Introduction
Lung cancer is a major cause of mortality from
malignant diseases due to its high incidence, malignant behavior
and lack of major advancements in treatment strategy (1). There is a large body of accumulated
evidence that the epidermal growth factor receptor (EGFR) and its
family members are heavily involved in the development and
progression of numerous human tumors, including lung cancer
(2,3). The EGFR tyrosine kinase inhibitor
(TKI), gefitinib, was approved for the treatment of non-small cell
lung cancer (NSCLC) in Japan in 2002. Two original studies have
revealed that EGFR mutation status at the tyrosine kinase
(TK) domain in NSCLC patients was correlated with a good response
to gefitinib (4,5). From the results of the Iressa
Pan-Asia Study (IPASS), EGFR mutations are the strongest
predictive biomarker for progression-free survival (PFS) and tumor
response to first-line gefitinib therapy for NSCLC (6).
The EGFR gene is highly polymorphic and its
expression and activity are significantly affected by various
polymorphisms (7–9). As for interethnic differences in CA
repeat length in intron 1, a length of less than 17 in Japanese
individuals is less frequent than in Caucasians (10). However, the frequency of
EGFR mutations is higher in the Japanese population than in
other ethnic groups. In intron 1, the −216G/T and −191C/A
polymorphisms in the EGFR promoter are associated with
altered promoter activity and gene expression (8). CA simple sequence repeats (CA-SSRs)
in intron 1 (rs45559542) (8,12,13),
−216G/T (rs712829) (8,12) and D994D (rs2293347) (11) polymorphisms have been reported to
influence clinical outcomes in gefitinib-treated NSCLC patients. In
addition, the 8227G/A polymorphism (rs763317) located in intron 1
has been reported to be associated with smoking status and gender
in lung adenocarcinomas in the Taiwanese population (14).
To determine the EGFR polymorphism status and
its correlation with clinicopathological features in lung carcinoma
in the Japanese population, we investigated EGFR gene status
using TaqMan single nucleotide polymorphism (SNP) genotyping
assays. These findings were analyzed in relation to the
clinicopathologic features of lung cancer.
Materials and methods
Patients and treatment
The study group included 261 lung cancer patients
who had undergone surgery at the Nagoya City University Hospital,
Japan, between 1997 and 2011. Thirty-three patients were treated
with gefitinib for the recurrence of lung cancer following surgery.
We also investigated polymorphisms for 13 NSCLC patients who had
been treated with gefitinib for the recurrence of cancer at the
Kinki-chuo Chest Medical Center, Osaka Japan. The lung tumors were
classified according to the general rule for clinical and
pathological recording of lung cancer in Japan, as well as
according to the WHO classification. All tumor samples were
immediately frozen and stored at −80°C until assayed.
The clinical and pathological characteristics of the
274 lung cancer patients were as follows: 194 (70.8%) were male and
80 were female; 192 were diagnosed as adenocarcinoma and 82 were
diagnosed as other types of carcinoma (63 squamous cell carcinomas,
6 adenosquamous carcinomas, 6 large cell carcinomas, 3 carcinoids,
3 pleomorphic carcinomas, 1 adenoid cystic carcinoma and 1
carcinosarcoma); 187 (68.2%) were smokers (current or former
smoker) and 87 were non-smokers (Table
I). Written informed consent was obtained from the patients and
the Institutional Ethics Committee of the Nagoya City University
approved the study.
 | Table IClinical and pathological
characteristics of the 274 lung cancer patients. |
Table I
Clinical and pathological
characteristics of the 274 lung cancer patients.
| Patients (n=274)
|
|---|
| No. | % |
|---|
| Age (years) | | |
| ≤60 | 78 | 28.5 |
| >60 | 196 | 71.5 |
| Gender | | |
| Male | 194 | 70.8 |
| Female | 80 | 29.2 |
| Smoking status | | |
| Non-smoker | 87 | 31.8 |
| Smoker | 187 | 68.2 |
| Pathological
subtype | | |
| Adeno | 192 | 70.1 |
| Other | 82 | 29.9 |
| EGFR
mutation | | |
| Positive | 81 | 29.9 |
| Negative | 190 | 70.1 |
Genotyping assays for the EGFR
polymorphism
Genomic DNA was extracted from peripheral blood
(n=109) taken prior to surgery or from adjacent normal lung tissues
taken at surgery using the Wizard SV Genomic DNA Purification
system (Promega Corp., Madison, WI, USA) according to the
manufacturer’s instructions. EGFR mutation statuses at the
kinase domain were investigated using the TaqMan PCR assay (Applied
Biosystems, Foster City, CA, USA). The results of the TaqMan PCR
assay have been previously reported (15).
TaqMan SNP genotyping assays (Applied Biosystems)
were used for genotyping 4 polymorphisms in the EGFR gene
(−216G/T, −191C/A, 8227G/A, assay ID: C_2310200_10; and D994D,
assay ID: C_15970737_20; Table II)
according to the manufacturer’s instructions (16). The cycling conditions for the
TaqMan SNP assays were as follows: 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min, with a 1-min
extension at 25°C following the last cycle. The R521K (rs11543848,
also assigned as R497K in the literature) polymorphism was examined
by the PCR-RFLP method as described previously (17). Sixty-four lung cancer samples were
analyzed for EGFR gene amplification using fluorescence
in situ hybridization (FISH) and the results have been
previously reported (18).
 | Table IIGenotyping approach for polymorphism
analysis of the EGFR gene. |
Table II
Genotyping approach for polymorphism
analysis of the EGFR gene.
| Primer
sequences | −216G/T
(rs712829) | −191C/A
(rs712830) |
|---|
| VIC-MGB | AGCCTCCGCCCCC | CCTCGGCCGCGTCG |
| FAM-MGB | CAGCCTCCTCCCCC | CCTCGGCCGCGGCG |
| Forward primer |
CCCGCGCGAGCTAGAC |
CCCCGCACGGTGTGA |
| Reverse primer |
GGGCGCTCACACCTG |
GGCTAGCTCGGGACTCC |
Statistical analyses
Statistical analyses were carried out using the
Mann-Whitney U test for unpaired samples and Wilcoxon’s signed rank
test for paired samples. Linear relationships between variables
were determined by means of simple linear regression. Correlation
coefficients were determined by rank correlation using Spearman’s
test and the χ2 test. The overall survival (OS) of lung
cancer patients was examined using the log-rank test. All analyses
were performed using the StatView software package (Abacus Concepts
Inc, Berkeley, CA, USA) and differences were considered significant
when p<0.05.
Results
EGFR gene mutation and amplification
statuses
Of the 274 patients, 42 had the deletion-type
EGFR mutations in exon 19; 35 had the missense point
mutations (5 G719S, 29 L858R and 1 L861Q) in exon 18 or 21; and 4
had exon 20 insertion mutations (15,20).
Sixty-four samples were studied for EGFR gene amplification
using FISH analyses. According to the criteria by Cappuzzo et
al, 21 were FISH-positive and 43 were FISH-negative (19).
EGFR polymorphisms in Japanese lung
cancers
In our Japanese cohort, there was no −191C/A
polymorphism and we did not perform any further analyses for this
polymorphism. For rs712829 (−216G/T), 255 patients were GG, 19 were
GT and no TT was found. For rs2293347 (D994D), 125 patients were
CC, 110 were CT and 39 were TT. For rs11543848 (R497K), 93 were AA,
135 were GA and 46 were GG. No correlation existed between these 3
SNPs (−216G/T, GG vs. GA+AA; D994D, CC+CT vs. TT; R497K, GG vs.
GA+AA) and clinicopathological features of the lung cancers.
Of the 274 patients, 87 had the 8227G/A EGFR
variant (9 AA and 78 GA). Of these, 64 were male and 23 were
female, 24 were non-smokers, 59 were smokers and 4 were unknown.
Adenocarcinomas were significantly more frequent in GG-type
patients (139/187, 74.3%) than in the GA- or AA-type patients
(53/87, 60.9%, p=0.0331). However, the polymorphism did not
correlate with gender (p=0.5687), smoking (non-smokers vs. smokers,
p=0.3325), or EGFR mutation (p=0.1539) statuses of lung
cancer (Table III). EGFR
gene amplification as identified by FISH positivity was not
correlated with polymorphism statuses, including D994D (p=0.5884),
−216G/T (p>0.9999), R497K (p=0.2043) and 8227G/A
(p>0.9999).
 | Table IIIAssociation of the EGFR
8227G/A polymorphism with clinicopathological data of 274 lung
cancer patients. |
Table III
Association of the EGFR
8227G/A polymorphism with clinicopathological data of 274 lung
cancer patients.
| GG
| GA+AA
| |
|---|
| Factors | No. | % | No. | % | p-value |
|---|
| Age (years) | | | | | |
| ≤60 | 52 | 27.8 | 26 | 29.9 | 0.7741 |
| >60 | 135 | 72.2 | 61 | 70.1 | |
| Gender | | | | | |
| Male | 130 | 69.5 | 64 | 73.6 | 0.5687 |
| Female | 57 | 30.5 | 23 | 26.4 | |
| Smoking status | | | | | |
| Non-smoker | 63 | 33.7 | 24 | 27.6 | 0.3325 |
| Smoker | 124 | 66.3 | 59 | 72.4 | |
| Pathological
subtype | | | | | |
| Adeno | 139 | 74.3 | 53 | 60.9 | 0.0331 |
| Others | 48 | 25.7 | 34 | 39.1 | |
| EGFR
mutation | | | | | |
| Positive | 61 | 32.6 | 20 | 23.8 | 0.1539 |
| Negative | 126 | 67.4 | 64 | 76.2 | |
Correlation between clinical course of
lung cancer patients and EGFR polymorphisms
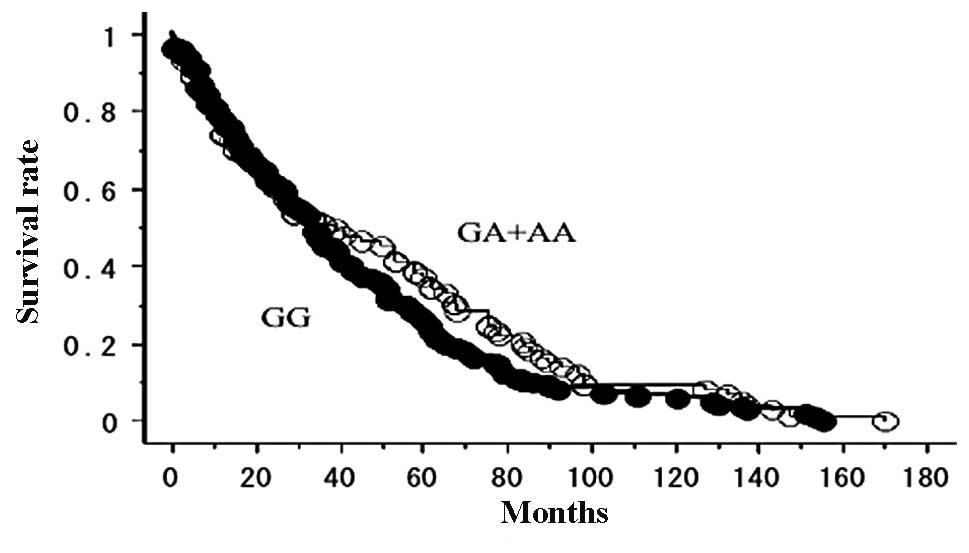
The OS of the 225 lung cancer patients who did not
receive gefitinib, with follow-up until June 30, 2011, was studied
in reference to the EGFR polymorphism status. The prognosis
was not significantly different between the EGFR 8227G/A
types (GA+AA, 23/73 were deceased; GG, 53/152 were deceased;
p=0.1753; Fig. 1). No significant
association was observed between the other 3 SNPs (−216G/T, D994D
and R497K) and disease outcome (data not shown).
Correlation between clinical course of
gefitinib-treated lung cancer patients and EGFR polymorphism
The OS of 46 gefitinib-treated lung cancer patients,
with follow-up until June 30, 2011, was studied in reference to the
EGFR polymorphism status. In this analysis, 12 patients had
EGFR 8227GA or AA types. Of the 46 patients, 31 had
EGFR mutations and 11 were EGFR 8227GA or AA. There
was a tendency towards higher EGFR mutation ratio in the
8227GA- or AA-type patients compared with GG-type patients
(p=0.0702). Other clinical backgrounds, including gender
(p=0.3071), smoking (p=0.4893) and pathological status (p=0.3059)
were not correlated with 8227G/A polymorphism status. The prognosis
following gefitinib therapy was significantly better for the
EGFR GA- or AA-type patients (5/12 were deceased; mean
survival, 1,014 days) when compared with the 8227GG-type patients
(26/34 were deceased; mean survival, 607 days; log-rank test,
p=0.0448; Fig. 2). Using the
multivariate analysis, EGFR mutation (p=0.0316; hazard
ratio, 2.174) but not 8227G/A (p=0.2232; hazard ratio, 1.587) was
the independent prognostic factor for gefitinib-treated
patients.
There was no association between the other 3
polymorphisms (−216G/T, p=0.7599; D994D, p=0.1813; and R497K,
p=0.885) and prognosis for the gefitinib-treated patients.
Discussion
In this study, gefitinib-treated patients with an A
allele at the EGFR 8227G/A site were found to have a better
prognosis compared with GG-type patients. However, there was no
association between the other 3 polymorphisms (−216G/T, D994D and
R497K) and prognosis following gefitinib therapy.
Previous studies have suggested that −216G/T
(8,12) and D994D (11) polymorphisms are associated with
clinical outcome of gefitinib therapy. In intron 1, CA-SSR of
EGFR has been the most studied polymorphism. CA-SSR has been
associated with EGFR gene expression and has been reported
to correlate with clinical outcome of gefitinib therapy (8,12,13,21).
Shorter CA repeats have been associated with higher transcription
levels of EGFR and have been reported to be correlated with
better clinical outcome of gefitinib therapy. Tiseo et al
revealed that patients with the CA-16 genotype had a longer
survival compared with those with other genotypes (13). Liu et al found that the
−216G/T polymorphism and CA-19 genotype are found more frequently
in patients with exon 19 deletions (22). On the other hand, Suzuki et
al reported that the EGFR protein expression level was
significantly higher in the shorter CA repeats group than in the
longer allele group, but its length was not associated with
EGFR somatic mutations (23). In a Japanese cohort, Ichihara et
al reported that patients with a short CA-SSR1 had a prolonged
OS as compared with those with a longer CA-SSR, but this difference
was not significant in patients with a drug-sensitive EGFR
mutation (p=0.13) (24). They
found that FISH status, CA-SSR1 length and the SNP status in the
promoter region (−216G/T or −191G/A) had no association with
responsiveness to gefitinib in cases of lung cancer in Japanese
individuals, similar to our results. One explanation for the
results is that the variant forms of the SNPs, −216 G/T (6.6%) and
−191G/A (0.6%), were less frequent in East Asians than in
individuals of European descent (60.3 and 37%, respectively)
(25). As for the D994D
polymorphism, using direct sequencing, our group has previously
revealed that the polymorphism did not affect the gefitinib
sensitivity in Japanese individuals (26). This polymorphism is located in exon
25 and a synonymous SNP does not change the amino acid sequence of
the protein, so it does not influence the biological function of
the protein itself. Ma et al revealed that the D994D
polymorphism did affect PFS but not OS following gefitinib therapy
(11).
The 8227G/A polymorphism is also located in intron
1, but there have been few studies examining this SNP (14,27).
Jou et al revealed that the EGFR 8227G/A polymorphism
was associated with lung cancer, especially in non-smoking female
lung adenocarcinoma patients in the Taiwanese population (14). Thus, this variation may lead to the
different modifications of cancer genes, including EGFR, in
tumorigenic pathways among different histological subtypes, gender
and ethnicity. The 8227G/A SNP is located in intron 1, 6.9 kb
downstream of the CA-SSR1 polymorphism. Additional functional
analyses of this SNP are needed to better understand the mechanism
by which the 8227G/A SNP of EGFR affects lung cancer. In our
analysis, although the 8227G/A polymorphism in intron 1 was not
correlated with EGFR somatic mutations, the GA or AA type
was associated with longer survival of the gefitinib-treated
patients. The underlying mechanisms remain unclear, but it may be
that intron 1 of EGFR is associated with sensitivity to EGFR
TKIs in lung cancer patients, and is correlated with certain
biomarkers other than EGFR mutations. The sample size of the
present study was too small to address this hypothesis. The extact
effect of the polymorphism on survival time of patients treated
with or without EGFR TKIs needs further clinical investigation with
a larger sample size.
In summary, the 8227G/A polymorphism of EGFR
may influence OS in gefitinib-treated lung cancer patients.
Acknowledgements
The authors thank Mrs. Miki Mochizuki
for her excellent technical assistance. This study was supported by
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (JSPS) (Nos. 23659674, 21390394 and 21591820)
and a grant for cancer research from the Program for developing the
supporting system for upgrading education and research (2009) from
the Ministry of Education, Culture, Sports, Science and Technology
of Japan.
References
|
1
|
Ginsberg RJ, Kris K and Armstrong G:
Cancer of the lung. Principles and Practice of Oncology. DeVita VT,
Hellman S and Rosenberg SA: 4th edition. JB Lippincott Co;
Philadelphia, PA: pp. 673–682. 1993
|
|
2
|
Nicholson RI, Gee JM and Harper ME: EGFR
and cancer prognosis. Eur J Cancer. 37:S9–S15. 2001. View Article : Google Scholar
|
|
3
|
Onn A, Choe DH, Herbst RS, et al:
Synchronous overexpression of epidermal growth factor receptor and
HER2-neu protein is a predictor of poor outcome in patients with
stage I non-small cell lung cancer. Clin Cancer Res. 10:136–143.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paez JG, Jänne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fukuoka M, Wu Y-L, Thongprasert S, et al:
Biomarker analyses and first overall survival results from a phase
III, randomized, open-label, first-line study of gefitinib versus
carboplatin/paclitaxel in clinically selected patients with
advanced non-small-cell lung cancer in Asia (IPASS). J Clin Oncol.
29:2866–2874. 2011. View Article : Google Scholar
|
|
7
|
Liu W, Innocenti F, Wu MH, et al: A
functional common polymorphism in a Sp1 recognition site of the
epidermal growth factor receptor gene promoter. Cancer Res.
65:46–53. 2005.PubMed/NCBI
|
|
8
|
Liu W, Wu X, Zhang W, et al: Relationship
of EGFR mutations, expression, amplification, and polymorphisms to
epidermal growth factor receptor inhibitors in the NCI60 cell
lines. Clin Cancer Res. 13:6788–6795. 2007. View Article : Google Scholar
|
|
9
|
Brandt B, Meyer-Staeckling S, Schmidt H,
et al: Mechanisms of EGFR gene transcription modulation:
relationship to cancer risk and therapy response. Clin Cancer Res.
12:7252–7260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu W, Innocenti F, Chen P, et al:
Interethnic difference in the allelic distribution of human
epidermal growth factor receptor intron 1 polymorphism. Clin Cancer
Res. 9:1009–1012. 2003.PubMed/NCBI
|
|
11
|
Ma F, Sun T, Shi Y, et al: Polymorphisms
of EGFR predict clinical outcome in advanced non-small-cell lung
cancer patients treated with Gefitinib. Lung Cancer. 66:114–119.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu G, Gurubhagavatula S, Zhou W, et al:
Epidermal growth factor receptor polymorphisms and clinical
outcomes in non-small-cell lung cancer patients treated with
gefitinib. Pharmacogenomics J. 8:129–138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tiseo M, Capelletti M, De Palma G, et al:
Epidermal growth factor receptor intron-1 polymorphism predicts
gefitinib outcome in advanced non-small cell lung cancer. J Thorac
Oncol. 3:1104–1111. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jou YS, Lo YL, Hsiao CF, et al:
Association of an EGFR intron 1 SNP with never smoking female lung
adenocarcinoma patients. Lung Cancer. 64:251–256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Endo K, Konishi A, Sasaki H, et al:
Epidermal growth factor receptor gene mutation in non-small cell
lung cancer using highly sensitive and fast TaqMan PCR assay. Lung
Cancer. 50:375–384. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De la Vega FM, Lazaruk KD, Rhodes MD and
Wenz MH: Assessment of two flexible and compatible SNP genotyping
platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping
System. Mutat Res. 573:111–135. 2005.PubMed/NCBI
|
|
17
|
Sasaki H, Okuda K, Shimizu S, et al: EGFR
R497K polymorphism is a favorable prognostic factor for advanced
lung cancer. J Cancer Res Clin Oncol. 135:313–318. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sasaki H, Shimizu S, Okuda K, et al:
Epidermal growth factor receptor gene amplification in surgical
resected Japanese lung cancer. Lung Cancer. 64:295–300. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cappuzzo F, Hirsch FR, Rossi E, et al:
Epidermal growth factor receptor gene and protein and gefitinib
sensitivity in non-small-cell lung cancer. J Natl Cancer Inst.
97:643–655. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sasaki H, Endo K, Takada M, et al: EGFR
exon 20 insertion mutation in Japanese lung cancer. Lung Cancer.
58:324–328. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nie Q, Wang Z, Zhang GC, et al: The
epidermal growth factor receptor intron1 (CA) n microsatellite
polymorphism is a potential predictor of treatment outcome in
patients with advanced lung cancer treated with Gefitinib. Eur J
Pharmacol. 570:175–181. 2007. View Article : Google Scholar
|
|
22
|
Liu W, He L, Ramírez J, et al: Functional
EGFR germline polymorphisms may confer risk for EGFR somatic
mutations in non-small cell lung cancer, with a predominant effect
on exon 19 microdeletions. Cancer Res. 71:2423–2427. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Suzuki M, Kageyama S, Shinmura K, et al:
Inverse relationship between the length of the EGFR CA repeat
polymorphism in lung carcinoma and protein expression of EGFR in
the carcinoma. J Surg Oncol. 98:457–461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ichihara S, Toyooka S, Fujiwara Y, et al:
The impact of epidermal growth factor receptor gene status on
gefitinib-treated Japanese patients with non-small-cell lung
cancer. Int J Cancer. 120:1239–1247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nomura M, Shigematsu H, Li L, et al:
Polymorphisms, mutations, and amplification of the EGFR gene in
non-small cell lung cancers. PLos One. 4:715–727. 2007.PubMed/NCBI
|
|
26
|
Sasaki H, Okuda K, Takada M, et al: A
novel EGFR mutation D1012H and polymorphism at exon 25 in Japanese
lung cancer. J Cancer Res Clin Oncol. 134:1371–1376. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Girard N, Lou E, Azzoli CG, et al:
Analysis of genetic variants in never-smokers with lung cancer
facilitated by an internet-based blood collection protocol: a
preliminary report. Clin Cancer Res. 16:755–763. 2010. View Article : Google Scholar : PubMed/NCBI
|