Introduction
The fate of hematopoietic stem cells is regulated by
Notch signaling. In bone marrow, Notch proteins, such as Notch1, on
hematopoietic stem cells are activated by the binding of Notch
ligands, such as Jagged1, on stromal cells (1). Notch activation is also involved in
the growth of leukemia cells (2),
particularly T-cell acute lymphoblastic leukemia (T-ALL) cells.
Gene mutations resulting in the activation of Notch1 are present in
half of T-ALL cases (3).
Previously, we reported, based on data obtained from
immunoblot analyses, that half of the samples of acute myeloid
leukemia (AML) cells express Notch1 and/or Jagged1 proteins
(4). We also demonstrated that
Notch activation induced by Notch ligand stimulation affects the
growth of AML cells (5,6). The detection of Notch and Jagged
proteins in leukemia cells aids the characterisation of leukemia
cases. However, the immunoblot analysis is not suitable for
clinical examination since it is time-consuming.
Flow cytometry (FCM) is suitable for clinical
examinations since it is currently performed as a routine
examination in hospital laboratories. The expression patterns of
Notch and Jagged proteins in normal blood cells and leukemia cells
have not been well-characterised by FCM. In the present study, we
examined the expression of Notch1 and Jagged1 proteins on the
surface of normal blood cells, normal bone marrow cells and various
leukemia cells.
Materials and methods
Cells
Four AML, four T-ALL and three B-cell lymphoma cell
lines were used for this study (Table
I). NB4 (7) was provided by Dr
M. Lanotte (Institut Universitaire d’Hématologie, Paris, France).
TMD7 and TMD8 were established in our laboratory. The T-ALL cell
lines had NOTCH1 mutations, and were provided by Dr A.
Harashima and Dr K. Orita (Fujisaki Cell Center, Okayama, Japan).
The other cell lines were supplied by the Japanese Cancer Research
Resources Bank (Tokyo, Japan).
 | Table IExpression of Notch1 and Jagged1
proteins in leukemia/lymphoma cell lines analysed by flow
cytometry. |
Table I
Expression of Notch1 and Jagged1
proteins in leukemia/lymphoma cell lines analysed by flow
cytometry.
| Lineage | Cell line (type) | Notch1a | Jagged1a |
|---|
| AML | THP1 (FAB M5) | 1,011 | 69 |
| TMD7 (FAB M2) | 3,024 | 1,114 |
| NB4 (FAB M3) | 799 | 23 |
| HL60 (FAB M2) | 1,621 | 167 |
| T-ALL | Jurkat | 445 | 249 |
| DND-41 | 252 | 13 |
| ALL-SIL | 4,319 | 0 |
| KOPT-K1 | 849 | 6 |
| B-lymphoma | TMD8 (DLBCL) | 539 | 407 |
| Daudi (BL) | 751 | 134 |
| MD901 (BL) | 226 | 422 |
Normal blood samples were obtained from 10 healthy
volunteers. Normal bone marrow cells were obtained from bone marrow
aspirates with no malignant cells from eight patients with B-cell
lymphoma in clinical stage I or II prior to treatment, with
informed consent. Blood samples from seven AML patients, five
patients with mature T-cell neoplasms and 10 patients with B-cell
chronic lymphocytic leukemia (B-CLL) were used with informed
consent. The study was approved by the Ethics Review Boards in our
University (Tokyo Medical and Dental University, Tokyo, Japan). The
profiles of the patients are shown in Table II.
 | Table IIClinical profiles of the patients. |
Table II
Clinical profiles of the patients.
| No. | Lineage | Profile in Fig. 3 | Symbol |
|---|
| 1 | AML | inv(16) | Open circle |
| 2 | | t(15;17) | Closed circle |
| 3 | |
Myelodysplasia-related changes | Open triangle |
| 4 | | FAB M0 | Closed triangle |
| 5 | | FAB M2 | Open square |
| 6 | | FAB M2 | Closed square |
| 7 | | FAB M5 | Open rhombus |
| 8 | Mature T-cell
neoplasm | T-prolymphocytic
leukemia | Open circle |
| 9 | T-prolymphocytic
leukemia | Closed circle |
| 10 | Adult T-cell
leukemia | Open triangle |
| 11 | Sézary syndrome | Closed triangle |
| 12 | Sézary syndrome | Open square |
| 13–22 | B-CLL | Typical CLL | Various |
FCM
The cells were first incubated with Fc receptor
saturation reagent (Beckman-Coulter, Brea, CA, USA) for 15 min. The
cells were then stained with phycoerythrin (PE)-labeled anti-human
Notch1 antibody (clone 527425P; R&D Systems, Minneapolis, MN,
USA), fluorescein isothiocyanate (FITC)-labeled anti-human Jagged1
antibody (clone J1G53-3; Enzo Life Science, Plymouth Meeting, PA,
USA) or each isotype control antibody. Samples were then incubated
with lysing solution (BD Biosciences, Franklin Lakes, NJ, USA),
washed with phosphate-buffered saline and analysed using a
FACSCalibur cytometer (BD Biosciences). The gates for lymphocytes,
monocytes, granulocytes, erythroblasts (in bone marrow samples) and
leukemia cells (in leukemia samples) were set in the side scatter
vs. forward scatter cytograms or the side scatter vs. CD45 PerCP
cytograms. Data were analysed using CellQuest software (BD
Biosciences). Data were presented as the percentage increase in
mean fluorescence intensity (% increase in MFI), calculated by the
equation: [(geometric MFI of relevant antibody - geometric MFI of
control)/geometric MFI of control] ×100%. The cut-off 20% increase
in MFI was used to divide into positive and negative for
convenience.
Results
Notch1 and Jagged1 expression in
leukemia/lymphoma cell lines
We first examined the expression of Notch1 and
Jagged1 proteins in 11 leukemia/lymphoma cell lines since we had
previously detected the expression of these proteins by immunoblot
analysis (4). The fluorescence
histograms from the representative cell lines are shown in Fig. 1. The percentage increase in MFI of
all the cell lines examined is shown in Table I. The cell lines examined expressed
Notch1 protein, and eight cell lines expressed Jagged1 protein. The
intensities were found to vary among the cells. Among the four
T-ALL cell lines, three cell lines did not express Jagged1.
Notch1 and Jagged1 expression in normal
blood cells and bone marrow cells
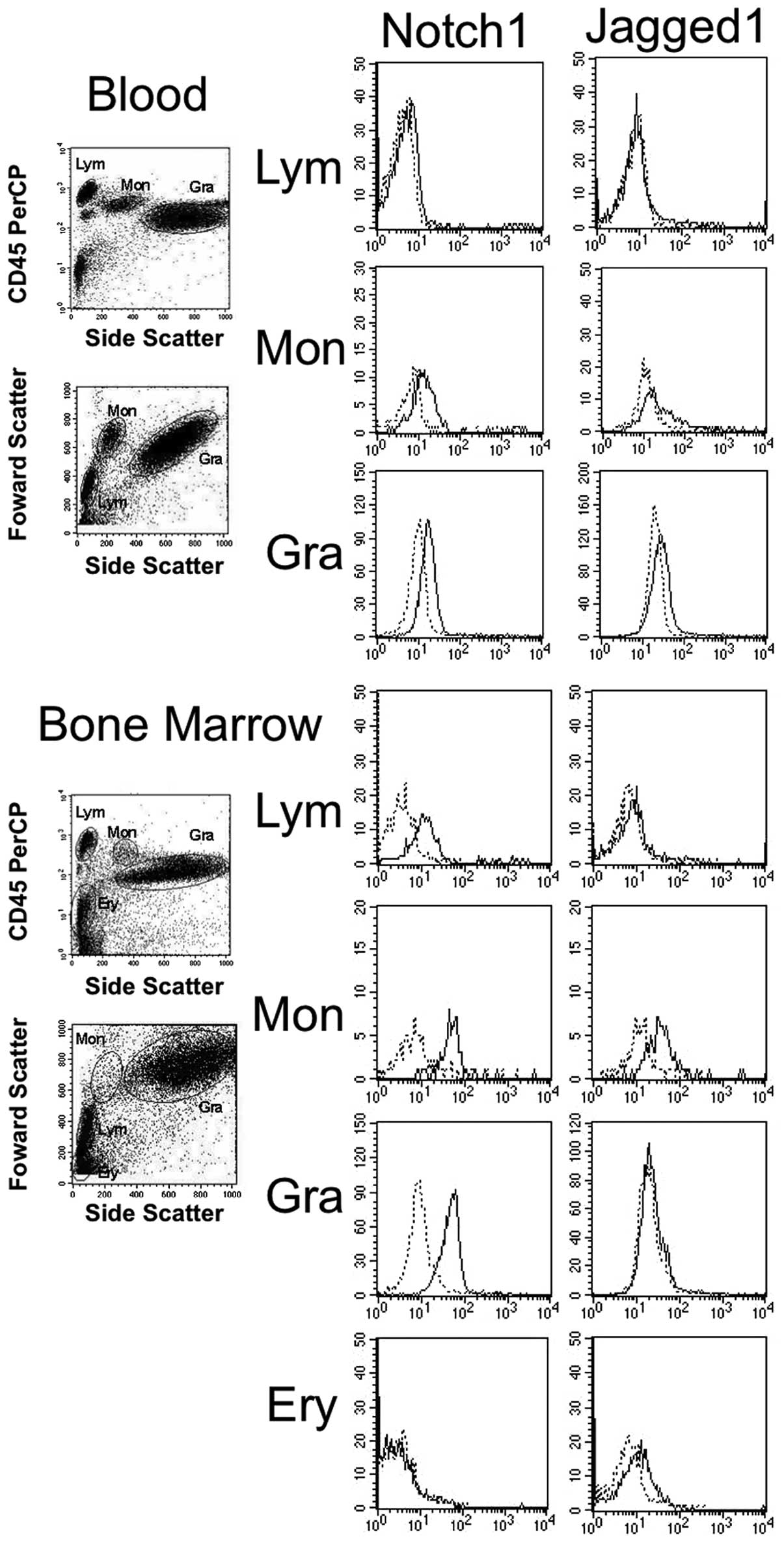
The expression of Notch1 and Jagged1 from the
representative samples of normal blood and bone marrow aspirates is
shown in Fig. 2. The average
values of the % increase in MFI of all samples examined are shown
in Table III. Notch1 is expressed,
in order of increasing intensity, in monocytes, granulocytes,
lymphocytes and erythroblasts. The expression of Jagged1 is
relatively strong in monocytes. The Notch1 and Jagged1 expression
in each cell fraction in blood was found to be weaker than that in
the same fraction in bone marrow aspirates.
 | Table IIIExpression of Notch1 and Jagged1
proteins in normal blood cells and bone marrow cells analysed by
flow cytometry. |
Table III
Expression of Notch1 and Jagged1
proteins in normal blood cells and bone marrow cells analysed by
flow cytometry.
| Source | Cells | Notch1a | Jagged1a |
|---|
| Blood | Lymphocytes | 27 | 13 |
| Monocytes | 210 | 82 |
| Granulocytes | 170 | 40 |
| Bone marrow | Lymphocytes | 186 | 29 |
| Monocytes | 659 | 298 |
| Granulocytes | 414 | 81 |
| Erythroblasts | 74 | 70 |
Notch1 and Jagged1 in leukemia
samples
The intensity of expression of Notch1 and Jagged1 in
leukemia cells from 22 patients is shown in Fig. 3. Of seven AML samples, three were
found to express both Notch1 and Jagged1, one expressed Jagged1
only and three expressed neither of the proteins. No mature T-cell
neoplasm samples expressed either of the proteins. All B-CLL
samples expressed both Notch1 and Jagged1. The intensities of the
expression of B-CLL cells were much stronger than those of normal
lymphocytes.
Discussion
The present study provides a flow cytometric
analysis of Notch1 and Jagged1 expression in normal blood cells and
various leukemia cells. Thus far, flow cytometric detection of
Notch proteins in monocytes (8),
eosinophils (9) and B-ALL cells
(10) has been reported. In these
studies, permeabilised cells were mainly used for the analysis.
Therefore, we aimed to detect Notch1 and Jagged1 in
non-permeabilised cells using leukemia/lymphoma cell lines in which
Notch1 and Jagged1 expression had been examined by immunoblot
analysis. We confirmed that the expression of these proteins was
detected in non-permeabilised cells.
We then obtained expression profiles of Notch1 and
Jagged1 in each cell fraction in normal blood and bone marrow
samples. The results showed that Notch1 expression is strong in
monocytes and granulocytes, and weak in lymphocytes. We confirmed
that this tendency is also reflected in mRNA expression levels in
each cell fraction according to quantitative RT-PCR (data not
shown). We also found that the levels of Notch1 and Jagged1
expression are stronger in bone marrow cells than in the equivalent
cells in blood. The mechanism of this phenomenon has not yet been
identified.
We identified the expression of Notch1 and Jagged1
in various leukemia cells. Since the expression of these genes in
T-ALL (3) and B-ALL cells
(10) had previously been
reported, we focused on AML, mature T-cell neoplasm and B-CLL
cells. Certain AML samples expressed Notch1 and/or Jagged1 while
other samples expressed neither protein. This observation is
similar to the results of our previous study of immunoblot analyses
(4). It is well known that Notch1
is crucial in the development of T-ALL (3). By contrast, mature T-cell neoplasm
cells did not express Notch1 or Jagged1. This suggests that Notch
signaling may not be important for the growth of mature T-cell
neoplasm cells. Notably, all the B-CLL samples expressed high
levels of both Notch1 and Jagged1. This suggests that Notch
signaling is important for the growth of B-CLL cells. This finding
is also useful for distinguishing between normal B-lymphocytes and
B-CLL cells.
The present study suggests that Notch1 and Jagged1
expression is detected by FCM. To clarify the expression patterns
in various types of leukemia, data from more patient samples should
be obtained. Subsequently, we predict that the examination of
Notch1 and Jagged1 expression is likely to be useful for the
characterisation of individual cases, detection of minimal residual
diseases and selection of patients that would most benefit from a
novel molecular-targeted therapy using Notch inhibitors in the
future.
Acknowledgements
We thank Drs N. Murakami, M. Yamamoto,
T. Fukuda and O. Miura (Tokyo Medical and Dental University) for
their assistance in obtaining samples from the patients. This study
was supported in part by a Grant-in-Aid for Scientific Research (C)
from the Japan Society for the Promotion of Science (no.
18690522).
References
|
1.
|
Suzuki T and Chiba S: Notch signaling in
hematopoietic stem cells. Int J Hematol. 82:285–294. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Leong KG and Karsan A: Recent insights
into the role of Notch signaling in tumorigenesis. Blood.
107:2223–2233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Weng AP, Ferrando AA, Lee W, Morris JP IV,
Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT and Aster
JC: Activating mutations of NOTCH1 in human T cell acute
lymphoblastic leukemia. Science. 306:269–271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Tohda S and Nara N: Expression of Notch1
and Jagged1 proteins in acute myeloid leukemia cells. Leuk
Lymphoma. 42:467–472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Tohda S, Murata-Ohsawa M, Sakano S and
Nara N: Notch ligands, Delta-1 and Delta-4 suppress the
self-renewal capacity and long-term growth of two myeloblastic
leukemia cell lines. Int J Oncol. 22:1073–1079. 2003.PubMed/NCBI
|
|
6.
|
Tohda S, Kogoshi H, Murakami N, Sakano S
and Nara N: Diverse effects of the Notch ligands Jagged1 and Delta1
on the growth and differentiation of primary acute myeloblastic
leukemia cells. Exp Hematol. 33:558–563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lanotte M, Martin-Thouvenin V, Najman S,
Balerini P, Valensi F and Berger R: NB4, a maturation inducible
cell line with t(15;17) marker isolated from a human acute
promyelocytic leukemia (M3). Blood. 77:1080–1086. 1991.PubMed/NCBI
|
|
8.
|
Ohishi K, Varnum-Finney B, Flowers D,
Anasetti C, Myerson D and Bernstein ID: Monocytes express high
amounts of Notch and undergo cytokine specific apoptosis following
interaction with the Notch ligand, Delta-1. Blood. 95:2847–2854.
2000.PubMed/NCBI
|
|
9.
|
Radke AL, Reynolds LE, Melo RC, Dvorak AM,
Weller PF and Spencer LA: Mature human eosinophils express
functional Notch ligands mediating eosinophil autocrine regulation.
Blood. 113:3092–3101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Nwabo Kamdje AH, Mosna F, Bifari F, Lisi
V, Bassi G, Malpeli G, Ricciardi M, Perbellini O, Scupoli MT,
Pizzolo G and Krampera M: Notch-3 and Notch-4 signaling rescue from
apoptosis human B-ALL cells in contact with human bone
marrow-derived mesenchymal stromal cells. Blood. 118:380–389.
2011.PubMed/NCBI
|