2 min,
followed by 40 cycles of 95°C for 15 sec and 62°C for 1 min. Data
were analyzed by MxPro 3.2 according to the manufacturer’s
instructions. The analysis of miR-related genes (C-MYC and
LIN-28) was carried out by quantitative real-time RT-PCR.
The reactions were performed at 95°C for 2 min, followed by 40
cycles of 95°C for 20 sec and 60°C for 1 min and β-actin was
used as an internal control. The primer sequences for qPCR and
RT-PCR are shown in Table I.
 | Table I.Primer sequences for real-time
RT-PCR. |
Table I.
Primer sequences for real-time
RT-PCR.
| Genes | Forward primer
sequences (5′→3′) | Reverse primer
sequences (5′→3′) |
|---|
|
let-7a-1 |
CGATTCAGTGAGGTAGTAGGTTGT |
TATGGTTGTTCTGCTCTCTGTCTC |
| let-7g |
CGCCAGTTGAGGTAGTAGTTTGTA |
TATGGTTGTTCTGCTCTCTGTCTC |
| U6
snRNA |
CTCGGTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| C-MYC |
CCACCAGCAGCGACTCTGA |
GCAGAAGGTGATCCAGACTC |
| LIN-28 |
AGGCGGTGGAGTTCACCTTTAAGA |
AGCTTGCATTCCTTGGCATGATGG |
| β-actin |
CAGAAGGAGATTACTGCTCTGGCT |
TACTCCTGCTTGCTGATCCACATC |
Cell viability assay
At 24, 48, 72 and 96 h post-treatment using
different concentrations of tea catechins, 20 μl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT,
5 mg/ml) was added to each well, and the cells were incubated for 4
h at 37°C in 5% CO2. After incubation, the medium was
carefully removed from the plates, and 150 μl of dimethyl sulfoxide
(DMSO) was added to solubilize the formazan produced from MTT by
the viable cells. Absorbance was measured at 490 nm by using an
automatic microplate reader (Labsystems, Helsinki, Finland).
Flow cytometry
Twenty-four hours post-stimulation by tea
cate-chins, the cells were harvested and washed with cold PBS and
then fixed in 70% ice-cold ethanol for 24 h. After washing with
PBS, the cells were incubated with 1 μg/ml propidium iodide (PI)
for 30 min at room temperature before FACS Calibur system
(Becton-Dickinson, USA) analysis. The data were collected and
processed using the ModFit LT FACS analysis software.
Western blot analysis
Forty-eight hours after tea catechin treatment, the
cells were washed three times with cold PBS and lysed in ice-cold
lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100,
100 μg/ml PMSF). Thirty minutes later, the cell pellets were
collected by scraping. They were then transferred to new tubes and
centrifuged at 12,000 rpm for 15 min at 4°C. The supernatant was
used for protein quantitative analysis by the Bradford procedure
(Bio-Rad, USA) and then for western blot analysis. The proteins
were resolved on 12% SDS-polyacrylamide gels, transferred onto PVDF
membranes and incubated with appropriate antibodies according to
the manufacturer’s instructions. The rabbit polyclonal antibody of
anti-β-actin (Beijing Bioss Biotechnology, Beijing, China) was used
at a 1:500 dilution, the rabbit monoclonal anti-LIN-28 antibody
(Epitomics, Inc., USA) was used at a 1:5,000 dilution, and the
secondary antibody of IgG-HRP (Sigma, USA) was used at a 1:6,000
dilution. Color development reaction of HRP was performed using the
BeyoECL Plus system (Beyotime, Haimen, China). The Quantity One
analysis program (Bio-Rad) was used to obtain the quantitative
data.
Statistical analysis
The experimental data are expressed as the mean ± SD
of three independent experiments, and all experiments were
performed in triplicate. The statistical significance between
different groups was determined using a two-tailed Student’s
t-test. A P-value of <0.05 was considered to indicate a
statitically significant difference.
Results
let-7 expression is upregulated after tea
catechin treatment
To determine whether let-7a-1 and let-7g are
involved in the response of lung cancer cells to tea catechin
treatment, NCI-H446 and MSTO-211H cells were incubated with
different doses of tea catechins before miR analysis. The qRT-PCR
results of miRs showed that both of the mature let-7a-1 and let-7g
were upregulated from the dose of 50 μg/ml, and their induced
expression levels were markedly higher than those of the control
groups (Fig. 1A and B). Moreover,
tea catechins upregulated let-7g in a dose-dependent manner in both
cell lines (Fig. 1B). Taken
together, these results demonstrated that tea catechins upregulated
the expression of the tumor-suppressor miRs, let-7a-1 and
let-7g.
Tea catechins repress let-7 target and
regulatory gene expression
For further confirmation of the effect of tea
catechins on let-7 target and regulatory gene expression, we used
qRT-PCR to detect the let-7 target gene C-MYC and its
regulatory gene LIN-28, at the mRNA level. C-MYC and
LIN-28 mRNA expression was significantly reduced 48 h after
treatment of the NCI-H446 and MSTO-211H cells with tea catechins
(Fig. 2A and B). To identify the
effect of tea catechins on the expression of let-7 regulatory
protein LIN-28, we further detected the LIN-28 expression at the
protein level by western blot analysis and found that LIN-28
protein was decreased after tea catechin treatment (Fig. 2C and D). The results revealed that
tea catechins not only affected the let-7 target C-MYC
expression, but also repressed let-7 regulatory LIN-28
expression.
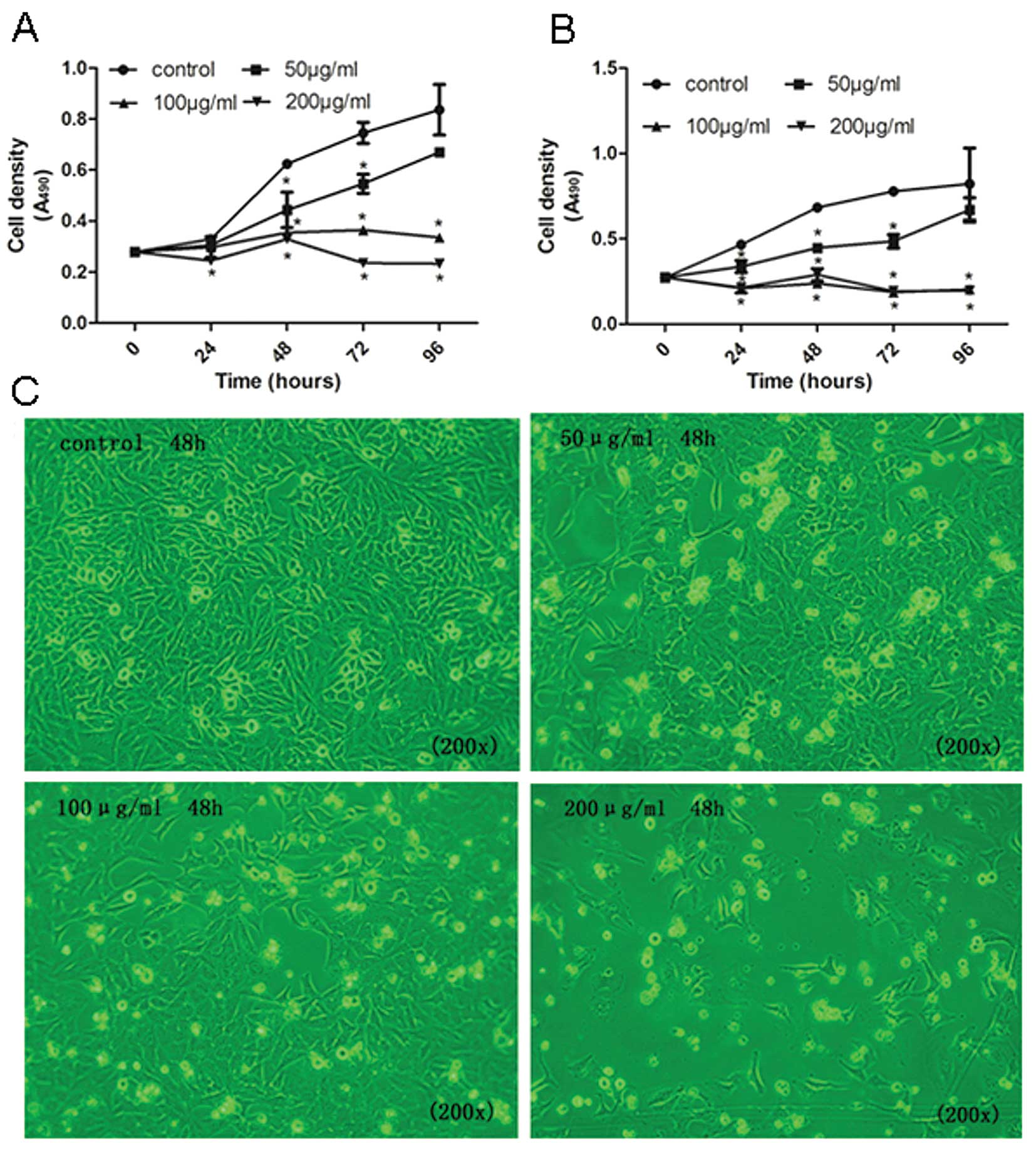
Tea catechins inhibit cell proliferation
through a let-7 signaling pathway
To determine the cancer-preventive effect of the
upregulation of let-7 by tea catechins on cell proliferation, we
investigated the effect of tea catechins on cell growth and the
cell cycle. The MTT results showed that tea catechins significantly
suppressed NCI-H446 and MSTO-211H cell proliferation (Fig. 3). Additionally, the inhibitory
effects were markedly exhibited in a dose- and time-dependent
manner. At doses of 50, 100 and 200 μg/ml of tea catechins, the
percentages of inhibition were 28.9, 43.13 and 47.25% in NCI-H446
cells, respectively (Fig. 3A) as
well as 34.72, 65.23 and 57.69% in MSTO-211H cells, respectively
(Fig. 3B). Furthermore, treatment
of the cells with tea-catechin resulted in a significant
dose-dependent decrease in cell density by suppression of NCI-H446
cell growth (Fig. 3C).
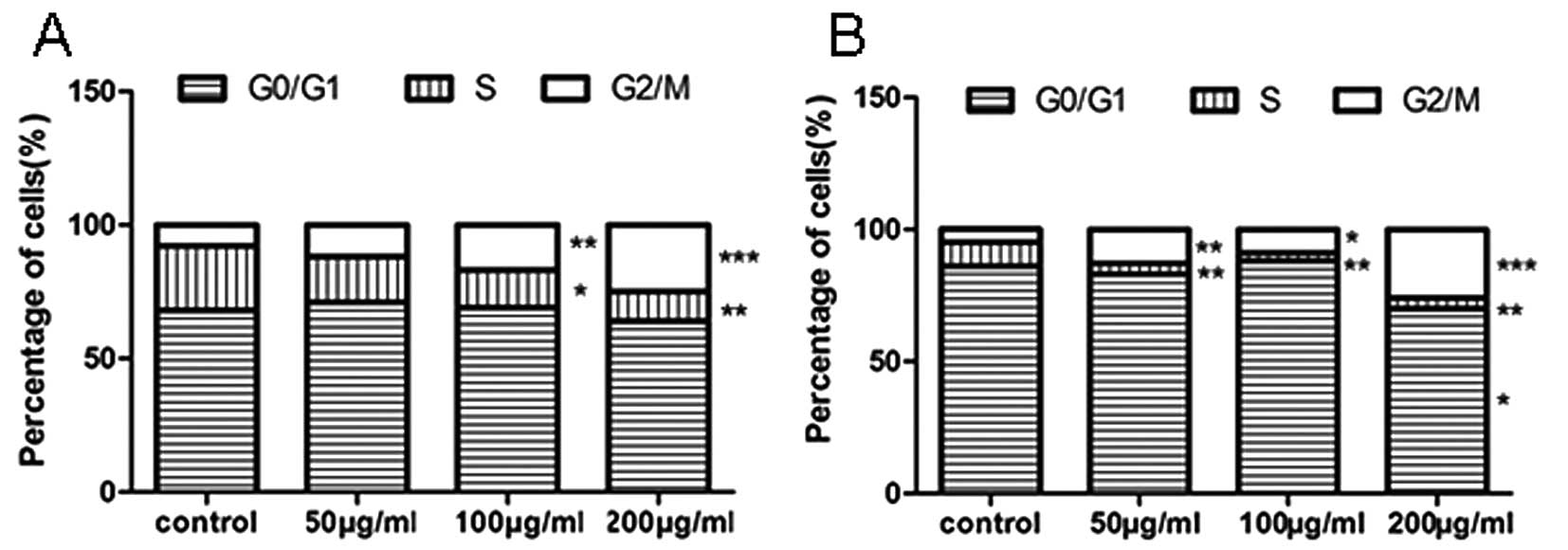
Tea catechins induce cell cycle arrest at
the G2/M phase
We further investigated whether the upregulation of
let-7 and the downregulation of the C-MYC/LIN-28 pathway by tea
catechins affect the cell cycle. The DNA content analysis of the
cell cycle was accomplished using flow cytometry. The results
showed that the cell cycle was arrested at the G2/M phase, and the
number of cells in the S phase was significantly decreased at an
appropriate dose of tea catechins in NCI-H446 and MSTO-211H lung
cancer cells (Fig. 4A and B).
These results indicate that treatment with tea catechins results in
cell cycle arrest.
Discussion
The present study reveals that tea catechins
significantly suppress cell growth by upregulating let-7 and
downregulating the C-MYC/LIN-28 signaling pathway in lung cancer
cells and mesothelioma cells which overexpress the C-MYC
protooncogene. Tea catechins were found to exhibit inhibitory
effects against the formation and development of various tumors.
More specifically, tea catechins have been reported to alter the
expression of C-MYC in lung carcinogenesis (29). Their anti-cancer effects include
suppressing cell proliferation, promoting apoptosis, modulating
signaling transduction and inhibiting cell invasion or metastasis
(30). Interestingly, tea
catechins also inhibit non-cancer (human embryonic lung fibroblast)
cell proliferation (unpublished data from our own group), which
indicates the suppression of cell growth is a non-specific effect.
The in vitro cytotoxicity of tea catechins to cancer and
normal cells has also been reported in previous studies (31,32).
Although the effects of tea catechins on inhibiting human cancer
cell proliferation have been reported in several studies (18,30),
the precise mechanism of suppressing cancer cell growth is unclear.
miRs are small non-coding RNA and are known to be important in the
regulation of numerous cellular events (33). In particular, the let-7 miR family
is an important class of cell regulatory factors of cellular
growth. Our results also showed that let-7 is a tumor-suppressor
miR and reduces C-MYC and LIN-28 expression in lung cancer cells.
Recently, it has been shown that glia dedifferentiation and retinal
regeneration are regulated through a LIN-28-dependent, let-7 miR
signaling pathway (34). Moreover,
the previous study indicates that LIN-28 functions as a negative
regulator of let-7 miRs, and C-MYC suppresses mature let-7 miR
expression through promotion of LIN-28 expression (15). Based on these studies, since C-MYC
and LIN-28 regulate let-7 expression in cell differentiation and
regeneration, here we confirmed our hypothesis that tea catechins
suppress lung cancer cell growth via upregulation of let-7; through
the inhibition of C-MYC and LIN-28 expression.
Recent studies have demonstrated that natural
agents, such as B-DIM, isoflavone, genistein and curcumin, can
alter the expression of specific miRs and thereby inhibit tumor
growth (35–37). In addition, Zhang et al
(38) reported that curcumin
down-regulates miR-186 and therefore induces cell apoptosis in lung
cancer cells. The main component of tea catechins, EGCG, was found
to upregulate miR-16 and downregulate its targeted gene
Bcl-2 to induce apoptosis in human cancer cells (23). This previous study also
demonstrated that EGCG modified the expression of various miRs in
human hepatocellular carcinoma HepG2 cells. In particular, 13 miRs
were upregulated and 48 miRs were downregulated. Among them, the
upregulated miRs included let-7a, let-7b and let-7c (23). Izzotti et al (27,28)
found that exposure to cigarette smoke alters miR expression in the
lungs of rats. Our study demonstrated that expression of the tumor
suppressors let-7a/7g was upregulated after tea catechin treatment.
The properties and functions of these miRs may contribute to the
differential anticancer effects of tea catechins. These emerging
studies suggest that various agents affect mechanisms through miRNA
pathways, and these support our finding that tea catechins alter
the expression of various miRs and their targets or expression of
related genes.
In conclusion, our study revelaed that tea catechins
modulate the expression of various miRs in lung cancer cells;
expression of let-7a-1 and let-7g was upregulated. The expression
levels of let-7 target C-MYC and regulatory protein LIN-28 were
reduced after treatment with a high dose of tea catechins. Based on
the fact that let-7 overexpression suppresses cancer cell
proliferation and tea catechins prevent cancers, our results found
that tea catechins suppress cell growth via increased expression of
let-7 miR and decreased expression of the targets, C-MYC and
LIN-28.
Acknowledgements
We are grateful to Mrs Qiong Liu and
Mrs Yanping Le for their technical assistance in flow cytometry and
microscopy. This study was supported by research grants from the
Key Scientific Research Fund of Zhejiang Provincial Education
Department (Z201119414), the Natural Science Foundation of Zhejiang
Province (Y12C060009), the Natural Science Foundation of Ningbo
(201201A6110009), Zhejiang Provincial Research Project
(2012F81G2070010), the Scientific Innovation Team Project of Ningbo
(2011B82014), the Scientific Research Foundation of Graduate School
of Ningbo University (G11JA007) and the K.C. Wong Magna Fund at
Ningbo University.
References
|
1.
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: the impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Rodriguez E and Lilenbaum RC: Small cell
lung cancer: past, present, and future. Curr Oncol Rep. 12:327–334.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Osada H and Takahashi T: let-7 and
miR-17-92: small-sized major players in lung cancer development.
Cancer Sci. 102:9–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Bandi N, Zbinden S, Gugger M, et al:
miR-15a and miR-16 are implicated in cell cycle regulation in a
Rb-dependent manner and are frequently deleted or down-regulated in
non-small cell lung cancer. Cancer Res. 69:5553–5559. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Wang QZ, Xu W, Habib N and Xu R: Potential
uses of microRNA in lung cancer diagnosis, prognosis, and therapy.
Curr Cancer Drug Targets. 9:572–594. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Cimmino A, Calin GA, Fabbri M, et al:
miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Valeri N, Gasparini P, Fabbri M, et al:
Modulation of mismatch repair and genomic stability by miR-155.
Proc Natl Acad Sci USA. 107:6982–6987. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liu X, Sempere LF, Ouyang H, et al:
MicroRNA-31 functions as an oncogenic microRNA in mouse and human
lung cancer cells by repressing specific tumor suppressors. J Clin
Invest. 120:1298–1309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
He XY, Chen JX, Zhang Z, Li CL, Peng QL
and Peng HM: The let-7a microRNA protects from growth of lung
carcinoma by suppression of k-Ras and c-Myc in nude mice. J Cancer
Res Clin Oncol. 136:1023–1028. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Wong TS, Man OY, Tsang CM, et al: MicroRNA
let-7 suppresses nasopharyngeal carcinoma cell proliferation
through downregulating c-Myc expression. J Cancer Res Clin Oncol.
137:415–422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lan FF, Wang H, Chen YC, et al: Hsa-let-7g
inhibits proliferation of hepatocellular carcinoma cells by
downregulation of c-Myc and upregulation of p16(INK4A). Int J
Cancer. 128:319–331. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Dangi-Garimella S, Yun J, Eves EM, et al:
Raf kinase inhibitory protein suppresses a metastasis signalling
cascade involving LIN28 and let-7. EMBO J. 28:347–358. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Balentine DA, Wiseman SA and Bouwens LC:
The chemistry of tea flavonoids. Crit Rev Food Sci Nutr.
37:693–704. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Dube A, Nicolazzo JA and Larson I:
Chitosan nanoparticles enhance the intestinal absorption of the
green tea catechins (+)-catechin and (-)-epigallocatechin gallate.
Eur J Pharm Sci. 41:219–225. 2010.
|
|
18.
|
Higdon JV and Frei B: Tea catechins and
polyphenols: health effects, metabolism, and antioxidant functions.
Crit Rev Food Sci Nutr. 43:89–143. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Fujiki H, Yoshizawa S, Horiuchi T, et al:
Anticarcinogenic effects of (-)-epigallocatechin gallate. Prev Med.
21:503–509. 1992. View Article : Google Scholar
|
|
20.
|
Liao S, Umekita Y, Guo J, Kokontis JM and
Hiipakka RA: Growth inhibition and regression of human prostate and
breast tumors in athymic mice by tea epigallocatechin gallate.
Cancer Lett. 96:239–243. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hirose M, Mizoguchi Y, Yaono M, Tanaka H,
Yamaguchi T and Shirai T: Effects of green tea catechins on the
progression or late promotion stage of mammary gland carcinogenesis
in female Sprague-Dawley rats pretreated with 7,12-dimethylbenz(a)
anthracene. Cancer Lett. 112:141–147. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Fix LN, Shah M, Efferth T, Farwell MA and
Zhang B: MicroRNA expression profile of MCF-7 human breast cancer
cells and the effect of green tea polyphenon-60. Cancer Genomics
Proteomics. 7:261–277. 2010.PubMed/NCBI
|
|
23.
|
Tsang WP and Kwok TT: Epigallocatechin
gallate up-regulation of miR-16 and induction of apoptosis in human
cancer cells. J Nutr Biochem. 21:140–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Esquela-Kerscher A, Trang P, Wiggins JF,
et al: The let-7 microRNA reduces tumor growth in mouse models of
lung cancer. Cell Cycle. 7:759–764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
He X, Duan C, Chen J, Ou-Yang X, Zhang Z,
Li C and Peng H: Let-7a elevates p21(WAF1) levels by targeting of
NIRF and suppresses the growth of A549 lung cancer cells. FEBS
Lett. 583:3501–3507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Izzotti A, Calin GA, Arrigo P, Steele VE,
Croce CM and De Flora S: Downregulation of microRNA expression in
the lungs of rats exposed to cigarette smoke. FASEB J. 23:806–812.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Izzotti A, Calin GA, Steele VE, Cartiglia
C, Longobardi M, Croce CM and De Flora S: Chemoprevention of
cigarette smoke-induced alterations of microRNA expression in rat
lungs. Cancer Prev Res (Phila). 3:62–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Manna S, Mukherjee S, Roy A, Das S and
Panda CK: Tea polyphenols can restrict benzo[a]pyrene-induced lung
carcinogenesis by altered expression of p53-associated genes and
H-ras, c-myc and cyclin D1. J Nutr Biochem. 20:337–349. 2009.
|
|
30.
|
Yang CS and Wang X: Green tea and cancer
prevention. Nutr Cancer. 62:931–937. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Babich H, Krupka ME, Nissim HA and
Zuckerbraun HL: Differential in vitro cytotoxicity of
(-)-epicatechin gallate (ECG) to cancer and normal cells from the
human oral cavity. Toxicol In Vitro. 19:231–242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Yang CS, Wang X, Lu G and Picinich SC:
Cancer prevention by tea: animal studies, molecular mechanisms and
human relevance. Nat Rev Cancer. 9:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Yang N, Coukos G and Zhang L: MicroRNA
epigenetic alterations in human cancer: one step forward in
diagnosis and treatment. Int J Cancer. 122:963–968. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Ramachandran R, Fausett BV and Goldman D:
Ascl1a regulates Müller glia dedifferentiation and retinal
regeneration through a Lin-28-dependent, let-7 microRNA signalling
pathway. Nat Cell Biol. 12:1101–1107. 2010.PubMed/NCBI
|
|
35.
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Sun Q, Cong R, Yan H, et al: Genistein
inhibits growth of human uveal melanoma cells and affects
microRNA-27a and target gene expression. Oncol Rep. 22:563–567.
2009.PubMed/NCBI
|
|
37.
|
Sun M, Estrov Z, Ji Y, Coombes KR, Harris
DH and Kurzrock R: Curcumin (diferuloylmethane) alters the
expression profiles of microRNAs in human pancreatic cancer cells.
Mol Cancer Ther. 7:464–473. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Zhang J, Du Y, Wu C, et al: Curcumin
promotes apoptosis in human lung adenocarcinoma cells through
miR-186* signaling pathway. Oncol Rep. 24:1217–1223.
2010. View Article : Google Scholar : PubMed/NCBI
|