Introduction
Chlorotoxin (CTX) is a scorpion-derived bioactive
peptide with 36 amino acids and four disulfide bonds. It was first
isolated from the venom of Leiurus quinquestriatus in 1993
and was named chlorotoxin due to its ability to block
small-conductance chloride channels (1). Following its discovery, CTX was
observed to preferentially bind to malignant gliomas and tumors of
neuroectodermal origin (2,3). Cell surface matrix
metalloproteinase-2 (4) and
annexin A2 (5) have been
identified as the molecular targets of CTX on the tumor cells.
Radionuclide iodine-131-labeled, chemically synthesized CTX
(commercial name 131I-TM601) has been designated as an
orphan drug for the treatment of malignant gliomas and melanomas by
the US Food and Drug Administration (FDA). Other fluorescent
dye-labeled and nanoparticle-labeled CTXs have also shown great
potential for the diagnosis and treatment of malignant tumors
(6–14).
Due to its small size, CTX is a promising
tumor-targeting peptide and significant quantities of mature CTX
are needed for therapeutic and experimental purposes. In the
present study, we established an efficient approach for the
preparation of mature CTX through the recombinant expression of
designed CTX precursors in Escherichia coli (E. coli)
and subsequent in vitro enzymatic and oxidative
refolding.
Materials and methods
Materials
The oligonucleotide primers were chemically
synthesized at Biosune Biotechnology Co. Ltd. (Shanghai, China) and
enterokinase was purchased from New England Biolabs, Inc. (Ipswich,
MA, USA). Agilent reverse-phase columns (Agilent Technologies,
Santa Clara, CA, USA), including an analytical column (Zorbax
300SB-C18; 4.6×250 mm) and a semi-preparative column (Zorbax
300SB-C18; 9.4×250 mm) were used in the experiments. The peptide
was eluted from the columns with an acetonitrile gradient composed
of solvent A and solvent B. Solvent A was 0.1% aqueous
trifluoroacetic acid (TFA), and solvent B was acetonitrile
containing 0.1% TFA. The elution gradient was as follows: 0 min,
10% solvent B; 3 min, 10% solvent B; 53 min, 60% solvent B; 55 min,
100% solvent B; 56 min, 100% solvent B and 57 min, 10% solvent B.
The flow rate for the analytical column was 0.5 ml/min, while that
for the semi-preparative column was 1.0 ml/min. The eluted peptide
was detected by UV absorbance at 280 and 214 nm.
Gene construction, recombinant expression
and purification of 6xHis-CTX
The gene of 6xHis-CTX was constructed from two
chemically synthesized DNA primers. Subsequent to annealing,
elongation by T4 DNA polymerase and cleavage by the restriction
enzymes NdeI and EcoRI, the DNA fragment was ligated
into a pET vector pretreated with the same restriction enzymes. Its
sequence was confirmed using DNA sequencing. The expression
construct pET/6xHis-CTX was then transformed into the E.
coli strain BL21 Star™ (DE3), prior to the transformed cells
being cultured in liquid Luria-Bertani (LB) medium, containing 10
g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl medium (with 100
μg/ml ampicillin), to an optical density at 600 nm
(OD600) of 1.0 at 37°C, with vigorous shaking (250 rpm).
Following induction by 1.0 mM isopropyl thio-β-D-galactoside (IPTG)
at 37°C for 6–8 h, the E. coli cells were harvested by
centrifugation (5,000 × g, 10 min), resuspended in lysis buffer (50
mM Tris-HCl, pH 8.5; 0.5 M NaCl) and lysed using sonication.
Subsequent to further centrifugation (10,000 × g, 15 min), the
inclusion body pellet was resuspended in solubilizing buffer (50 mM
Tris-HCl, 6 M guanidine chloride; pH 8.5) and S-sulfonated by the
addition of solid sodium sulfite and sodium tetrathionate to the
final concentrations of 200 mM and 150 mM, respectively. The
S-sulfonation reaction was carried out at 4°C with gentle shaking
for 2–3 h. Following centrifugation (10,000 × g, 15 min), the
supernatant was loaded onto an Ni2+ column that was
pre-equilibrated with the washing buffer (50 mM Tris-HCl, 3 M
guanidine chloride; pH 8.5). The S-sulfonated precursor was then
eluted from the column by a step-wise increase of imidazole
concentration in the washing buffer. The eluted S-sulfonated
6xHis-CTX was subsequently further purified using C18 reverse-phase
high-performance liquid chromatography (HPLC) and lyophilized. The
molecular mass was measured using electro-spray mass
spectrometry.
Gene construction, recombinant expression
and purification of glutathione transferase (GST)-6xHis-CTX
The coding region of 6xHis-CTX was amplified by
polymerase chain reaction (PCR) using pET/6xHis-CTX as the
template. The amplified DNA fragment was digested by the
restriction enzymes BamHI and XhoI and subsequently
ligated into a pGEM-4T-1 vector, providing the GST tag, pretreated
with the same restriction enzymes. Its sequence was confirmed by
DNA sequencing. Following this, the construct pGEM/GST-6xHis-CTX
was transformed into the E. coli strain BL21 Star™ (DE3) and
the transformed cells were cultured in liquid terrific broth (TB)
medium, containing 12 g/l tryptone, 24 g/l yeast extract, 0.4%
(V/V) glycerol, 17 mM KH2PO4, 72 mM
K2HPO4;(with 100 μg/ml ampicillin), to
an OD600 of ∼5.0 at 37°C, with vigorous shaking (250
rpm). Subsequent to overnight induction with 1.0 mM IPTG at 37°C,
the E. coli cells were harvested by centrifugation (5,000 ×
g, 10 min), resuspended in lysis buffer (50 mM phosphate, pH 7.4;
0.5 M NaCl) and lysed using a French press. The soluble
GST-6xHis-CTX in the supernatant was then subjected to
S-sulfonation by the addition of solid sodium sulfite and sodium
tetrathionate to final concentrations of 200 and 150 mM,
respectively. Following shaking at 4°C for 2–3 h, the S-sulfonated
sample was loaded onto an Ni2+ column that was
pre-equilibrated with the washing buffer (50 mM Tris, 150 mM NaCl;
pH 8.0). The S-sulfonated GST-6xHis-CTX was eluted from the column
by a step-wise increase of imidazole concentration in the washing
buffer. The eluted GST-6xHis-CTX fraction was concentrated by
ultrafiltration for enzymatic digestion in the next step.
Enterokinase cleavage of the S-sulfonated
CTX precursors and in vitro refolding
The S-sulfonated CTX precursors (6xHis-CTX and
GST-6xHis-CTX) were digested by enterokinase (peptide:enzyme molar
ratio 105:1) in the digestion buffer (10 mM Tris-HCl, 50
mM NaCl, 10 mM CaCl2; pH 8.0) at 25°C overnight. For
6xHis-CTX, the digestion mixture was directly subjected to in
vitro refolding. For GST-6xHis-CTX, the digestion mixture was
first subjected to gel filtration (Sephadex G-50 column, Sinopharm
Chemical Reagent Co., Ltd., China) and the sulfonated CTX fraction
was collected and used for refolding. For oxidative refolding, the
S-sulfonated CTX was initially treated with 10 mM dithiothreitol
(DTT) at room temperature for 15 min, prior to being 10-fold
diluted into the pre-incubated refolding buffer (0.5 M L-arginine,
1.0 mM EDTA and 2.0 mM oxidized glutathione; pH 8.5). The refolding
reaction was carried out at 4°C for 1–2 h. Following this, the
refolding mixture was acidified to pH 3.0 using TFA and subjected
to a C18 reverse-phase HPLC. The eluted, refolded CTX fraction was
manually collected, lyophilized and analyzed using mass
spectrometry.
Activity assay of the folded CTX
The activity of the recombinant CTX was evaluated
using a tumor cell invasion assay in matrigel. U251-MG cells (Cell
Bank in Shanghai Institutes For Biological Sciences, CAS, China)
were cultured in Dulbecco’s modified Eagle’s medium (DMEM)
supplemented with 10% fetal bovine serum and antibiotics. Following
trypsin digestion, the cells were washed with phosphate-buffered
saline (PBS), resuspended in DMEM without fetal bovine serum and
seeded into the matrigel-covered invasion chamber (5×104
cells/chamber, 8-μm pores; BD Biosciences, Franklin Lakes,
NJ, USA). The chambers were then placed in 24-well plates
containing DMEM with 10% fetal bovine serum. The cells were
cultured at 37°C for 1 h, prior to indicated concentrations of CTX
being added into the chamber. The cells were subsequently
continuously cultured at 37°C for 20 h. Following this, the cells
at the inner side of the invasion chamber were scraped off and the
migrated cells at the outside of the chamber were stained with
crystal violet and counted under a microscope.
Statistical analysis
Sigmaplot 12.1 from Systat Software Inc. (http://www.sigmaplot.com/) was used for statistical
analysis. A t-test was used to analyze statistically significant
differences.
Results
Recombinant expression and purification
of the designed CTX precursors
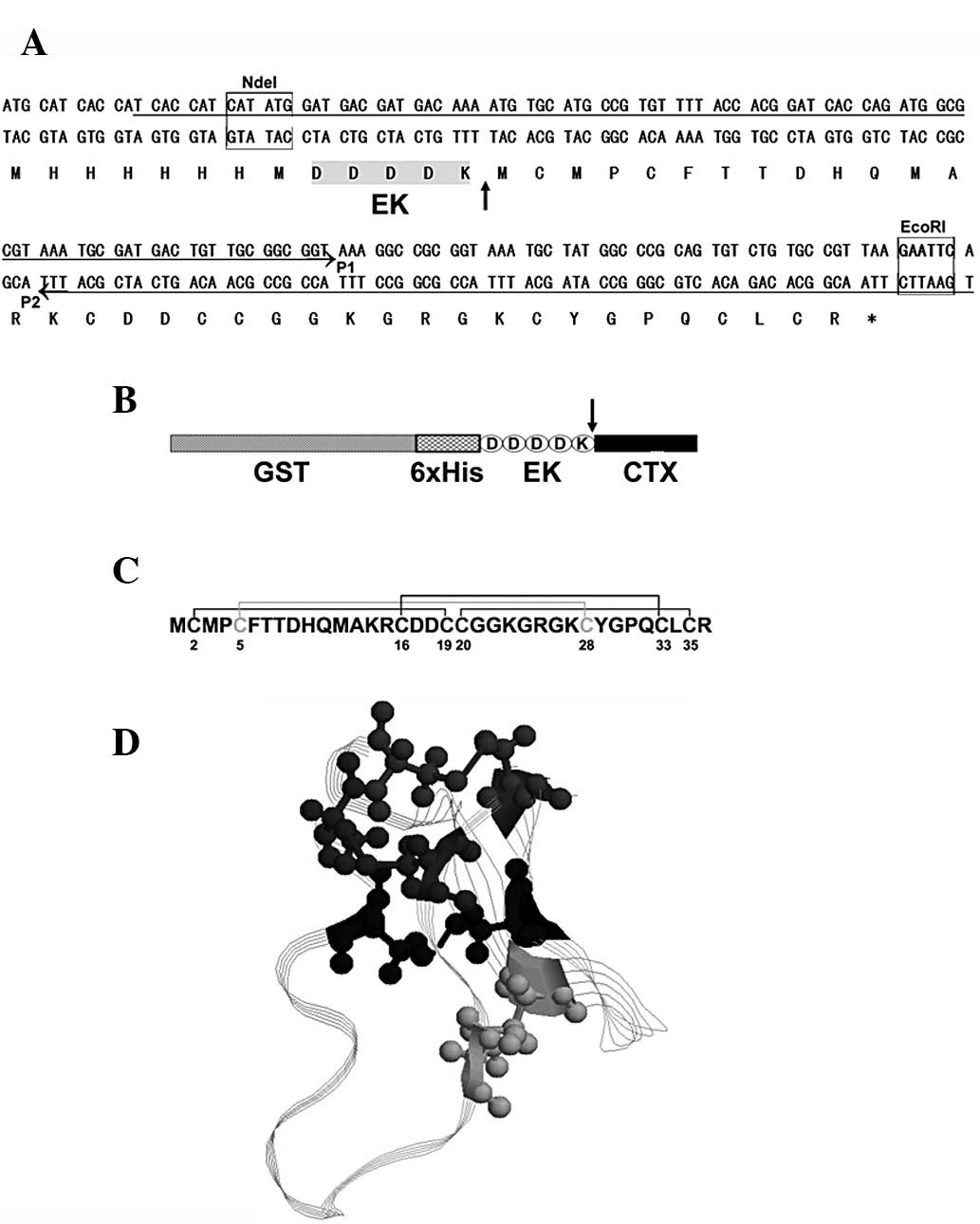
To prepare mature CTX using heterologous expression
in E. coli, a 6xHis-CTX precursor was designed, as shown in
Fig. 1A. A 6xHis-tag, to
facilitate purification, was present at the precursor N-terminal.
An enterokinase cleavage site (DDDDK) was introduced between the
6xHis-tag and the mature CTX to enable the removal of the tag
following purification. To improve its expression level, E.
coli-biased codons were used in the synthetic gene. A
GST-6xHis-CTX precursor was also constructed, as shown in Fig. 1B. In this precursor, a large
GST-tag was fused at the N-terminal in order to stabilize the small
CTX, which was prone to degradation in E. coli.
6xHis-CTX was recombinantly expressed in the E.
coli strain BL21 Star™ (DE3) under IPTG induction. As analyzed
using tricine sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE), a band with a molecular weight of ∼8
kDa was significantly increased subsequent to induction (Fig. 2A, inner panel). Once the E.
coli cells had been lysed by sonication, the precursor was
shown to be present in the pellet due to the formation of inclusion
bodies (data not shown). The precursor was solubilized by guanidine
chloride and S-sulfonated, which broke the inter-chain disulfide
cross-linking and reversibly modified the eight cysteine residues
of the precursor with negatively charged sulfonate moieties. Our
later experiments showed that the precursor refolded into a mixture
of disulfide isomers due to the presence of the tag; therefore, the
removal of the 6xHis-tag was a prerequisite for the efficient
oxidative folding of the recombinant CTX. Furthermore, the enzyme
enterokinase was not able not cleave the CTX precursor with
disulfide bonds due to steric hindrance; thus, S-sulfonation was a
necessary step for efficient enzyme cleavage and oxidative
refolding. The S-sulfonated precursor was then purified using an
immobilized metal-ion affinity chromatography (Ni2+
column), as shown in Fig. 2A. The
eluted precursor fraction (indicated by an asterisk) was further
purified using C18 reverse-phase HPLC.
 | Figure 2.(A) Purification of the S-sulfonated
6xHis-chlorotoxin (CTX) precursor using immobilized metal ion
affinity chromatography. The peak of the S-sulfonated precursor is
indicated by an asterisk. Inner panel, tricine sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of
the 6xHis-CTX expression in E. coli. M, marker; Lane 1,
prior to isopropyl thio-β-D-galactoside (IPTG) induction; Lane 2,
following IPTG induction. The band of 6xHis-CTX is indicated by an
asterisk. (B) SDS-PAGE analyses of the glutathione transferase
(GST)-6xHis-CTX precursor at different purification stages. (Ba)
SDS-PAGE analysis of GST-6xHis-CTX expression: (−) prior to, and
(+) subsequent to IPTG induction. (Bb) SDS-PAGE analysis of the
solubility of GST-6xHis-CTX. T, total lysate; S, supernatant; P,
pellet. (Bc) SDS-PAGE analysis of Ni2+ column-purified
GST-6xHis-CTX. The band of GST-6xHis-CTX is indicated by an
asterisk. F, flow-through; E, eluted fraction by 100 mM
imidazole. |
GST-6xHis-CTX was also heterologously expressed in
the E. coli strain BL21 Star™ (DE3). As analyzed using
SDS-PAGE, a 30 kDa band that was consistent with the expected
molecular weight of the precursor was significantly increased
following IPTG induction (Fig.
2Ba). Once the E. coli cells had been lysed using a
French press, ∼60% of the precursor was present in the supernatant
(Fig. 2Bb). The soluble
GST-6xHis-CTX was then subjected to S-sulfonation and purified
using immobilized metal ion affinity chromatography
(Ni2+ column). The S-sulfonation step was necessary for
the efficient enterokinase cleavage of the precursor. As analyzed
using SDS-PAGE (Fig. 2Bc), the
S-sulfonated GST-6xHis-CTX was eluted from the Ni2+
column by 100 mM imidazole. The eluted precursor fraction was
subjected to ultrafiltration in order to concentrate the precursor
and partially remove the salt.
Enterokinase cleavage and in vitro
refolding
The S-sulfonated 6xHis-CTX precursor, eluted from
the Ni2+ column, was analyzed using C18 reverse-phase
HPLC, as shown in Fig. 3A. The
measured molecular mass of the eluted peak (indicated by an
asterisk) was 6,318.0 Da, which was consistent with the expected
value (6,319.0 Da) of the S-sulfonated precursor. The purified
S-sulfonated precursor was then subjected to sequential
enterokinase digestion and in vitro refolding. As analyzed
using HPLC (Fig. 3B), two major
peaks appeared. The first peak had a measured molecular mass of
1,692.0 Da, which was consistent with the expected value (1,691.8
Da) of the N-terminal 6xHis-tag. The second peak had a measured
molecular mass of 3,996.0 Da, which was consistent with the
expected value (3,996.8 Da) of mature CTX. The final yield of
mature CTX was 150–200 μg per liter of culture.
The S-sulfonated GST-6xHis-CTX was also subjected to
enterokinase digestion in order to remove the GST-tag and
6xHis-tag. As shown in Fig. 3C
(inner panel, a), a band with the expected molecular weight of the
S-sulfonated CTX (indicated by an asterisk) appeared following
enterokinase digestion. The digestion mixture was then subjected to
gel filtration and two peaks were eluted from the Sephadex G-50
cloumn (data not shown). As analyzed using tricine SDS-PAGE
(Fig. 3C, inner panel b), the
second eluted peak was the S-sulfonated CTX fraction. Subsequent to
refolding, a major peak appeared on the HPLC (Fig. 3C) with a measured molecular mass
(3,998.0 Da) that was consistent with the theoretical value
(3,996.8 Da) of the mature folded CTX. The final yield of mature
CTX was 2 mg per liter of culture.
Activity assay of the recombinant
CTX
The activity of the recombinant CTX was assessed by
evaluating its ability to inhibit cell invasion in matrigel. As
shown in Fig. 4A, the
glioma-derived U251-MG cell migration through the matrigel was
significantly inhibited by 5.6 μM mature CTX. This
inhibitory effect was concentration-dependent. The maximum
inhibition reached ∼60%, with an IC50 value of ∼1.5
μM (Fig. 4B), which
suggested that the recombinant CTX was biologically active.
Discussion
CTX is a disulfide-rich peptide with 36 amino acids
and four disulfide bonds. Although CTX may be prepared through
chemical synthesis and subsequent oxidative folding, the final
yield of this process is insufficient (15,16).
Therefore, the present study attempted to prepare CTX using
recombinant expression in E. coli. To facilitate
purification and improve the expression level, a 6xHis-tag alone or
with a large GST-tag was fused to the N-terminus of the CTX.
However, it was demonstrated that the 6xHis-CTX was not able to
fold into a unique disulfide isomer (data not shown), which was
most likely due to the disturbance of the N-terminal extension
(6xHis-tag and the enterokinase cleavage site). In addition, the
folded 6xHis-CTX (a mixture of disulfide isomers) was not
efficiently digested by enterokinase, most likely due to steric
hindrance, since one disulfide bond was in the proximity of the
cleavage site. Therefore, an S-sulfonation approach was employed,
by which the eight cysteine residues of the CTX precursors were
reversibly modified by sulfonate moieties. The S-sulfonated CTX
precursors were highly soluble in the enzyme digestion buffer and
were efficiently cleaved by enterokinase. Subsequent to the removal
of the N-terminal tag, the S-sulfonated CTX was efficiently
refolded in vitro with ∼80% yield under optimized
conditions. In addition, it was demonstrated that the use of the
precursor (GST-6xHis-CTX) in the expression significantly improved
the final yield of mature CTX (versus that with 6xHis-CTX).
Therefore, the present study provided an efficient approach for the
preparation of active CTX and its analogs for further
investigation.
Acknowledgements
This study was supported by the
Chinese Major Scientific and Technological Special Project for
Major New Drug Creation (2009ZX09103-656).
References
|
1.
|
DeBin JA, Maggio JE and Strichartz GR:
Purification and characterization of chlorotoxin, a chloride
channel ligand from the venom of the scorpion. Am J Physiol.
264:C361–C369. 1993.PubMed/NCBI
|
|
2.
|
Soroceanu L, Gillespie Y, Khazaeli MB and
Sontheimer H: Use of chlorotoxin for targeting of primary brain
tumors. Cancer Res. 58:4871–4879. 1998.PubMed/NCBI
|
|
3.
|
Lyons SA, O’Neal J and Sontheimer H:
Chlorotoxin, a scorpion-derived peptide, specifically binds to
gliomas and tumors of neuroectodermal origin. Glia. 39:162–173.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Deshane J, Garner CC and Sontheimer H:
Chlorotoxin inhibits glioma cell invasion via matrix
metalloproteinase-2. J Biol Chem. 278:4135–4144. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kesavan K, Ratliff J, Johnson EW, et al:
Annexin A2 is a molecular target for TM601, a peptide with
tumor-targeting and anti-angiogenic effects. J Biol Chem.
285:4366–4374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Huang R, Han L, Li J, et al:
Chlorotoxin-modified macromolecular contrast agent for MRI tumor
diagnosis. Biomaterials. 32:5177–5186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kievit FM, Veiseh O, Fang C, Bhattarai N,
Lee D, Ellenbogen RG and Zhang M: Chlorotoxin labeled magnetic
nanovectors for targeted gene delivery to glioma. ACS Nano.
4:4587–4594. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Orndorff RL and Rosenthal SJ: Neurotoxin
quantum dot conjugates detect endogenous targets expressed in live
cancer cells. Nano Lett. 9:2589–2599. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Sun C, Veiseh O, Gunn J, et al: In vivo
MRI detection of gliomas by chlorotoxin-conjugated
superparamagnetic nanoprobes. Small. 4:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Sun C, Fang C, Stephen Z, et al:
Tumor-targeted drug delivery and MRI contrast enhancement by
chlorotoxin-conjugated iron oxide nanoparticles. Nanomedicine
(Lond). 3:495–505. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Veiseh M, Gabikian P, Bahrami SB, et al:
Tumor paint: a chlorotoxin:Cy5.5 bioconjugate for intraoperative
visualization of cancer foci. Cancer Res. 67:6882–6888. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Veiseh O, Kievit FM, Gunn JW, Ratner BD
and Zhang M: A ligand-mediated nanovector for targeted gene
delivery and transfection in cancer cells. Biomaterials.
30:649–657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Veiseh O, Gunn JW, Kievit FM, Sun C, Fang
C, Lee JS and Zhang M: Inhibition of tumor-cell invasion with
chlorotoxin-bound superparamagnetic nanoparticles. Small.
5:256–264. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Veiseh O, Sun C, Fang C, et al: Specific
targeting of brain tumors with an optical/magnetic resonance
imaging nanoprobe across the blood-brain barrier. Cancer Res.
69:6200–6207. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Jacoby DB, Dyskin E, Yalcin M, et al:
Potent pleiotropic anti-angiogenic effects of TM601, a synthetic
chlorotoxin peptide. Anticancer Res. 30:39–46. 2010.PubMed/NCBI
|
|
16.
|
The MICAD Research Team: 131I-Chlorotoxin.
Molecular Imaging and Contrast Agent Database (MICAD). National
Center for Biotechnology Information (US); 2004–2013
|