Introduction
Lung carcinoma is one of the most common and lethal
types of malignancy. Due to the high occurrence and mortality rates
of lung cancer, the treatment of the disease has always been a
significant issue (1).
Furthermore, the lack of clear characteristic symptoms in patients
with early-stage lung cancer presents a formidable therapeutic
challenge. To date, radiotherapy and chemotherapy have been the two
major treatment methods (2,3).
However, lung cancer may become resistant to radiotherapy, causing
the radiation treatment to be ineffective. Therefore, the
exploration of new therapies to enhance the radiosensitivity of
lung carcinoma may be of vital clinical significance (4). Ionizing radiation (IR) has been
demonstrated to evoke a series of biochemical events inside the
cell, including cell cycle arrest, DNA damage and repair, signal
transduction and apoptosis (5).
However, the mechanism by which cancer cells escape from IR-induced
events remains to be elucidated.
The modification of proteins with small
ubiquitin-related modifier (SUMO) modulates the substrate’s
activation, function and subcellular localization (6). SUMOylation is catalyzed by
SUMO-specific activating (E1), conjugating (E2) and ligating (E3)
enzymes. SUMOylation is a dynamic process that is reversed by a
family of SUMO-specific proteases (SENPs) (7). These enzymes are critical in
maintaining a balance between the level of unmodified and
SUMOylated proteins that mediate SUMOylation-dependent cellular
function (8). In mammalian cells,
to date, six SENPs have been identified (SENP1, -2, -3, -5, -6 and
-7), which have different substrate specificities and subcellular
localizations (9). Among the SENP
family, most is understood about SENP1, which is important in
placental development and erythropoiesis (10,11).
Furthermore, SENP1 has also been revealed to be involved in the
development and progression of several types of cancer (12,13).
However, while SENP1-specific inhibitors have been designed
(14,15), whether SENP1 is a potential drug
target for cancer treatment remains unclear.
The present study was conducted to identify markers
of radioresistance that may serve as future targets for modulation
to enhance the efficacy of radiotherapy. The results of the study
showed that the inhibition of SENP1 markedly enhanced the
radiosensitivity of lung carcinoma by promoting IR-induced cell
cycle arrest, γ-H2AX expression and apoptosis. Thus, these data
suggest that SENP1 may be a promising target for enhancing the
efficacy of lung carcinoma radiotherapy.
Materials and methods
Tissue samples
Primary lung carcinoma and adjacent non-tumor lung
tissues were collected during routine therapeutic surgery conducted
at the Thoracic Department of the Xuanwu Hospital of Capital
Medical University (CMU; Beijing, China). All samples were obtained
with informed consent from the patient and with approval from the
Thoracic Department of the Xuanwu Hospital of CMU. This study was
approved by the ethics committee of Xuanwu Hospital of Capital
Medical University (Beijing, China).
Cell culture
The human lung carcinoma cell line A549 was cultured
in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen Inc.,
Carlsbad, CA, USA) supplemented with glutamine, penicillin,
streptomycin and 10% fetal bovine serum (FBS; Invitrogen Inc.).
H460 cells were cultured in RPMI media (Invitrogen Inc.) with 10%
FBS. The cells were maintained in a humidified incubator with 5%
CO2.
Quantitative polymerase chain reaction
(PCR)
RNA was extracted using the mirVana™ miRNA Isolation
kit (Applied Biosystems, Invitrogen Life Technologies, Carlsbad,
CA, USA). cDNA was synthesized from total RNA using the Taqman
miRNA High-Capacity cDNA Reverse Transcription kit (Applied
Biosystems) with primers specific to SENP1 or 18S, an endogenous
control. Quantitative PCR was performed using the Taqman microRNA
PCR system (Applied Biosystems) according to the manufacturer’s
instructions. Briefly, cDNA was combined with Taqman Universal PCR
Master mix and probes specific for SENP1 or 18S (Applied
Biosystems). PCR was performed in 96-well optical plates. SENP1 Ct
values were normalized to 18S Ct values and the relative expression
was calculated using the −Δ ΔCt method. Primers for SENP1 (forward:
TTGGCCAGAGTGCAAATGG; reverse: TCGGCTGTTTCTTGATTTTTGTAA) and the
housekeeping 18S rRNA (Applied Biosystems) were used.
Plasmids and transfection
Human SENP1 was amplified from 293T cDNA library and
subcloned into pcDNA-3.1-Myc plasmid. The transient transfections
were performed by Lipofectamine 2000 (Invitrogen, Shanghai, China),
according to the manufacturer’s instructions.
RNA interference
Two 21-nucleotide SENP1 small interfering RNAs
(siRNAs; si-1: AACTACATCTTCGTGTACCTC and si-2:
CTAAACCATCTGAATTGGCTC) and nonspecific siRNA were synthesized
(Dharmacon, Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
SENP1 and nonspecific siRNA oligos were then inserted into a
pSuppressorNeo vector (Imgenex Corp., San Diego, CA, USA),
according to the manufacturer’s instructions. Following this, the
A549 and H460 cells were transfected with the siRNA plasmid using
Lipofectamine 2000 (Invitrogen Life Technologies).
Flow cytometry
Apoptotic cells were assessed in a flow cytometer
(Becton Dickinson, BD Biosciences, Franklin Lakes, NJ, USA) using a
fluorescein isothiocyanate (FITC)-labeled Annexin-V and propidium
iodide (PI) kit (BD Pharmingen, BD Biosciences).
Western blot analysis
Cells were lysed in sample solution. Following this,
the proteins were separated on 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels,
transferred to nitrocellulose membranes. The membranes were
incubated with the primary antibodies at 4°C overnight, prior to
being incubated with horseradish peroxidase-conjugated secondary
antibodies (Cell signaling Technology, Inc., Danvers, MA, USA) for
1 h at room temperature for detection using the SuperSignal™ West
Pico Chemiluminescent Substrate kit (Pierce, Rockford, IL, USA).
Anti-β-actin and anti-SENP1 antibodies were purchased from Santa
Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), while anti-cleavage
caspase 3 and anti-γ-H2AX antibodies were obtained from Cell
Signaling Technology, Inc.
Bromodeoxyuridine (BrdU) assays
Cell proliferation was evaluated using an
enzyme-linked immunosorbent assay to determine the incorporation of
BrdU during DNA synthesis, in accordance with the manufacturer’s
instructions (BrdU Cell Proliferation Assay kit; Beyotime Institute
of Biotechnology, Shanghai, China). Briefly, 2×103 cells
were plated per well onto 96-well plates. Following 36 h of
transfection, cell proliferation was measured. The absorbance of
the plates at 450 nm was read using a spectrometer.
Colony-forming assay
A colony-forming assay was performed to determine
the radiosensitivity of cells. A549 cells were digested with 0.25%
trypsin, pelleted and then resuspended in 1 ml fresh media. The
trypan blue dye exclusion method was used to determine the cell
viability. Cells were planted at a density of 4×105
cells/ml in six-well dishes and allowed to attach overnight.
Following this, cells were transfected with SENP1 or nonspecific
siRNAs for 24 h and then irradiated (0–10 Gy). Ten days subsequent
to irradiation, cells were fixed using methyl alcohol and stained
with Giemsa staining buffer. A population of >50 cells was
counted as one colony, and the number of colonies was expressed as
a percentage of the number of mock-irradiated cells. The survival
curves were plotted using linear regression analyses. The number of
clones was examined using macroscopic observation and the colony
forming efficiency (CFE) was calculated with the following formula:
CFE = clone number/number of seeded cells.
Radiation exposure
The cells were seeded into 96-well plates and were
treated with a range of radiation doses (0–10 Gy) using a 250 kV
orthovoltage unit the following day (Philips, Amsterdam, The
Netherlands).
Statistical analysis
All the results were derived from at least three
independent experiments and are presented as the mean ± standard
error of the mean. The Student’s t-test was used to compare the
differences between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
SENP1 is overexpressed in lung cancer
tissues
SENP1 has been shown to be overexpressed in several
types of cancer. To determine whether SENP1 was overexpressed in
lung cancer, the mRNA levels of SENP1 in 10 paired lung cancer and
adjacent non-tumor lung tissues were assessed. As depicted in
Fig. 1A, the mRNA levels of SENP1
were significantly increased in the cancer tissues when compared
with the levels in adjacent normal tissues. Western blot analysis
with an anti-SENP1 antibody was then employed to examine the SENP1
protein expression in clinical samples, including five primary lung
cancer samples and their adjacent normal tissues. The results
showed that SENP1 was overexpressed in the lung cancer samples
(Fig. 1B).
SENP1 regulates lung cancer cell
proliferation
To further determine the potential effects of SENP1
in lung cancer cells, H460 cells were transfected with empty vector
(Con) or Myc-SENP1 (Fig. 2A). As
shown in Fig. 2B, SENP1
overexpression led to a significant increase in the cell number of
the H460 cells (Fig. 2B).
Moreover, BrdU analysis also suggested that forced SENP1 expression
promoted cell proliferation (Fig.
2C). Following this, A549 cells were transfected with siRNA
targeting SENP1 to knockdown endogenous SENP1 expression (Fig. 2D and E). As a result of the
downregulation of SENP1, there was a marked reduction in the cell
number and proliferation of the A549 cells (Fig. 2F and G).
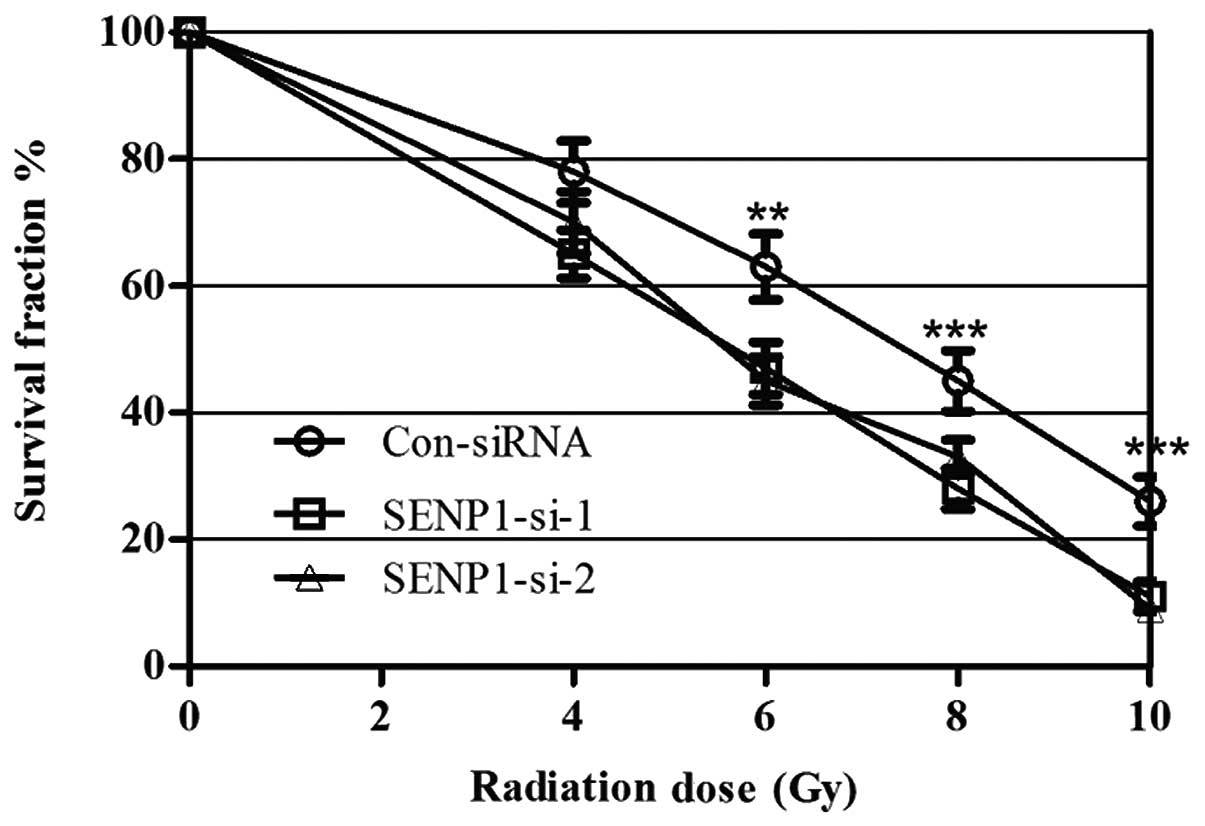
SENP1-silencing sensitizes lung cancer
cells to radiation
To determine whether SENP1 was required for lung
cancer cell radioresistance, A549 cells were treated with different
doses of IR, from 0 to 10 Gy, following transfection with SENP1
siRNA or nonspecific siRNA. Cell proliferation was subsequently
assessed using a colony-forming assay. IR treatment exhibited a
dose-dependent inhibitory effect on the growth of the A549 cells,
and the SENP1 depletion enhanced this effect, suggesting that
inhibition of SENP1 increased A549 cell radio-sensitivity (Fig. 3). Furthermore, identical
experiments were performed using H460 cells, which provided similar
results to those in the A549 cell line, indicating that SENP1
depletion may increase the radiosensitivity of lung cancer
cells.
Inhibition of SENP1 enhances IR-induced
cell cycle arrest, γ-H2AX expression and apoptosis
The underlying mechanisms that may have caused the
SENP1 depletion to increase the radiosensitivity of lung cancer
cells were investigated. Since cells in the G1 and G2 stage are
sensitive to IR (16), cell cycle
analysis was performed to detect whether the cell cycle changed
when SENP1-depleted cells were treated with IR. Unlike the
nonspecific-siRNA-transfected cells, the SENP1-siRNA-transfected
cells treated with IR showed an increased percentage of cells in
the G1 stage, suggesting that the enhanced radiosensitivity of the
A549 cells may have been due to changes in the cell cycle induced
by SENP1 inhibition (Fig. 4A).
γ-H2AX has been demonstrated to be positively associated with tumor
radiosensitivity (17). Therefore,
western blotting was performed to analyze the γ-H2AX expression in
the A549 cells following different treatments.
SENP1-siRNA-transfected cells treated with IR showed the highest
protein level of γ-H2AX (Fig. 4B),
suggesting that increased γ-H2AX expression may be one of the
underlying mechanisms responsible for the increase in the
radiosensitivity of lung cancer cells when SENP1 is inhibited.
Furthermore, the ability of SENP1 inhibition to increase IR-induced
lung cancer cell apoptosis was investigated.
SENP1-siRNA-transfected cells treated with IR were observed to have
an increased apoptotic population (Fig. 4D) and an increased activation of
caspase 3 (Fig. 4C), suggesting
that the increased radiosensitivity may have resulted from an
increased rate of apoptosis.
Discussion
In the present study, it was demonstrated that SENP1
may be a regulator of lung cancer radioresistance. The results
showed that SENP1 mRNA and protein was significantly overexpressed
in lung cancer tissues when compared with their adjacent non-tumor
lung tissues. Moreover, the increased expression of SENP1 in A549
cells significantly increased cell proliferation compared with that
of the control A549 cells, and silencing the SENP1 expression in
the H460 cell line inhibited cell proliferation. Thus, these data
suggested that SENP1 was involved in the regulation of lung cancer
cell proliferation.
Following this, the ability of SENP1 inhibition to
affect lung cancer radioresistance was examined. It was shown that
SENP1 depletion significantly sensitized lung cancer cells to IR
and that SENP1 depletion enhanced IR-induced lung cancer cell cycle
arrest at the G0/G1 stage. Consistent with this observation, a
previous study of SENP1 in colon cancer showed that silencing the
expression of SENP1 upregulated the expression of several CDK
inhibitors, such as p16, p19, p21 and p27 (18). In addition to these cell cycle
regulators, silencing the expression of SENP1 also significantly
increased IR-induced γ-H2AX expression, which has been revealed to
be positively associated with tumor radiosensitivity. Notably,
SENP1 inhibition did not result in lung cancer cell apoptosis, as
demonstrated by fluorescence-activated cell sorting (FACS) assay
and the caspase 3 activation assay. However, silencing the
expression of SENP1 enhanced IR-induced lung cancer cell apoptosis
and caspase 3 activation, which may have contributed to the
IR-induced lung cancer cell sensitivity.
SENP1 is a deSUMOylation enzyme that has been
demonstrated to target a number of key proteins for deSUMOylation
(10,19–22).
SENP1 has also been revealed to be involved in the development and
progression of several types of cancer. In the present study, it
was identified that SENP1 was also overexpressed in lung cancer.
Recently, specific inhibitors of SENP1 have been designed, and
there is consequently a requirement to investigate whether these
inhibitors enhance the radiosensitivity of lung carcinoma. It may
be of interest to assess whether the deSUMOylation activity of
SENP1 is required for the radiosensitization induced by SENP1
knockdown. Further studies are necessary to identify the
SUMOylation target of SENP1 that contributes to the
radiosensitization induced by SENP1 knockdown.
References
|
1.
|
Sculier JP: Nonsmall cell lung cancer. Eur
Respir Rev. 22:33–36. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Saadeddin A: Radiotherapy for NSCLC:
review of conventional and new treatment techniques. J Infect
Public Health. 5(Suppl 1): S45–S49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Hennon MW and Yendamuri S: Advances in
lung cancer surgery. J Carcinog. 11:212012. View Article : Google Scholar
|
|
4.
|
Nadkar A, Pungaliya C, Drake K, et al:
Therapeutic resistance in lung cancer. Expert Opin Drug Metab
Toxicol. 2:753–777. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ryan JL: Ionizing radiation: the good, the
bad, and the ugly. J Invest Dermatol. 132:985–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Verger A, Perdomo J and Crossley M:
Modification with SUMO. A role in transcriptional regulation. EMBO
Rep. 4:137–142. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Wang Y and Dasso M: SUMOylation and
deSUMOylation at a glance. J Cell Sci. 122:4249–4252. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Yeh ET: SUMOylation and De-SUMOylation:
wrestling with life’s processes. J Biol Chem. 284:8223–8227.
2009.PubMed/NCBI
|
|
9.
|
Bawa-Khalfe T and Yeh ET: SUMO losing
balance: SUMO proteases disrupt SUMO homeostasis to facilitate
cancer development and progression. Genes Cancer. 1:748–752. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Cheng J, Kang X, Zhang S and Yeh ET:
SUMO-specific protease 1 is essential for stabilization of HIF1α
during hypoxia. Cell. 131:584–595. 2007.PubMed/NCBI
|
|
11.
|
Yu L, Ji W, Zhang H, et al: SENP1-mediated
GATA1 deSUMOylation is critical for definitive erythropoiesis. J
Exp Med. 207:1183–1195. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bawa-Khalfe T, Cheng J, Lin SH, Ittmann MM
and Yeh ET: SENP1 induces prostatic intraepithelial neoplasia
through multiple mechanisms. J Biol Chem. 285:25859–25866. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Bettermann K, Benesch M, Weis S and
Haybaeck J: SUMOylation in carcinogenesis. Cancer Lett.
316:113–125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Qiao Z, Wang W, Wang L, et al: Design,
synthesis, and biological evaluation of benzodiazepine-based
SUMO-specific protease 1 inhibitors. Bioorg Med Chem Lett.
21:6389–6392. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Chen Y, Wen D, Huang Z, et al:
2-(4-Chlorophenyl)-2-oxoethyl 4-benzamidobenzoate derivatives, a
novel class of SENP1 inhibitors: Virtual screening, synthesis and
biological evaluation. Bioorg Med Chem Lett. 22:6867–6870. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Hanai K, Babazono T, Nyumura I, et al:
Involvement of visceral fat in the pathogenesis of albuminuria in
patients with type 2 diabetes with early stage of nephropathy. Clin
Exp Nephrol. 14:132–136. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Taneja N, Davis M, Choy JS, et al: Histone
H2AX phosphorylation as a predictor of radiosensitivity and target
for radiotherapy. J Biol Chem. 279:2273–2280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Xu Y, Li J, Zuo Y, et al: SUMO-specific
protease 1 regulates the in vitro and in vivo growth of colon
cancer cells with the upregulated expression of CDK inhibitors.
Cancer Lett. 309:78–84. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Li X, Lee YK, Jeng JC, et al: Role for
KAP1 serine 824 phosphorylation and sumoylation/desumoylation
switch in regulating KAP1-mediated transcriptional repression. J
Biol Chem. 282:36177–36189. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Yang Y, Fu W, Chen J, et al: SIRT1
sumoylation regulates its deacetylase activity and cellular
response to genotoxic stress. Nat Cell Biol. 9:1253–1262. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Lindberg MJ, Popko-Scibor AE, Hansson ML
and Wallberg AE: SUMO modification regulates the transcriptional
activity of MAML1. FASEB J. 24:2396–2404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Van Nguyen T, Angkasekwinai P, Dou H, et
al: SUMO-specific protease 1 is critical for early lymphoid
development through regulation of STAT5 activation. Mol Cell.
45:210–221. 2012.PubMed/NCBI
|