Introduction
Systemic lupus erythematosus (SLE) is a prototypic
auto-immune disease, characterized by autoantibody production,
immune complex formation and multiple organ damage (1). Although the pathogenic mechanisms of
SLE are not yet fully understood, previous studies have indicated
that abnormalities of apoptosis may be involved in the development
of autoimmune disorders (2,3). SLE
patients demonstrated accelerated apoptosis of circulating
lymphocytes and delayed clearance of apoptotic cells (4). The excess of lymphocyte apoptosis and
deficient clearance of apoptotic cells may contribute to B-cell
hyperactivity and subsequent autoantibody overproduction (5,6).
Accordingly, FAS, as the major mediator of the induction of
apoptosis in activated lymphocytes, has received more attention in
SLE.
FAS (CD95/APO-1/TNFSF6), as a transmembrane receptor
among cell surface death receptors, belongs to the tumor necrosis
factor receptor (TNFR) superfamily. It is expressed on numerous
types of immune cells (7) and
plays a key role in the homeostasis of immune cells, regulation of
T lymphocytes, and elimination of infected and malignant cells.
When the FAS receptor is cross-linked, either by its natural ligand
(FASL) or by specific monoclonal antibodies, the target cell
undergoes apoptosis (8). Enhanced
or defective FAS-mediated apoptosis may result in an impaired
clearance of apoptotic cells or failure to eliminate autoreactive
cells (9), which is one of the
susceptibility factors of SLE development.
The human FAS gene, which is mapped to chromosome
10q24.1, consists of nine exons and eight introns (10). Previously, two single nucleotide
polymorphisms (SNPs) (−1377G>A, rs2234767 and −670A>G,
rs1800682) located in the promoter region of the FAS gene have been
examined for relevance in a number of autoimmune diseases including
SLE (11–16). These findings were not always
consistent and a number of issues are controversial. Until now,
little is known with regard to the correlation between SNPs in the
FAS gene and SLE susceptibility in the Chinese population. In the
present study, we investigated the association between the two SNPs
in the FAS promoter and SLE susceptibility in the Chinese
population. We also performed a meta-analysis on all eligible
published case-control studies including the present study, to
assess the association.
Materials and methods
Case-control study
Patients and healthy controls
A total of 552 patients with SLE were recruited from
Anhui Provincial Hospital and The First Affiliated Hospital of
Anhui Medical University. All patients fulfilled the 1997 revised
criteria of the American College of Rheumatology for the
classification of SLE (17). Their
mean age was 37.53±12.34 years. A total of 718 healthy blood donors
were included as controls, all of whom did not have SLE or other
autoimmune diseases. Their mean age was 36.54±16.70 years. All
patients and controls were ethnic Han Chinese. Patient written
consent was provided in order to participate.
DNA samples and genotyping
EDTA anti-coagulated venous blood samples were
obtained from all the participants. Genomic DNA was extracted from
peripheral blood lymphocytes by standard procedures using Flexi
Gene DNA kits (Qiagen, Hilden, Germany). All samples were stored at
−80°C prior to testing. A total of two SNPs were genotyped by the
TaqMan SNP Genotyping Assay kit (Applied Biosystems, Foster City,
CA, USA; catalogue nos. C_12123966_10 for rs2234767, C_9578811_10
for rs1800682). Real-time polymerase chain reaction (PCR) was
performed on the ABI PRISM 7300 (Applied Biosystems, Foster City,
CA, USA). The reaction conditions were initially denatured at 95°C
for 10 min followed by 45 cycles of denaturing at 95°C for 15 sec
and annealing/extension at 60°C for 1 min.
Statistical analysis
Data analysis was performed using SPSS 10.0 software
(SPSS Inc.; 2000). The Chi-square test or Fisher's exact test was
used to estimate the Hardy-Weinberg equilibrium (HWE) in both SLE
patients and healthy controls, as well as to compare the genotype
and allele frequencies between the two groups. Odds ratios (ORs)
and 95% confidence intervals (CIs) were also calculated. The
multiple-locus haplotypes comprising the two SNPs were estimated by
comparing the difference in the haplotype frequencies for the
overall subjects with the SHEsis software (18). In the two-tailed test a probability
level of <0.05 was considered to indicate a statistically
significant result. The power analysis was performed using the
statistical program G*Power (http://www.psycho.uniduesseldorf.de/aap/projects/gpower).
Meta-analysis
Identification of eligible
studies
All studies examining the association between FAS
polymorphism and SLE were fully considered and carefully selected.
Three electronic databases (PubMed, Embase and Web of Science) were
searched using the search terms ‘FAS’, ‘TNFSF6’, ‘CD95’, ‘APO-1’,
‘systemic lupus erythematosus’ and ‘SLE’. The most recent search
was updated in March 2012. We only recruited data from fully
published papers, not meeting or conference abstracts. The
inclusion criteria were as follows; i) independent case-control
design, ii) available allele frequency or genotype distribution
data and iii) sufficient published data for estimating OR with 95%
CI.
Meta-analysis methods
Meta-analysis was performed using the STATA software
package v.7.0 (Stata Corporation, College Station, TX, USA).
Cochran's Q-statistic and inconsistency (I2) values were
presented to assess the heterogeneity (19). When a significant statistic
(P<0.10 or I2>50%) indicated heterogeneity across
studies, the random effects model was used, and when heterogeneity
was not indicated across studies, the fixed effects model was used.
The Z test was used to determine the significance of overall effect
(20,21). Begg's funnel plot and Egger's test
were performed to analyze the publication bias (22). P<0.05 was considered to indicate
a statistically significant result.
Results
Case-control study
Association between FAS polymorphisms
and SLE
The allelic and genotypic frequencies of two SNPs in
the FAS gene in patients and controls are listed in Table I. Case-control comparison revealed
a significant association between SLE and the minor allele A at SNP
rs2234767 (P=0.033, OR=0.836, 95% CI, 0.709–0.986). Significant
differences in genotypic distribution were also observed in SLE
patients and controls (AG vs. GG, P=0.041, OR=0.779, 95% CI,
0.613–0.990; AG+AA vs. GG, P=0.024, OR=0.773, 95% CI, 0.618–0.967).
However, no significant differences were demonstrated in the
allelic and genotypic distributions at SNP rs1800682 between
patients and controls.
 | Table IAssociation between FAS polymorphisms
and SLE. |
Table I
Association between FAS polymorphisms
and SLE.
| Polymorphism | SLE patients n
(%) | Controls n (%) | P-value | OR (95% CI) |
|---|
| rs2234767 | | | | |
| Allele | | | | |
| G | 737 (66.8) | 900 (62.7) | | - |
| A | 367 (33.2) | 536 (37.3) | 0.033 | 0.836
(0.709–0.986) |
| Genotype | | | | |
| GG | 257 (46.6) | 289 (40.3) | | - |
| AG | 223 (40.4) | 322 (44.8) | 0.041 | 0.779
(0.613–0.990) |
| AA | 72 (13.0) | 107 (14.9) | 0.110 | 0.757
(0.537–1.066) |
| AG+AA | 295 (53.4) | 429 (59.7) | 0.024 | 0.773
(0.618–0.967) |
| rs1800682 | | | | |
| Allele | | | | |
| A | 675 (61.1) | 834 (58.1) | | - |
| G | 429 (38.9) | 602 (41.9) | 0.119 | 0.880
(0.750–1.033) |
| Genotype | | | | |
| AA | 219 (39.7) | 254 (35.4) | | - |
| AG | 237 (42.9) | 326 (45.4) | 0.175 | 0.843
(0.659–1.079) |
| GG | 96 (17.4) | 138 (19.2) | 0.184 | 0.807
(0.588–1.108) |
| AG+GG | 333 (60.3) | 464 (64.6) | 0.116 | 0.832
(0.662–1.047) |
Haplotype analysis
The association between the frequencies of
haplotypes and SLE was also estimated. A total of four (22) possible haplotypes (AG, GA, GG and
AA) were constructed from two SNPs. Analysis of the haplotypes
revealed that the haplotype GA was significantly associated with
SLE (P=0.039, OR=1.184, 95% CI, 1.009–1.391; Table II). Thus, the GA haplotype appeared
to indicate an increased risk of SLE. The HWE P-value in controls
was >0.05. Strong linkage disequilibrium (LD) was observed
between the two SNPs (D'=0.937, r2=0.708). We estimated
that our study has a statistical power of 94.6% by the statistical
program G*Power.
 | Table IIHaplotype analysis of FAS
polymorphisms in SLE patients and controls. |
Table II
Haplotype analysis of FAS
polymorphisms in SLE patients and controls.
| Haplotype | SLE patients n
(%) | Controls n (%) | χ2 | P-value | OR (95% CI) |
|---|
|
rs2234767-rs1800682 | | | | | |
| AG | 367 (33.2) | 502 (34.9) | 1.771 | 0.183 | 0.893
(0.757–1.055) |
| GA | 675 (61.1) | 800 (55.7) | 4.266 | 0.039 | 1.184
(1.009–1.391) |
| GG | 62 (5.6) | 100 (7.0) | 2.421 | 0.120 | 0.772
(0.557–1.070) |
| AA | 0 (0.0) | 34 (2.4) | - | - | - |
| Global | 1104 | 1436 | 5.176 | 0.075 | - |
Meta-analysis
Studies included in the
meta-analysis
In the meta-analysis, four studies (12,13,16)
were identified with regard to the SNP rs2234767 (three published
articles and the present study). These studies were included and up
to 996 patients with SLE and 1160 controls were combined. A total
of six studies (11–15) were identified with regard to the
SNP rs1800682 (five published articles and one current study). In
total 1,122 patients with SLE and 1,339 controls were recruited in
this meta-analysis. The pooled population originated from Asia and
Europe.
Meta-analysis of the FAS polymorphisms
and SLE susceptibility
A summary of the meta-analysis findings on the
association between the FAS polymorphisms and SLE is provided in
Table III. The meta-analysis
showed that the two SNPs were significantly associated with SLE
susceptibility rs2234767 A vs. G allele; P=0.004, OR=0.819, 95% CI,
0.715–0.938; rs1800682 G vs. A allele; P=0.034, OR=0.791, 95% CI,
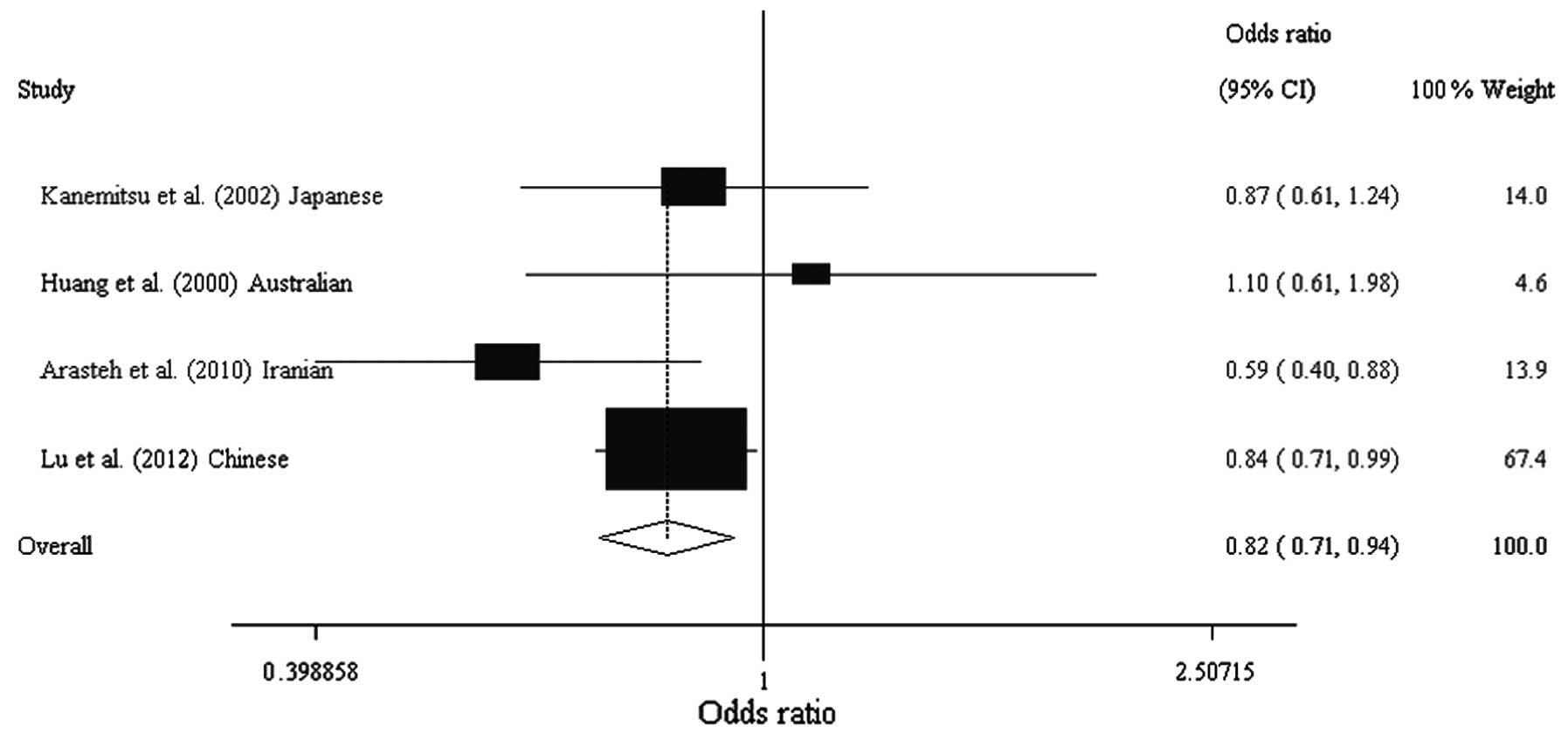
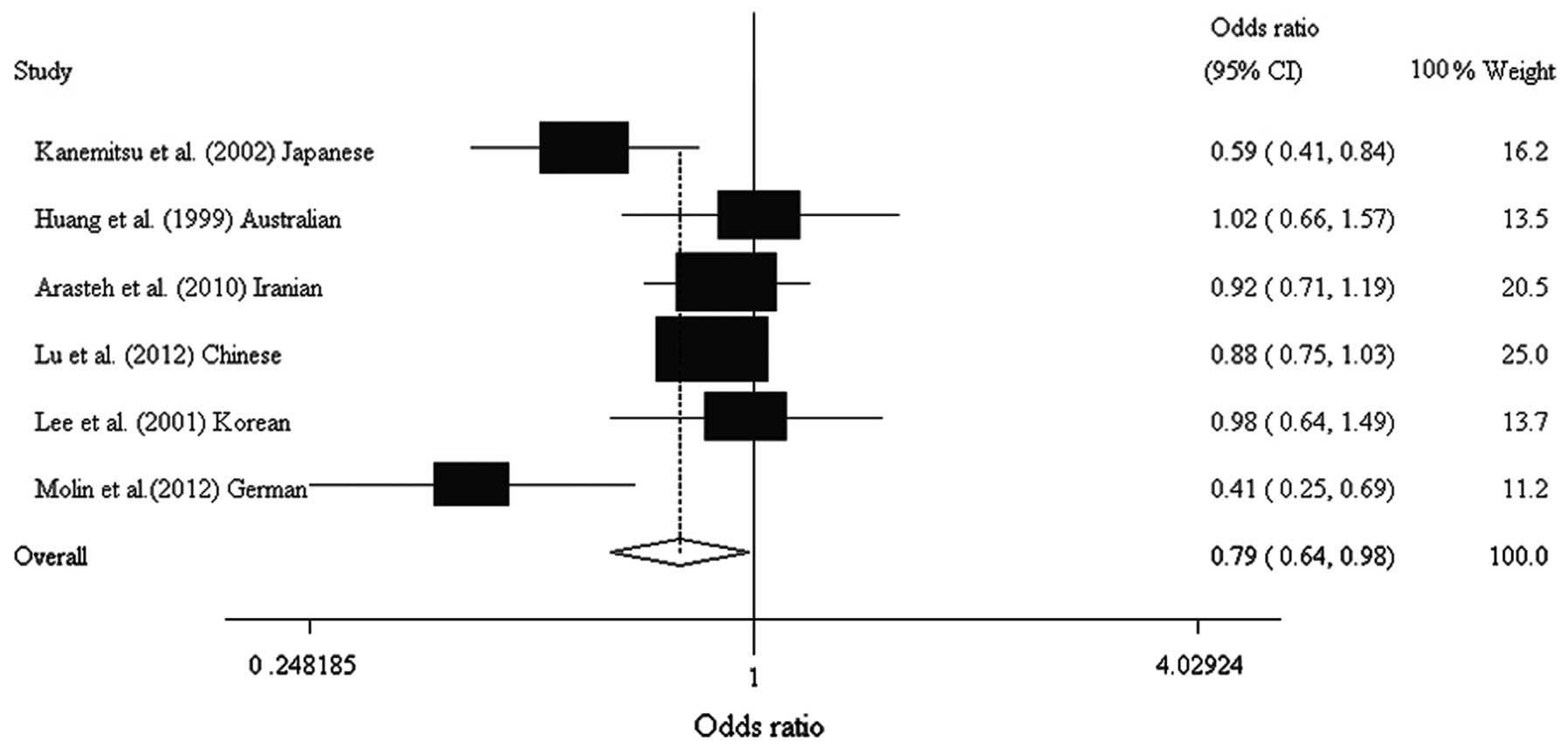
0.637–0.983; Table IV, Figs. 1 and 2). Analysis following stratification by
population indicated that the two SNPs were significantly
associated with SLE in Asian populations (rs2234767 A vs. G allele;
P=0.002, OR=0.805, 95% CI, 0.700–0.926; rs1800682 G vs. A allele;
P=0.012, OR=0.855, 95% CI, 0.758–0.966). The meta-analysis also
showed that the dominant effects of rs2234767 and rs1800682 were
associated with the susceptibility to SLE in overall (AG+AA vs. GG;
P=0.002, OR=0.751, 95% CI, 0.628–0.898; AG+GG vs. AA; P=0.015,
OR=0.646, 95% CI, 0.454–0.920, respectively) and Asian populations
(AG+AA vs. GG; P=0.001, OR=0.730, 95% CI, 0.606–0.880; AG+GG vs.
AA; P=0.034, OR=0.711, 95% CI, 0.518–0.975, respectively; Table IV).
 | Table IIICharacteristics of the individual
studies included in the meta-analysis. |
Table III
Characteristics of the individual
studies included in the meta-analysis.
| | | Patients (n)
| MAF
| | |
|---|
| Study (Ref.) | Nationality | Ethnicity | SLE | Control | SLE | Control | HWE | OR (95% CI) |
|---|
| rs2234767 | | | | | | | | |
| Kanemitsu et
al (13) | Japanese | Asian | 109 | 140 | 0.420 | 0.460 | 0.202 | 0.867
(0.607–1.239) |
| Huang et
al (16) | Australian | European
origin | 86 | 90 | 0.160 | 0.130 | 0.917 | 1.103
(0.615–1.978) |
| Arasteh et
al (12) | Iranian | Asian | 249 | 212 | 0.098 | 0.156 | 0.652 | 0.592
(0.399–0.879) |
| Lu et al
(Present study) | Chinese | Asian | 552 | 718 | 0.332 | 0.373 | 0.266 | 0.836
(0.709–0.986) |
| Total | | | 996 | 1160 | 0.269 | 0.326 | | 0.819
(0.715–0.938) |
| rs1800682 | | | | | | | | |
| Kanemitsu et
al (13) | Japanese | Asian | 109 | 140 | 0.450 | 0.590 | 0.094 | 0.588
(0.412–0.841) |
| Huang et
al (15) | Australian | European
origin | 79 | 86 | 0.490 | 0.490 | 0.825 | 1.021
(0.663–1.573) |
| Arasteh et
al (12) | Iranian | Asian | 249 | 212 | 0.484 | 0.505 | 0.272 | 0.920
(0.710–1.192) |
| Lu et al
(Present study) | Chinese | Asian | 552 | 718 | 0.389 | 0.419 | 0.070 | 0.880
(0.750–1.033) |
| Lee et al
(14) | Korean | Asian | 87 | 87 | 0.420 | 0.430 | 0.230 | 0.977
(0.638–1.495) |
| Molin et
al (11) | German | European
origin | 46 | 96 | 0.402 | 0.620 | 0.210 | 0.413
(0.248–0.686) |
| Total | | | 1122 | 1339 | 0.466 | 0.509 | | 0.791
(0.637–0.983) |
 | Table IVMeta-analysis of the association
between FAS polymorphisms and SLE. |
Table IV
Meta-analysis of the association
between FAS polymorphisms and SLE.
| | | Test of association
| Test of
heterogeneity
| |
|---|
| SNPs | Population | No. of studies | OR | 95% CI | P-value | I2 | P-value | Model | Publication bias
P-value |
|---|
| rs2234767 | | | | | | | | | |
| A vs. G
allele | Overall | 4 | 0.819 | 0.715–0.938 | 0.004 | 20.1 | 0.289 | F | 0.969 |
| Asian | 3 | 0.805 | 0.700–0.926 | 0.002 | 25.9 | 0.259 | F | 0.586 |
| European
origin | 1 | 1.103 | 0.615–1.978 | 0.742 | NA | NA | NA | NA |
| AG+AA vs. GG
(Dominant) | Overall | 4 | 0.751 | 0.628–0.898 | 0.002 | 0.0 | 0.462 | F | 0.917 |
| Asian | 3 | 0.730 | 0.606–0.880 | 0.001 | 0.0 | 0.490 | F | 0.587 |
| European
origin | 1 | 1.065 | 0.548–2.067 | 0.853 | NA | NA | NA | NA |
| AA vs. GG+AG
(Recessive) | Overall | 4 | 0.865 | 0.659–1.137 | 0.300 | 0.0 | 0.659 | F | 0.940 |
| Asian | 3 | 0.853 | 0.647–1.125 | 0.260 | 0.0 | 0.559 | F | 0.536 |
| European
origin | 1 | 1.590 | 0.259–9.758 | 0.616 | NA | NA | NA | NA |
| rs1800682 | | | | | | | | | |
| G vs. A
allele | Overall | 6 | 0.791 | 0.637–0.983 | 0.034 | 62.6 | 0.020 | R | 0.384 |
| Asian | 4 | 0.855 | 0.758–0.966 | 0.012 | 40.2 | 0.171 | F | 0.670 |
| European
origin | 2 | 0.656 | 0.270–1.594 | 0.352 | 85.9 | 0.008 | R | NA |
| AG+GG vs. AA
(Dominant) | Overall | 6 | 0.646 | 0.454–0.920 | 0.015 | 65.6 | 0.013 | R | 0.256 |
| Asian | 4 | 0.711 | 0.518–0.975 | 0.034 | 53.9 | 0.089 | R | 0.489 |
| European
origin | 2 | 0.469 | 0.112–1.958 | 0.299 | 84.9 | 0.010 | R | NA |
| GG vs. AA+AG
(Recessive) | Overall | 6 | 0.889 | 0.732–1.079 | 0.234 | 15.6 | 0.314 | F | 0.512 |
| Asian | 4 | 0.921 | 0.747–1.134 | 0.438 | 0.0 | 0.608 | F | 0.970 |
| European
origin | 2 | 0.675 | 0.237–1.926 | 0.463 | 71.3 | 0.062 | R | NA |
Evaluation of study quality and
heterogeneity
The distribution of rs2234767 and rs1800682
genotypes in control groups was consistent with the HWE in all
studies (Table III). We evaluated
that the meta-analysis had statistical powers of 99.6 and 99.9% for
rs2234767 and rs1800682, respectively, using the program
G*Power. Heterogeneity was demonstrated in the
meta-analysis for the rs1800682 but not for the rs2234767
polymorphism. Egger's regression test showed no evidence of
publication bias in this meta-analysis of FAS polymorphisms in any
of the studies included (Egger's regression test P>0.1; Table IV).
Discussion
Evidence has accumulated that FAS-mediated apoptosis
may be involved in the pathogenesis of SLE. The expression of FAS
on peripheral blood lymphocytes has been reported to be upregulated
in patients with SLE (23). The
variation of the FAS gene in the promoter region was described to
be associated with the differential expression of the gene in SLE
(24). The SNPs rs2234767 and
rs1800682 are located in the promoter of the FAS gene; the two SNPs
have been shown to interfere with the SP1 and STAT1 transcription
factor binding sites, respectively, affecting promoter activity and
in turn FAS gene expression (24,25).
Thus, based on the function of the two SNPs, variation may have an
effect on susceptibility to SLE.
In our case-control study, we analyzed the
frequencies of alleles and genotypes at the two SNPs in 552 SLE
patients and 718 healthy controls of the Chinese population. We
showed that the A allele frequency of rs2234767 is associated with
SLE susceptibility. Significant differences in the genotype
frequencies of AG vs. GG, AG+AA vs. GG at rs2234767 were observed.
However, on evaluation of allele and genotype distributions at
rs1800682, no significant differences were observed between
patients and controls. Analysis of the haplotypes revealed that
individuals carrying the haplotype GA had an increased risk of SLE.
The results of the haplotype analysis were concordant with the
suggestion that the rs2234767 G allele was mainly associated with
the rs1800682 A allele in SLE patients by Kanemitsu et al
(13). They observed that the
rs1800682 A allele with higher STAT1 binding activity may result in
the alteration of FAS gene expression. Previous studies have
suggested that STAT1 binding alone cannot be equated with a
biological function and that concomitant SP1 binding may influence
transcriptional activation (26).
This complex association may explain why the GA haplotype shows an
increased risk of SLE.
The repeated investigation of the association of FAS
polymorphisms with SLE in different populations supports the
involvement of this gene in SLE susceptibility. However, certain
conclusions are inconsistent. For example, studies by Molin et
al (11) and Kanemitsu et
al (13) demonstrated an
association between rs1800682 and SLE, however, the association was
not identified in Iranian (12),
Korean (14), Australian (15) populations and, in our present
study, in a Chinese population. In a study by Arasteh et al
(12), the SNP rs2234767 showed a
significant association with SLE, but this was not observed in
Japanese (13) and Australian
populations (16). Due to the
relatively small samples in these studies, the results were not
convincing and needed to be refined. Thereby we performed a
meta-analysis with all available studies (published and our present
results) to investigate the association between the FAS
polymorphisms and SLE. The result of our meta-analysis demonstrated
the association of the two SNPs with SLE susceptibility. Analysis
following stratification by population detected a significant
association with the FAS polymorphisms in Asian individuals. The
meta-analysis revealed a dominant effect associated with
susceptibility to SLE at the two SNPs overall and in Asian
populations.
The positive result of rs2234767 was presented by
our replication study and confirmed by meta-analysis. Although our
study did not find an association between rs1800682 and SLE in the
Chinese population, the association was identified in overall and
Asian populations in the meta-analysis. Heterogeneity was observed
in the meta-analysis for rs1800682 and SLE. One of the reasons for
this disparity may be due to population differences in genotype
distributions. In addition, we observed a strong LD between the two
SNPs in our study population, but a weaker LD was suggested in the
Japanese population (13).
Therefore the inconsistent genetic association with the two SNPs in
different populations, including the present study may be due to
different LD structure. In addition, SLE is a multi-factorial
disease; individual exposure to various environmental factors in
combination with genetic susceptibility may have contributed to the
conflicting results.
There are certain limitations of this meta-analysis
that should be discussed. First, the number of studies and the
number of subjects in the studies included in the meta-analysis by
the disease were small. Second, when we explored the association
between rs1800682 and SLE, heterogeneity was observed. Third, our
ethnicity-specific meta-analysis included data from individuals
with Asian and European origin, and thus the results are applicable
to only these groups. In the subgroup analysis, the majority of the
studies were performed in populations of Asian descent. Further
studies are therefore required in other ethnic populations. Despite
these limitations, our meta-analysis confirmed an association
between SLE and FAS gene polymorphisms.
Our investigation provided evidence that FAS gene
polymorphisms contributed to SLE susceptibility in the Chinese
population. The combined results of independent association studies
by meta-analysis showed significant association between FAS
polymorphisms and SLE. The investigation of the genetic basis of
SLE in other populations may advance the overall understanding of
the pathogenesis of this disease. Further studies are still
required in larger numbers of samples and other ethnic
populations.
Acknowledgements
This study was supported by grants
from the key program of National Natural Science Foundation of
China (30830089). We thank all participants for their participation
in this study.
References
|
1.
|
Akahoshi M, Nakashima H and Shirakawa T:
Roles of genetic variations in signaling/immunoregulatory molecules
in susceptibility to systemic lupus erythematosus. Semin Immunol.
18:224–229. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Kaplan MJ: Apoptosis in systemic lupus
erythematosus. Clin Immunol. 112:210–218. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Sheriff A, Gaipl US, Voll RE, Kalden JR
and Herrmann M: Apoptosis and systemic lupus erythematosus. Rheum
Dis Clin North Am. 30:505–527. 2004. View Article : Google Scholar
|
|
4.
|
Emlen W, Niebur J and Kadera R:
Accelerated in vitro apoptosis of lymphocytes from patients with
systemic lupus erythematosus. J Immunol. 152:3685–3692.
1994.PubMed/NCBI
|
|
5.
|
Ren Y, Tang J, Mok MY, et al: Increased
apoptotic neutrophils and macrophages and impaired macrophage
phagocytic clearance of apoptotic neutrophils in systemic lupus
erythematosus. Arthritis Rheum. 48:2888–2897. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Shoshan Y, Shapira I, Toubi E, et al:
Accelerated Fas-mediated apoptosis of monocytes and maturing
macrophages from patients with systemic lupus erythematosus:
relevance to in vitro impairment of interaction with IC3b-opsonized
apoptotic cells. J Immunol. 167:5963–5969. 2001. View Article : Google Scholar
|
|
7.
|
Leithäuser F, Dhein J, Mechtersheimer G,
et al: Constitutive and induced expression of APO-1, a new member
of the nerve growth factor/tumor necrosis factor receptor
superfamily, in normal and neoplastic cells. Lab Invest.
69:415–429. 1993.
|
|
8.
|
Papo T, Parizot C, Ortova M, et al:
Apoptosis and expression of soluble Fas mRNA in systemic lupus
erythematosus. Lupus. 7:455–461. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Navratil JS and Ahearn JM: Apoptosis and
autoimmunity: complement deficiency and systemic lupus
erythematosus revisited. Curr Rheumatol Rep. 2:32–38. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Inazawa J, Itoh N, Abe T and Nagata S:
Assignment of the human Fas antigen gene (FAS) to 10q24.1.
Genomics. 14:821–822. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Molin S, Weiss EH, Ruzicka T and Messer G:
The FAS/cd95 promoter single-nucleotide polymorphism −670 A/G and
lupus erythematosus. Clin Exp Dermatol. 37:425–427. 2012.PubMed/NCBI
|
|
12.
|
Arasteh JM, Sarvestani EK, Aflaki E and
Amirghofran Z: Fas gene polymorphisms in systemic lupus
erythematosus and serum levels of some apoptosis-related molecules.
Immunol Invest. 39:27–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kanemitsu S, Ihara K, Saifddin A, et al: A
functional polymorphism in fas (CD95/APO-1) gene promoter
associated with systemic lupus erythematosus. J Rheumatol.
29:1183–1188. 2002.PubMed/NCBI
|
|
14.
|
Lee YH, Kim YR, Ji JD, et al: Fas promoter
−670 polymorphism is associated with development of anti-RNP
antibodies in systemic lupus erythematosus. J Rheumatol.
28:2008–2011. 2001.
|
|
15.
|
Huang QR, Danis V, Lassere M, Edmonds J
and Manolios N: Evaluation of a new Apo-1/Fas promoter polymorphism
in rheumatoid arthritis and systemic lupus erythematosus patients.
Rheumatology (Oxford). 38:645–651. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Huang QR and Manolios N: Investigation of
the −1377 polymorphism on the Apo-1/Fas promoter in systemic lupus
erythematosus patients using allele-specific amplification.
Pathology. 32:126–130. 2000.
|
|
17.
|
Hochberg MC: Updating the American College
of Rheumatology revised criteria for the classification of systemic
lupus erythematosus. Arthritis Rheum. 40:17251997. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction, and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Mantel N and Haenszel W: Statistical
aspects of the analysis of data from retrospective studies of
disease. J Natl Cancer Inst. 22:719–748. 1959.PubMed/NCBI
|
|
21.
|
DerSimonian R and Laird N: Meta-analysis
in clinical trials. Control Clin Trials. 7:177–188. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Egger M, Davey Smith G, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Bijl M, Horst G, Limburg PC and Kallenberg
CG: Fas expression on peripheral blood lymphocytes in systemic
lupus erythematosus (SLE): relation to lymphocyte activation and
disease activity. Lupus. 10:866–872. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Huang QR, Morris D and Manolios N:
Identification and characterization of polymorphisms in the
promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol.
34:577–582. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Sibley K, Rollinson S, Allan JM, et al:
Functional FAS promoter polymorphisms are associated with increased
risk of acute myeloid leukemia. Cancer Res. 63:4327–4330.
2003.PubMed/NCBI
|
|
26.
|
Look DC, Pelletier MR, Tidwell RM, et al:
Stat1 depends on transcriptional synergy with Sp1. J Biol Chem.
270:30264–30267. 1995. View Article : Google Scholar : PubMed/NCBI
|