Introduction
Alcoholic cirrhosis (AC) is an end-stage alcoholic
liver disease (ALD) caused by long-term excessive drinking. It
manifests as chronic inflammation and progressive fibrosis of the
liver tissue and is the leading cause of mortality in chronic
alcoholics. A 48-month prospective study of 280 patients with
severe ALD in the United States revealed that 30% of the patients
had alcoholic fatty liver disease, that more than half of those
individuals developed cirrhosis and that two-thirds of the patients
with cirrhosis developed alcoholic hepatitis and ultimately
succumbed (1).
AC has a complex pathogenesis and has been indicated
to be the result of the external environment (alcoholism) and
genetic factors (2). Therefore, it
is important to study the genetic components of AC to identify
susceptible populations according to genotypes; such findings may
be significant for the prevention and treatment of AC. At present,
a number of genetic factors, including alcohol dehydrogenase,
aldehyde dehydrogenase (3),
apolipoprotein E (4) and
cytochrome 450-2E1 (CYP2E1) gene polymorphisms (5,6), as
well as superoxide dismutase gene dimorphisms (7), have been demonstrated to be
associated with AC susceptibility. These genetic studies have
focused on polymorphisms in enzymes involved in alcohol metabolism;
there have been few studies on genetic factors affecting hepatocyte
susceptibility to alcohol damage.
We previously demonstrated that hepatic
mitochondrial DNA (mtDNA) in patients with AC exhibited reduced
copy numbers, large deletions from bases 749 to 15,486 and a
downregulation of one of its encoding products, cytochrome c
oxidase 2. By contrast, no significant mtDNA damage was observed in
chronic alcoholics without AC. The results suggested that specific
mtDNA damage may be an important pathogenic factor in AC (8). mtDNA replication, transcription and
repair are regulated by nuclear genes (9), primarily mitochondrial transcription
factor A (mtTFA) (10) and nuclear
respiratory factor 1 (NRF-1) (11). mtTFA is a nuclear-encoded factor
that is central to mtDNA expression, regulation and repair
(10). Therefore, we hypothesized
that there may be mtTFA single nucleotide polymorphisms (SNPs)
unique to patients with AC that affect the expression of mtTFA and
its ability to repair damaged mtDNA in liver cells, thereby
affecting normal mitochondrial functions and contributing to the
pathogenesis of AC.
In this study, we aimed to screen SNPs in mtTFA in
patients with AC and analyze their impact on hepatic mtDNA copy
number and genetic susceptibility to AC. Our results are likely to
enhance the understanding of AC pathogenesis and provide a new
approach to the clinical prevention of the disease.
Materials and methods
Patients and clinical specimen
collection
Study subjects were enrolled from Daping Hospital,
Third Military Medical University (Chongqing, China), between June
2007 and June 2011. All experimental procedures were approved by
the Medical Ethics Committee of the Third Affiliated Hospital of
the Third Military Medical University. Written informed consent was
obtained from each subject prior to specimen collection. Due to the
fact that alcoholism is much more common in males than females in
this region, all specimens were collected from male subjects. Blood
(5 ml) was collected by venipuncture for liver function tests and
mtTFA SNP analysis. Liver biopsies were collected for pathological
examination and assessment of mtTFA expression and mtDNA copy
number. AC was diagnosed according to the 2006 ALD diagnostic
criteria established by the Chinese Medical Association (12), as follows: i) a long-term history
of heavy drinking and alcohol intake ≥40 g/day for more than five
consecutive years; ii) hypohepatia and portal hypertension,
cirrhosis confirmed by imaging and alcoholic liver injury confirmed
by a serum enzyme test; and iii) negative for hepatitis B and C
antigens and antibodies and DNA tests (to exclude patients with
cirrhosis due to other causes). Alcohol intake was calculated as
follows: Alcohol intake = alcoholic drink intake (ml) × alcohol
percentage × 0.8.
Three groups were assessed in this study. Group A
(normal subjects): No long-term alcohol consumption, no liver
diseases and no significant liver changes observed during upper
abdominal surgery for other diseases (e.g. gallbladder stones). A
total of 50 peripheral blood and 25 liver tissue specimens were
collected from this group. Group B (chronic alcoholics without AC):
Chronic alcoholism (confirmed according to the diagnostic criteria
listed previously), normal liver function test results and no
significant liver changes observed using imaging or during upper
abdominal surgery for other diseases. A total of 50 peripheral
blood and 26 liver tissue specimens were collected from this group.
Group C (patients with AC): The three inclusion criteria described
previously were met and liver scarring was confirmed during upper
abdominal surgery for AC or other diseases. A total of 50
peripheral blood and 22 liver tissue specimens were collected from
this group. For pathological examination, the liver specimens fixed
in 10% formalin were embedded in paraffin. Sections of 4 μm were
cut and stained with hematoxylin and eosin. Light microscopic
evaluation was performed on the sections in a blinded fashion to
assess liver parenchymal changes.
Analysis of mtTFA SNPs
Genomic DNA extraction
A Universal Genomic DNA Extraction kit (Takara
Biotechnology Ltd., Dalian, China) was used to extract total DNA
from peripheral venous blood, in accordance with the kit’s
instructions, and the extracted DNA was stored at −20°C.
Polymerase chain reaction (PCR)
primers
Fifteen primers (Table
I) were designed according to the full-length sequences of
mtTFA (gi: 224589801) to perform segmented full-length PCR
amplification of mtTFA.
 | Table IPrimer sequences for the segmented
full-length polymerase chain reaction (PCR) amplification of
mitochondrial transcription factor A (mtTFA). |
Table I
Primer sequences for the segmented
full-length polymerase chain reaction (PCR) amplification of
mitochondrial transcription factor A (mtTFA).
| No. | Forward | Reverse |
|---|
| 1 | TAC CTT CGA TTT TCT
AAA | TCA ACA AAC ATC CTA
CCT |
| 2 | GAT AAA TCC TTT CTT
GTC T | AAT ACA TTC TCG ATC
CAT |
| 3 | CGG TTA AGA TGA AGA
AGG | CCA ACT AAT TTA AAC
GTA AGT A |
| 4 | TAC TCC TTA GTT GTT
AGA T | AGT AAG TCA ACA AAC
CAT |
| 5 | AGA GCA CAT TTT CCA
CCT | CCA TAT CAA ACT CAC
CAT |
| 6 | TGA AAC AGT CAA ACC
AGG AG | GGG AGA ATG AAA GAG
GGA |
| 7 | GGA GCC AGA AAG ATA
CTA | TTT AAA GCT CCA TAG
TTG |
| 8 | AAT GCA ATA TCA CTC
CCT | GAA TCA CCC TTA GCT
TCT |
| 9 | CAG GAA AAG TCT AGA
GTG | AAA TCA AGA AGC AAC
AAT |
| 10 | TTC CAT ATC CCT AAA
TAA C | CAA TAA ATC CCT ATA
CCT T |
| 11 | GTA AAG GTG CAT GGG
AGA | AAT TAG CCA GGC TTG
GTG |
| 12 | CAA CCT CTT GAG TAG
CCA | TGA AAA GCA GAT GCA
TTA |
| 13 | ACA ATT AGC TTT CTT
TTG TC | CTG TGC TTC AGT GTT
TAG |
| 14 | ATA AAA CCT AAA GCT
ACA | GAA TGA AAT TGT TAC
AAG T |
| 15 | TTC TCC AGT CTG CCT
TTA | CCT TTA TCT GGG TTT
TCC |
PCR system and conditions
The Tiangen 2X Taq PCR MasterMix [Tiangen Biotech
(Beijing) Co., Ltd., Beijing, China] was used for PCR
amplification. The loading system included 1.5 μl template DNA, 1
μl each of forward and reverse primers (10 μM), 12.5 μl 2X Master
Mix and 9 μl H2O. The total reaction volume was 25 μl.
The amplification consisted of initial denaturation for 3 min at
94°C, 30 cycles of denaturation for 30 sec at 94°C, primer
annealing for 30 sec at 58°C and primer extension for 1 min at
72°C, prior to primer extension for 5 min at 72°C after 30 cycles.
A total of 5 μl PCR products were then mixed with 2.5 μl 6X loading
buffer and the mixture was subsequently analyzed by 1.2% agarose
gel electrophoresis (6 V/cm, 20 min).
All PCR products were purified prior to being
submitted to Sangon Biotechnology Co., Ltd. (Shanghai, China) for
sequencing. The resulting sequences were compared with genomic
sequences of normal subjects in GenBank (http://www.ncbi.nlm.nih.gov/gene/7019) to identify
SNPs in each group. Samples from the three groups were compared to
screen for mtTFA SNPs unique to patients with AC. Gene analysis was
performed using Cluxtal X (SNPStats, web tool for SNP analysis.
http://bioinfo.iconcologia.net/snpstats/start.htm) and
mtTFA SNP frequency was analyzed in patients with AC.
Detection of mtTFA expression in
hepatocytes by quantitative PCR (qPCR)
Due to the fact that liver tissue was only collected
from 22 patients with AC, we randomly selected 22 cases from the
normal controls and chronic alcoholics without AC.
Total RNA was isolated from liver cells using
guanidine isothiocyanate extraction and randomly selected samples
were evaluated using electrophoresis. Portions of the samples were
reverse transcribed into cDNA using PrimeScript Reverse
Transcriptase (Takara Biotechnology Ltd.), in accordance with the
manufacturer’s instructions. The PCR primers used were as follows:
mtTFA forward, 5′-AGATTCCAAGAAGCTAAG GGTGATT-3′ and reverse,
5′-TTTCAGAGTCAGACAGAT TTTTCCA-3′; β-actin forward,
5′-GATGACCCAGATCAT GTTTGAG-3′ and reverse, 5′-AGGGCATACCCCTCGTA
GAT-3′. SYBR Green qPCR Master Mix (2X) from Ruian Biotechnologies
Co., Ltd. (Shanghai, China) and the ABI 7500 Fast Real-Time PCR
system (Applied Biosystems, Life Technologies, Carlsbad, CA, USA)
were used for qPCR. Amplification was performed in 25-μl reaction
mixtures containing 12.5 μl SYBR Green qPCR Master Mix, 0.5 μl each
of forward and reverse primers (10 μM), 9.5 μl ddH2O and
2 μl template (cDNA). The following cycling protocol was used: 2
min at 95°C, followed by 40 cycles consisting of 10 sec at 95°C and
40 sec at 60°C. The relative quantitative mtTFA value was
represented by the ratio of mtTFA to β-actin. Three parallel
experiments were conducted for each sample.
Western blot analysis of mtTFA in
liver tissue
Denatured protein samples (50 mg) were subjected to
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE), transferred under semi-dry conditions onto
nitrocellulose (NC) membranes and visualized using Ponceau-S
staining. Following this, the NC membranes were blocked with
Tris-buffered saline solution (TBS) containing 5% non-fat dried
milk overnight, prior to being incubated with mtTFA antibody (1:400
dilution) at 4°C overnight and washed in TBS with Tween (TBST)
three times (10 min each). The membranes were subsequently
incubated with goat anti-rabbit immunoglobulin (Ig) G (PICPI31462 -
Pierce Peroxidase Affinity-Purified Polyclonal Antibodies - HRP,
Thermo Fisher Scientific Inc., Waltham, MA, USA; 1:5,000 dilution)
at room temperature for 1 h and washed a further three times in
TBST (10 min each). The blots were visualized using enhanced
chemiluminescence substrate (PICPI32209 - Pierce ECL Western
Blotting Substrate, Thermo Fisher Scientific Inc.) and X-ray films.
Bands were analyzed according to their optical density and the
values were normalized to β-actin bands.
Detection of hepatocyte mtDNA copy
number by qPCR
A Universal Genomic DNA Extraction kit (Takara
Biotechnology Ltd.) was used to extract total DNA from the liver
tissues, in accordance with the kit’s instructions. The extracted
DNA was stored at −20°C.
Due to the fact that the mtDNA displacement (D)-loop
region is highly conserved, it was used as a surrogate for the
mtDNA copy number. The nuclear β-globin served as an internal
control. The PCR primers used were as follows: mtDNA forward,
5′-TTGCACGGTACCATAAATACTTGAC-3′ and reverse,
5′-GAGTTGCAGTTGATGTGTGATAGTTG-3′; β-globin forward,
5′-CAACTTCATCCACGTTCACC-3′ and reverse, 5′-CAACTTCATCCACGTTCACC-3′.
The hepatic mtDNA copy number was determined using qPCR with a
SYBR® Premix Ex Taq™ II (Perfect Real-Time) kit from
Takara Biotechnology Ltd. The PCR system included 12.5 μl
SYBR® Premix Ex Taq™, 2 μl DNA template (~100 ng), l μl
each of forward and reverse primers (final concentration, 0.4
μmol/l) and 8.5 μl H2O. The total reaction volume was 25
μl. qPCR was performed using a Bio-Rad CFX96 real-time PCR system
(Bio-Rad, Hercules, CA, USA) at conditions of 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec, 55°C for 30 sec and 72°C
for 30 sec. The relative quantitative mtTFA value was represented
by the ratio of mtTFA to β-globin. Three parallel experiments were
conducted for each sample.
Statistical analysis
All data are presented as the mean ± standard
deviation (SD). Groups were compared using one-way analysis of
variance and Student-Newman-Keuls tests, with P<0.05 considered
to indicate a statistically significant difference.
Results
Study subject clinical
characteristics
Table II
summarizes the clinical and biochemical characteristics of the
three groups. There were no significant differences in age among
the three groups or in the duration of alcoholism (years) or daily
intake of alcohol (g) between groups B and C. The patients in group
C had significantly impaired liver function, as evidenced by
increased levels of total bilirubin and aspartate aminotransferase
(AST) and the decreased albumin level (Table II). Among the patients with AC,
there were 36 (72%) with a history of upper gastrointestinal
bleeding, 23 (46%) with ascites and 26 (52%) with
hypersplenism.
 | Table IIClinical and biochemical
characteristics of the study subjects. |
Table II
Clinical and biochemical
characteristics of the study subjects.
| Characteristic | Group A (Normal
controls) | Group B (Alcoholics
without AC) | Group C (AC) |
|---|
| Number of cases | 50 | 50 | 50 |
| Gender | Male | Male | Male |
| Age (years) | 50.35±11.96
(34–81) | 48.78±11.82
(30–73) | 52.98±12.13
(33–83) |
| Average daily alcohol
intake (g) | - | 109.80±44.65 | 114.40±55.11 |
| Years of
alcoholism | - | 24.90±10.08 | 25.80±12.26 |
| Total bilirubin
(μmol/l) | 18.32±9.74 | 17.78±7.60 | 34.32±16.43a |
| ALT (U/L) | 35.75±13.02 | 35.78±19.64 | 29.41±16.10 |
| AST (U/L) | 36.86±14.83 | 37.74±12.50 | 49.85±12.48a |
| Albumin (g/l) | 38.16±5.32 | 37.45±5.88 | 30.75±6.06a |
Eighteen patients underwent splenectomy and
pericardial devascularization and two patients in group C underwent
laparoscopic cholecystectomy during the liver function compensation
stage. A further two patients in group C received emergency
splenectomy and pericardial devascularization due to upper
gastrointestinal bleeding that was not controlled by conservative
non-surgical treatment. In groups A and B, 25 and 26 patients,
respectively, underwent laparoscopic cholecystectomy. Liver tissue
specimens were collected during surgery.
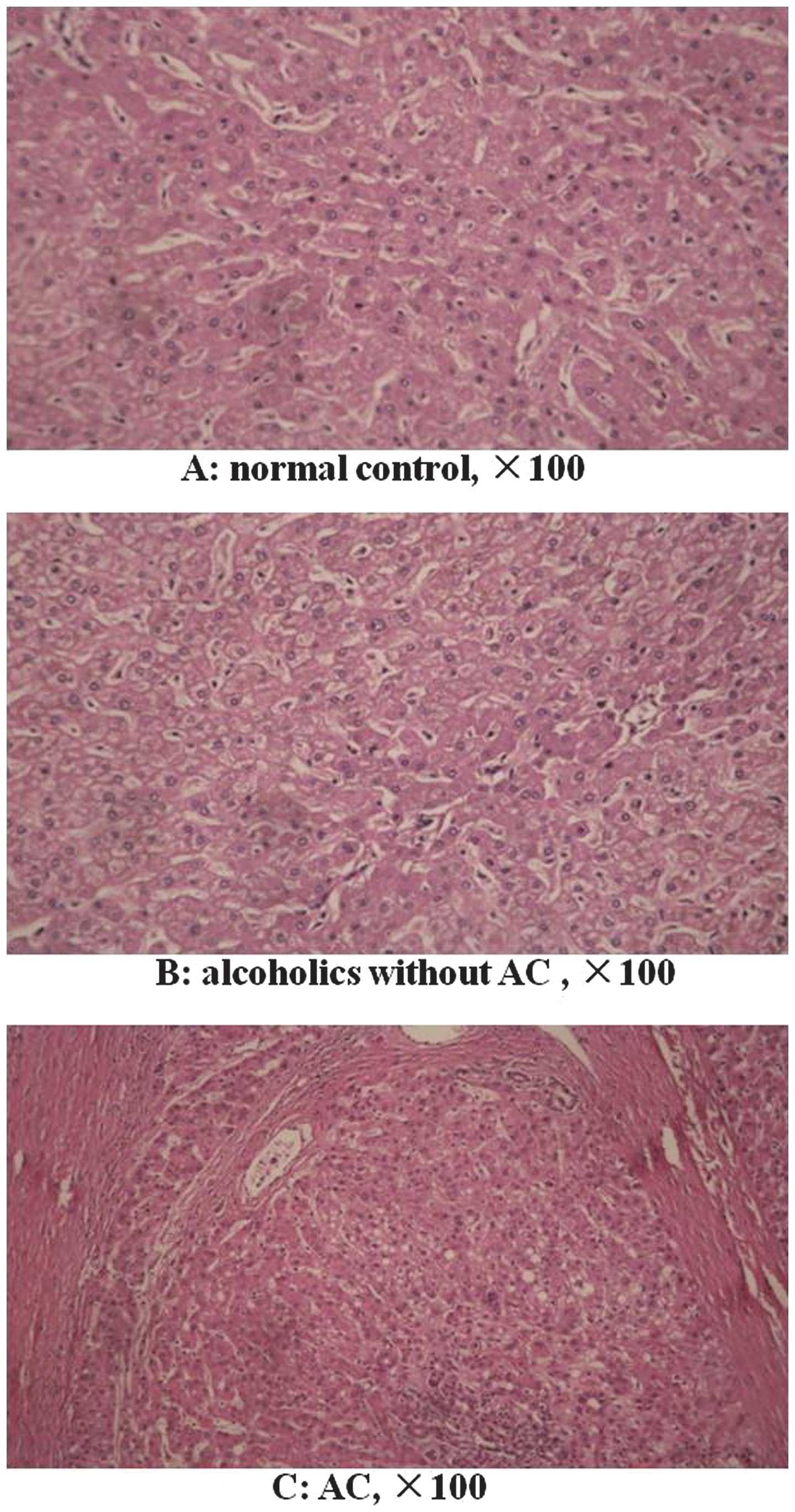
Pathological examination
All liver tissue specimens were pathologically
examined. There were no morphological cirrhotic changes in the
specimens from the normal control and alcoholics without AC groups
(Fig. 1A and B). In the normal
control group, certain patients showed a normal morphological liver
tissue structure, while a number had mild steatosis. In the
alcoholics without AC group, certain patients exhibited a normal
morphological liver tissue structure, while steatosis was observed
in others. In the AC group, all patients showed morphological
changes in the liver tissue, including typical pseudolobule
formation (Fig. 1C).
SNPs unique to patients with AC
The three groups were compared to screen 72 base
mutations unique to group C that were not present in groups A and
B. Among them, 18 SNPs occurred in patients with AC at frequencies
>10% (Table III).
 | Table IIISNPs of mtTFA unique to patients with
AC (frequency >10%). |
Table III
SNPs of mtTFA unique to patients with
AC (frequency >10%).
| Locus | Mutation | Number | Frequency (%) | dbSNP |
|---|
| 664 | G-A | 9 | 18 | |
| 678 | G-A | 7 | 14 | |
| 2542 | A-G | 5 | 10 | |
| 2557 | C-T | 9 | 18 | |
| 2582 | G-A | 16 | 32 | |
| 2596 | A-T | 5 | 10 | rs189223626 |
| 2601 | A-G | 7 | 14 | rs139675989 |
| 2641 | G-A | 5 | 10 | |
| 2665 | C-G | 5 | 10 | rs193210579 |
| 2930 | A-C | 8 | 16 | rs145188595 |
| 3278 | G-A | 11 | 22 | rs139514719 |
| 3361 | T-G | 5 | 10 | |
| 4985 | A-T | 10 | 20 | |
| 5068 | G-T | 5 | 10 | rs140714664 |
| 5106 | A-G | 5 | 10 | |
| 5217 | C-T | 7 | 14 | rs19060687 |
| 5223 | C-A | 8 | 16 | |
| 7648 | A-T | 6 | 12 | |
Table III shows
that mtTFA SNPs were present from bases 664 to 678, 2,542 to 3,361,
4,985 to 5,223 and at base 7,648 in patients with AC. Among the 18
SNPs present in the full-length 10,722-bp mtTFA, seven were known
in the SNP Database (dbSNP) and 11 were newly discovered. According
to the mtTFA gene information provided by GenBank, the SNPs at
3,278 and 3,361 were located in the same coding region, while the
remaining SNPs were located in non-coding regions.
mtTFA SNPs unique to patients with AC and
normal subjects
We also screened 28 base mutations unique to groups
A and C that were not present in group B. Five mutations occurred
in group C at frequencies >10% (Table IV); one was in the dbSNP and four
were novel. Due to the fact that group A did not have a long-term
history of alcohol abuse, if the individuals with these five
mutations in group A were to develop AC following alcohol abuse,
these mtTFA SNPs may be considered to be a contributing factor to
AC. If the individuals were not to develop AC, it may be that these
SNPs have other genetic influences. While our results suggest that
these five mtTFA SNPs may be specific to AC, further investigations
are required.
 | Table IVmtTFA SNPs unique to patients with AC
and normal subjects (frequency >10%). |
Table IV
mtTFA SNPs unique to patients with AC
and normal subjects (frequency >10%).
| | Number | Frequency (%) | |
|---|
| |
|
| |
|---|
| Locus | Mutation | Group A | Group C | Group A | Group C | dbSNP |
|---|
| 680 | G-A | 2 | 6 | 4 | 12 | |
| 2344 | C-T | 7 | 9 | 14 | 18 | rs184602713 |
| 2652 | A-G | 5 | 10 | 10 | 20 | |
| 3261 | G-T | 2 | 7 | 4 | 14 | |
| 6836 | C-G | 7 | 5 | 14 | 10 | |
mtTFA SNP distribution in patients with
AC
The number of unique mtTFA SNPs was calculated to be
between zero and six in each patient with AC. It was determined
that 70% had between two and four SNPs (Table V).
 | Table VSpecific mtTFA SNP distributions in
patients with AC (n=50). |
Table V
Specific mtTFA SNP distributions in
patients with AC (n=50).
| Number of SNPs | Number of cases
(%) |
|---|
| 0 | 5 (10) |
| 1 | 6 (12) |
| 2 | 13 (26) |
| 3 | 11 (22) |
| 4 | 11 (22) |
| 5 | 2 (4) |
| 6 | 2 (4) |
| Total | 50 (100) |
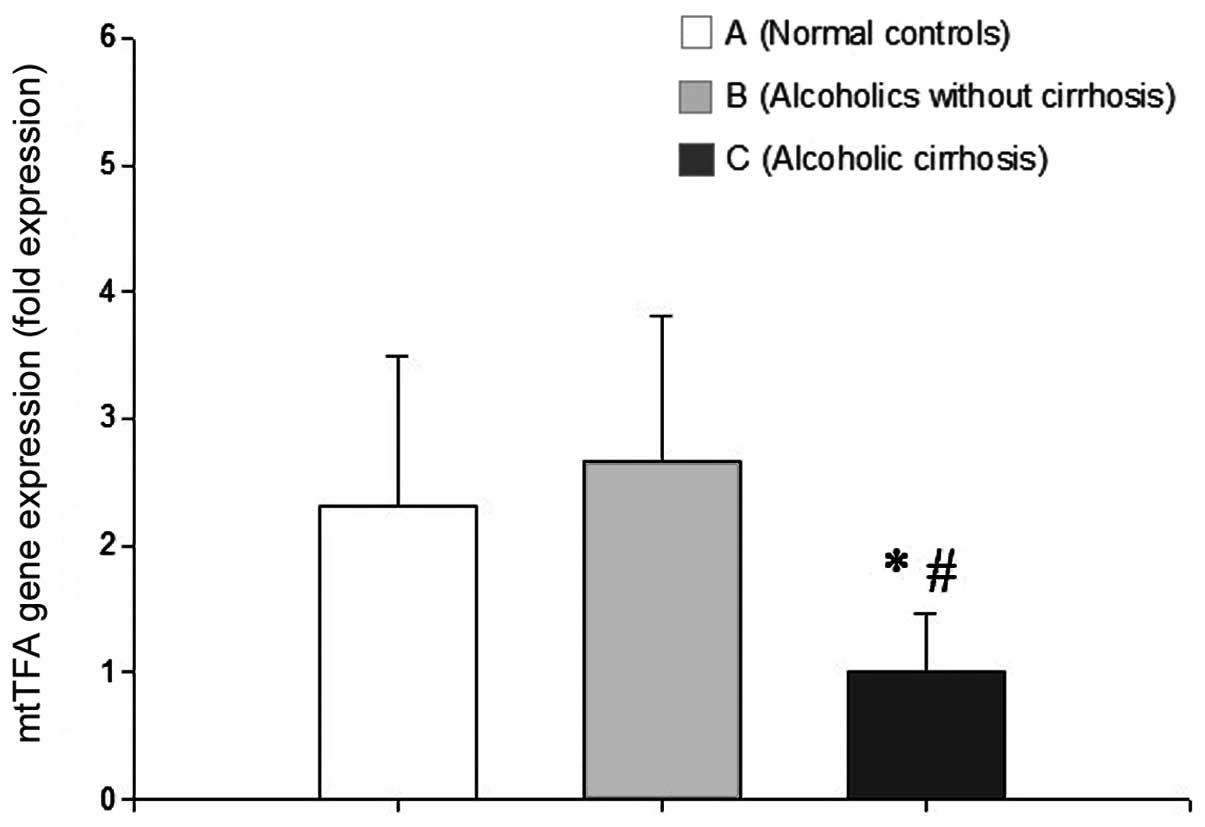
Analysis of mtTFA mRNA expression using
qPCR
No significant difference was identified in the
mtTFA mRNA level between the normal control and alcoholics without
AC groups (P>0.05). By contrast, mtTFA mRNA expression was
significantly lower in the AC group than in the normal control and
the alcoholics without AC groups (P<0.05 for each; Fig. 2).
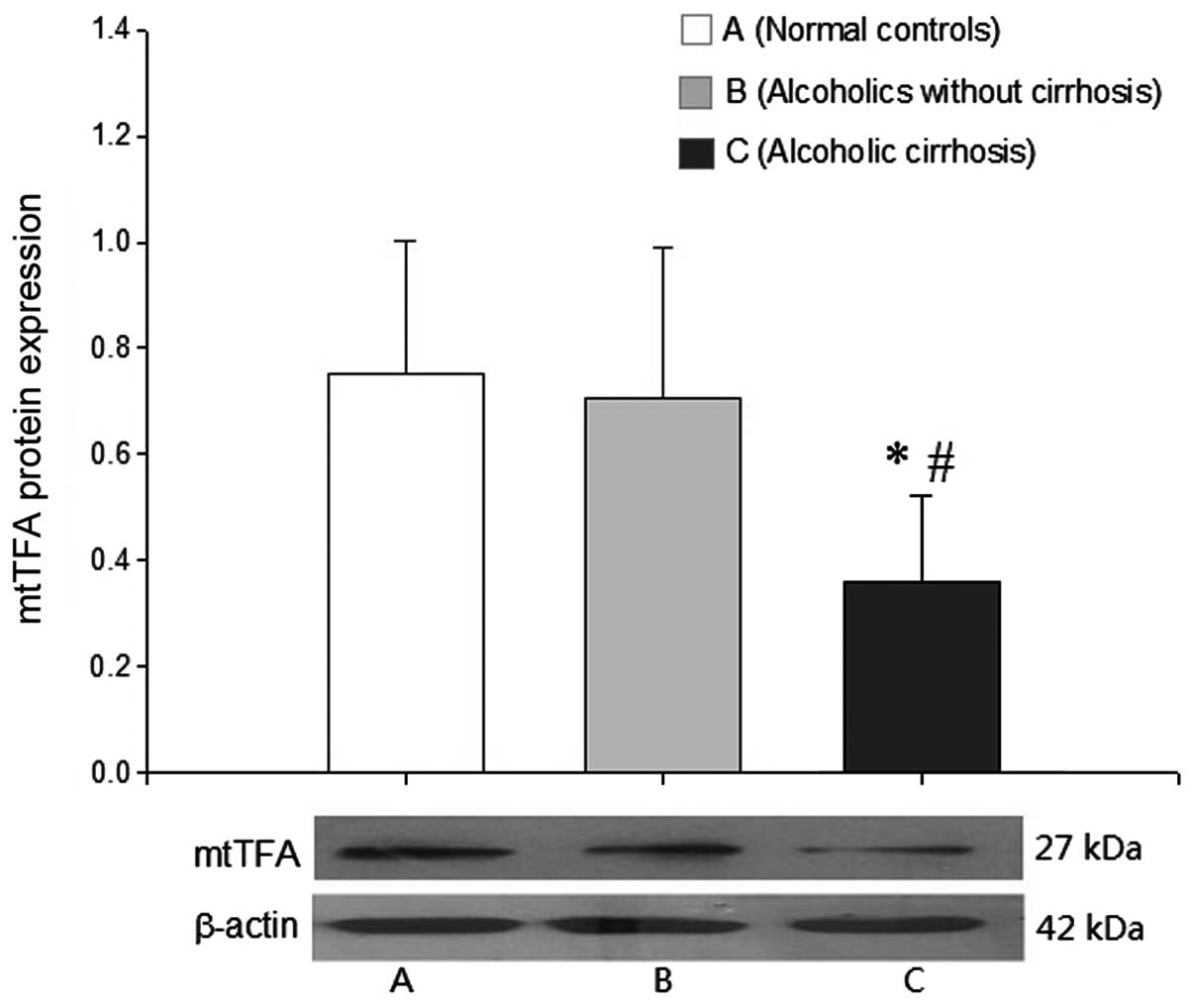
Analysis of mtTFA protein expression
using western blotting
Western blot analysis did not demonstrate a
significant difference in the level of mtTFA protein expression
between the normal control and the alcoholics without AC groups. By
contrast, the levels of mtTFA protein expression in the AC group
were significantly decreased compared with those in the other two
groups (P<0.05 for each; Fig.
3).
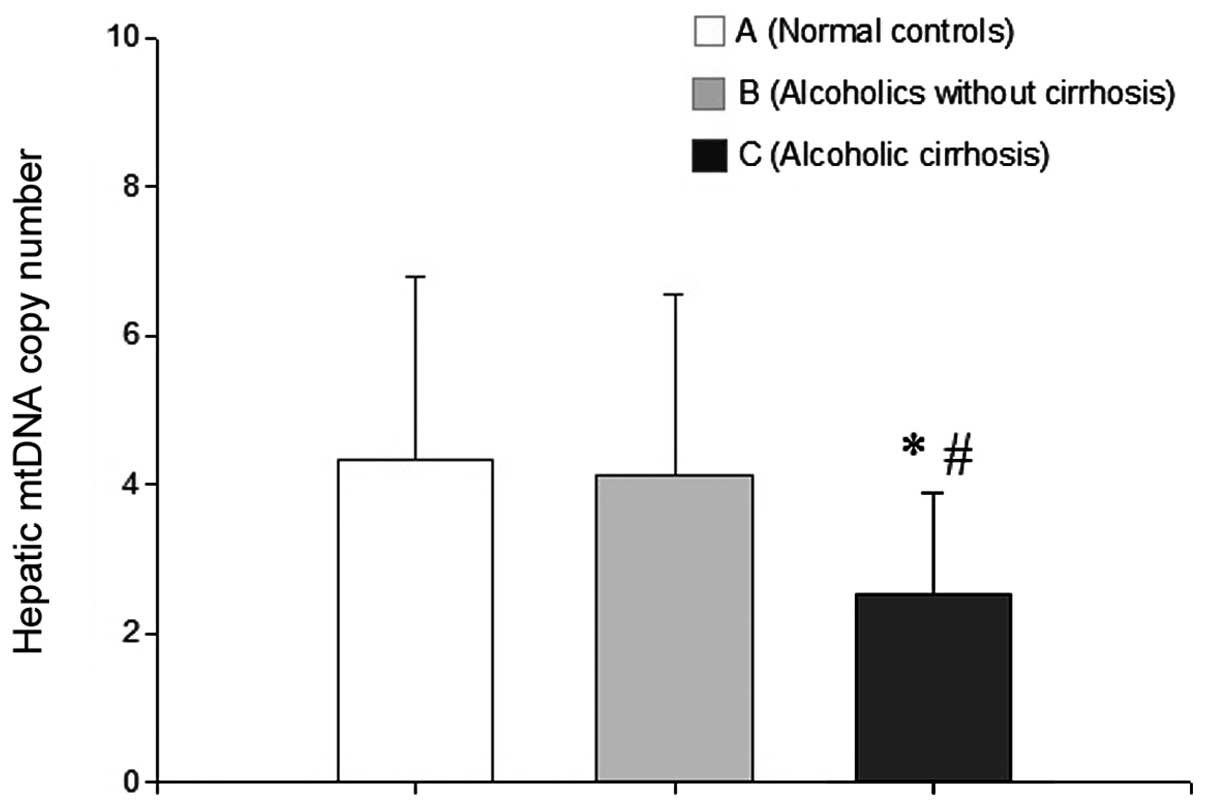
Analysis of mtDNA copy number using
qPCR
No significant difference in mtDNA copy number was
identified between the normal control and alcoholics without AC
groups. However, the mtDNA copy number was significantly lower in
the AC group than in the normal control and the alcoholics without
AC groups (P<0.05 for each; Fig.
4).
Discussion
mtTFA is a nuclear-encoded factor that exerts
powerful effects on mtDNA; it promotes mtDNA transcription and
expression (13), increases mtDNA
copy number (14), maintains
integrity and stability (15),
protects and repairs damaged mtDNA and restores mitochondrial
function (16). Therefore, mtTFA
genetic variation is likely to affect mtDNA number, structure and
repair, thereby altering mitochondrial structure and function. As a
result, this affects the extent of ethanol-induced liver cell
damage.
SNPs are DNA sequence variations that occur when a
single nucleotide differs among members of a biological species.
Since it was first described in 1994 (17), SNP analysis has become a focus in
fields that assess molecular markers. As a third generation genetic
marker, SNPs are characterized by their high density and
conservedness in the genome. Statistically, an SNP occurs every
kilobase and it is estimated that there are more than three million
SNPs in the entire three billion-base human genome. The majority of
SNPs are located in non-coding regions; however certain SNPs are
present in gene promoters and are thus able to increase or decrease
gene transcription activity, thereby affecting protein expression
and biological activity (18). A
number of SNPs located in protein coding regions may change the
amino acid sequence of key functional groups, which may
subsequently affect protein function (19) and ultimately influence
susceptibility to specific environments or pathogens.
The 10,722-bp human mtTFA gene is located on
chromosome 10. Based on the estimated frequency of SNPs, it is
predicted that there are ~10 SNPs in the mtTFA gene. If certain
SNPs affect normal mtTFA expression, they may also affect
mitochondrial function via their effects on mtDNA, thus leading to
cell dysfunction and damage. A genetic analysis of cattle revealed
that specific SNPs in the mtTFA gene promoter region caused a
decline in mtDNA copy number (20), while certain SNPs in cattle mtTFA
affected animal growth and development (21,22).
Clinical studies have demonstrated that mtTFA SNPs may cause a
shortage of intracellular mtDNA copies, which is a risk factor for
Alzheimer’s disease (23–25). However, the existence of specific
mtTFA SNPs in human patients with AC has not been described, to
date.
In this study, 18 mtTFA SNPs were identified in 50
patients with AC, including five possible AC-specific SNPs. These
SNPs occurred at a frequency of 0.168%, which is significantly
higher than ~0.1% in the normal population. Two of the SNPs were
located in the same coding region and were likely to affect normal
mtTFA expression. Although the remaining SNPs were located in
non-coding regions, they were present at particular base sequences.
Two were located at bases 664 to 678 (total length of 14 bp), eight
were located at bases 2,542 to 2,930 (total length of 389 bp) and
five were at bases 4,985 to 5,223 (total length of 239 bp). SNPs
occurring in such short base sequences at such a high frequency are
also likely to affect normal mtTFA expression, despite the fact
that they are in non-coding regions. In addition, the analysis of
mtTFA SNP distribution in patients with AC demonstrated that these
SNPs were not present in five patients (10%), suggesting that the
underlying causes of AC are complex and multifactorial (3–7).
mtTFA mRNA and protein levels were measured in liver
tissue using qPCR and western blotting, respectively, to
investigate whether mtTFA SNPs in patients with AC affected mtTFA
expression. The results showed that mRNA and protein levels were
significantly lower in the AC group than in the normal control and
alcoholics without AC groups (P<0.05 for each). Appropriate
mtTFA expression is required for mtDNA stability and mtTFA protein
level directly affects mtDNA copy number (26). Decreased expression of mtTFA
reduces the ability to repair mtDNA damage and results in a
consequent reduction in the mtDNA copy number. Notably, it was
observed that the mtDNA copy number was significantly reduced in
patients with AC.
There are a number of reasons why long-term alcohol
abuse may result in mtDNA damage. Oxidative stress is believed to
be important in AC pathogenesis and development, and mitochondria
are particularly vulnerable to oxidative stress (27). Excessive oxidative stress injury
may induce apoptosis, the main mechanism of progressive hepatic
injury, ultimately leading to cirrhosis (28). Furthermore, the liver is the organ
that is primarily responsible for metabolizing ethanol. Hepatocytes
oxidize ethanol to acetaldehyde via alcohol dehydrogenase,
subsequently producing acetic acid via acetaldehyde dehydrogenase
and ultimately carbon dioxide and water (29). A high plasma ethanol concentration
following heavy drinking may also activate the microsomal ethanol
oxidizing system (MEOS), which catalyzes acetaldehyde production.
However, ethanol-induced MEOS activity fails to oxidize ethanol to
produce ATP and also increases oxygen and nicotinamide adenine
dinucleotide phosphate (NADPH) consumption. This results in cell
hypoxia and increased levels of oxygen free radicals (29), which are the most important factor
causing mtDNA damage. In addition, acetaldehyde produced from
ethanol metabolism may damage the antioxidant defense system and
directly bind to DNA, thus inhibiting its repair (30).
The factors mentioned previously may all lead to
hepatic mtDNA damage. Specific mtTFA SNPs may decrease mtTFA
expression, resulting in a reduced ability to effectively repair
mtDNA damaged by chronic alcoholism and a reduction in hepatic
mtDNA copy number. Moreover, we previously demonstrated that
hepatic mtDNA in patients with AC exhibited reductions in copy
number and large deletions, in addition to a downregulation of one
of its encoding products (8).
In conclusion, specific mtTFA SNPs may decrease
mtTFA expression, resulting in an inability to effectively repair
mtDNA damaged by chronic alcoholism. These changes may increase
susceptibility to AC.
Acknowledgements
This study was supported by the National Natural
Science Fund of China (project number: 30801114).
References
|
1
|
Chedid A, Mendenhall CL, Gartside P,
French SW, Chen T and Rabin L: Prognostic factors in alcoholic
liver disease. VA Cooperative Study Group. Am J Gastroenterol.
86:210–216. 1991.PubMed/NCBI
|
|
2
|
Stickel F and Osterreicher CH: The role of
genetic polymorphisms in alcoholic liver disease. Alcohol Alcohol.
41:209–224. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cichoz-Lach H, Partycka J, Nesina I,
Celinski K, Slomka M and Wojcierowski J: Alcohol dehydrogenase and
aldehyde dehydrogenase gene polymorphism in alcohol liver cirrhosis
and alcohol chronic pancreatitis among Polish individuals. Scand J
Gastroenterol. 42:493–498. 2007. View Article : Google Scholar
|
|
4
|
Hernández-Nazará ZH, Ruiz-Madrigal B,
Martínez-López E, Roman S and Panduro A: Association of the epsilon
2 allele of APOE gene to hypertriglyceridemia and to early-onset
alcoholic cirrhosis. Alcohol Clin Exp Res. 32:559–566.
2008.PubMed/NCBI
|
|
5
|
Cichoz-Lach H, Partycka J, Nesina I,
Celiński K and Slomka M: The influence of genetic polymorphism of
CYP2E1 on the development of alcohol liver cirrhosis. Wiad Lek.
59:757–761. 2006.(In Polish).
|
|
6
|
Khan AJ, Ruwali M, Choudhuri G, Mathur N,
Husain Q and Parmar D: Polymorphism in cytochrome P450 2E1 and
interaction with other genetic risk factors and susceptibility to
alcoholic liver cirrhosis. Mutat Res. 664:55–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nahon P, Sutton A, Pessayre D, et al:
Genetic dimorphism in superoxide dismutase and susceptibility to
alcoholic cirrhosis, hepatocellular carcinoma, and death. Clin
Gastroenterol Hepatol. 3:292–298. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang C, Liang X, Liu H, Guo L, Pi R and
Yang J: Changes in mitochondrial DNA and its encoded products in
alcoholic cirrhosis. Int J Clin Exp Med. 5:245–250. 2012.PubMed/NCBI
|
|
9
|
Polimeno L, Margiotta M, Marangi L, et al:
Molecular mechanisms of augmenter of liver regeneration as
immunoregulator: its effect on interferon-gamma expression in rat
liver. Dig Liver Dis. 32:217–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang D, Kim SH and Hamasaki N:
Mitochondrial transcription factor A (TFAM): roles in maintenance
of mtDNA and cellular functions. Mitochondrion. 7:39–44. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Choi YS, Kim S, Kyu Lee H, Lee KU and Pak
YK: In vitro methylation of nuclear respiratory factor-1 binding
site suppresses the promoter activity of mitochondrial
transcription factor A. Biochem Biophys Res Commun. 314:118–122.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
No authors listed. Fatty liver and
alcoholic liver disease study group of the Chinese Liver Disease
Association. Guidelines for diagnosis and treatment of alcoholic
liver diseases. Chinese Journal of Hepatology. 14:164–166.
2006.
|
|
13
|
Garstka HL, Schmitt WE, Schultz J, et al:
Import of mitochondrial transcription factor A (TFAM) into rat
liver mitochondria stimulates transcription of mitochondrial DNA.
Nucleic Acids Res. 31:5039–5047. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ekstrand MI, Falkenberg M, Rantanen A, et
al: Mitochondrial transcription factor A regulates mtDNA copy
number in mammals. Hum Mol Genet. 13:935–944. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alam TI, Kanki T, Muta T, et al: Human
mitochondrial DNA is packaged with TFAM. Nucleic Acids Res.
31:1640–1645. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suliman HB, Carraway MS and Piantadosi CA:
Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J
Respir Crit Care Med. 167:570–579. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nikiforov TT, Rendle RB, Goelet P, et al:
Genetic Bit Analysis: a solid phase method for typing single
nucleotide polymorphisms. Nucleic Acids Res. 22:4167–4175. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Srivastava P, Kapoor R and Mittal RD:
Association of single nucleotide polymorphisms in promoter of
matrix metalloproteinase-2, 8 genes with bladder cancer risk in
Northern India. Urol Oncol. 31:247–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu Y and Hein DW: Functional effects of
single nucleotide polymorphisms in the coding region of human
N-acetyltransferase 1. Pharmacogenomics J. 8:339–348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang Z, Kunej T, Michal JJ, et al:
Significant associations of the mitochondrial transcription factor
A promoter polymorphisms with marbling and subcutaneous fat depth
in Wagyu x Limousin F2 crosses. Biochem Biophys Res Commun.
334:516–523. 2005. View Article : Google Scholar
|
|
21
|
Ayres DR, Souza FR, Mercadante ME, et al:
Evaluation of TFAM and FABP4 gene polymorphisms in three lines of
Nellore cattle selected for growth. Genet Mol Res. 9:2050–2059.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clempson AM, Pollott GE, Brickell JS,
Bourne NE, Munce N and Wathes DC: Polymorphisms in the autosomal
genes for mitochondrial function TFAM and UCP2 are associated with
performance and longevity in dairy cows. Animal. 5:1335–1343. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Günther C, von Hadeln K, Müller-Thomsen T,
et al: Possible association of mitochondrial transcription factor A
(TFAM) genotype with sporadic Alzheimer disease. Neurosci Lett.
369:219–223. 2004.PubMed/NCBI
|
|
24
|
Laumet G, Chouraki V, Grenier-Boley B, et
al: Systematic analysis of candidate genes for Alzheimer’s disease
in a French, genome-wide association study. J Alzheimers Dis.
20:1181–1188. 2010.
|
|
25
|
Zhang Q, Yu JT, Wang P, et al:
Mitochondrial transcription factor A (TFAM) polymorphisms and risk
of late-onset Alzheimer’s disease in Han Chinese. Brain Res.
1368:355–360. 2011.
|
|
26
|
Rantanen A, Jansson M, Oldfors A and
Larsson NG: Downregulation of Tfam and mtDNA copy number during
mammalian spermatogenesis. Mamm Genome. 12:787–792. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Castro Mdel R, Suarez E, Kraiselburd E, et
al: Aging increases mitochondrial DNA damage and oxidative stress
in liver of rhesus monkeys. Exp Gerontol. 47:29–37. 2012.PubMed/NCBI
|
|
28
|
Miñana JB, Gómez-Cambronero L, Lloret A,
et al: Mitochondrial oxidative stress and CD95 ligand: a dual
mechanism for hepatocyte apoptosis in chronic alcoholism.
Hepatology. 35:1205–1214. 2002.PubMed/NCBI
|
|
29
|
Lieber CS: Ethanol metabolism, cirrhosis
and alcoholism. Clin Chim Acta. 257:59–84. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Seitz HK and Stickel F: Risk factors and
mechanisms of hepatocarcinogenesis with special emphasis on alcohol
and oxidative stress. Biol Chem. 387:349–360. 2006. View Article : Google Scholar : PubMed/NCBI
|