Introduction
Toll-like receptors (TLRs) are key regulators of the
innate and adaptive immune responses, and are activated by specific
pathogen-associated molecular patterns (1). The activation of TLRs initiates a
signaling cascade and induces the expression of various important
pro-inflammatory cytokines and finally leads to the eradication of
infecting microbes (2).
The expression of TLR has been shown for several
immune cells, whereby the amount of expression and the combination
of TLR differs from one type to another (3). In a human peripheral blood
mononuclear cell (PBMC) survey, TLR1 was expressed in all cell
types examined, while high expression of TLR2 was characteristic
for monocytes (4). Among all TLR
members, TLR1 usually interacts physically and functionally with
TLR2, which appears to be involved in the discrimination of subtle
changes in the lipid portion of lipoproteins (5).
The present study examined the in vitro
responsiveness of PBLs from normal healthy volunteers to the TLR1/2
agonist in order to determine which types of immunomodulatory
molecules are involved in the activation of the TLR1/2 pathway and
in the promotion of the inflammatory status of PBLs.
Materials and methods
Isolation and stimulation of PBLs
Mixed PBLs were isolated from the blood of normal
healthy volunteers using gradient centrifugation (800 × g for 20
min; Sigma-Aldrich, Oakville, ON, Canada), according to the
manufacturers’ protocol. The PBLs (2×106) were treated
with 100 ng/ml TLR1/2 agonist, Pam3Cys. The study was approved by
the ethics committee of West China Hospital, Sichuan University,
Chengdu, China. Written informed consent was obtained from all
participants.
Reverse transcription and quantification
PCR
At 4 h post-stimulation, total RNA was isolated
using an RNeasy mini kit (Qiagen, Dusseldorf, Germany) from the
Pam3Cys-treated and untreated groups. cDNA was synthesized using
the ReverTra Ace quantitative polymerase chain reaction (qPCR) kit
(FSQ-101; Toyobo, Kagoshima, Japan). The reverse transcription
conditions were 65°C for 5 min, followed by 37°C for 15 min and
98°C for 5 min.
qPCR was performed using RealMaster Mix (SYBR Green;
FP202; Tiangen, Beijing, China). The qPCR was performed in an
iCycler iQTM Optical Module (Beckman Coulter, Fullerton, CA, USA)
under the following conditions: One cycle at 95°C for 30 sec, then
40 cycles at 95°C for 30 sec, 58°C for 30 sec and 72°C for 30 sec,
followed by a melt curve from 55 to 95°C in 0.5°C increments and
10-sec intervals. The primers used are listed in Table I. All tests were conducted three
times.
 | Table IList of primers for qPCR analysis. |
Table I
List of primers for qPCR analysis.
| Gene | Forward primer | Reverse primer | GenBank number |
|---|
| c-myc |
CAAGACTCCAGCGCCTTCTC |
GTTGAGTAACGAGCTGACCCC | AM393287 |
| CXCL2 |
AGGTGAAGTCCCCCGGAC |
GCCCATTCTTGAGTGTGGCT | NM_002089 |
| CXCL6 |
GCTGAGAGTAAACCCCAAAACG | GGAGCACTGCGGGCC | NM_002993 |
| CCL26 |
CCAAGACCTGCTGCTTCCAA |
GAATTCATAGCTTCGCACCCA | NM_006072 |
| CCL28 |
CTCGCCATCGTGGCCTT |
GCAATGGGAAGTATGGCTTCTG | AF220210 |
| IFN-β |
CAGCAATTTTCAGTGTCAGAAGCT |
TCATCCTGTCCTTGAGGCAGT | M28622 |
| IL-1β |
ACGAATCTCCGACCACCACT |
CCATGGCCACAACAACTGAC | M15330 |
| IL-6 |
GACCCAACCACAAATGCCA |
GTCATGTCCTGCAGCCACTG | M14584 |
| IL-8 |
CTGGCCGTGGCTCTCTTG |
CCTTGGCAAAACTGCACCTT | NM_000584 |
| IL-15 |
GACCCCACCAAAGCTGGAC |
TCACAGTGCTGCTGTCTGCTG | M90391 |
| IP-10 |
TGAAATTATTCCTGCAAGCCAA |
CAGACATCTCTTCTCACCCTTCTTT | NM_001565 |
| ITGB3 |
TGCCGCCCTGCTCATCTGGA |
TCCTGCAATCGTGGCACAGGC | NM_000212 |
| MIP-1α |
AGCTGACTACTTTGAGACGAGCAG |
CGGCTTCGCTTGGTTAGGA | NM_002983 |
| MIP-1β |
CTGCTCTCCAGCGCTCTCA |
GTAAGAAAAGCAGCAGGCGG | NM_002984 |
| RANTES |
GACACCACACCCTGCTGCT |
TACTCCTTGATGTGGGCACG | NM_002985 |
| PTEN |
ACCATAACCCACCACAGC |
CAGTTCGTCCCTTTCCAG | NM_058074 |
| GAPDH |
GAAGGTGAAGGTCGGAGTC |
GAAGATGGTGATGGGATTTC | J04038 |
Antibody array
Conditioned media from TLR1/2-treated and untreated
PBLs were analyzed for protein expression using RayBio Human
Antibody Array C Series 1000 (RayBiotech; Norcross, GA, USA),
according to the manufacturer’s instructions. Blots were analyzed
with ImageJ software (National Institutes of Health, Bethesda, MD,
USA).
Western-blot analysis
Proteins of PBLs were extracted using a standard
mammalian protein extraction reagent (Pierce, Rockford, IL, USA)
containing protease inhibitor (Roche Applied Science, Indianapolis,
IN, USA). Lysates were clarified by centrifugation at 13,000 × g
for 10 min at 4°C. Protein concentration was measured using a Micro
BCA Protein Assay kit (Pierce). The total protein at 20 μg was
loaded on 15% SDS-polyacrylamide gels and transferred to
nitrocellulose membranes (Invitrogen, Carlsbad, CA, USA). The
membranes were blocked with 5% skimmed dried milk in Tris-buffered
saline (TBS) containing 0.2% Tween-20 (TBST) for 1 h at room
temperature, and then incubated at 4°C overnight with the primary
antibodies (Table II). Next, the
membranes were washed in TBST (3 times, 60 min) and incubated with
secondary antibody conjugated to horseradish peroxidase (1:5000;
Abcam, Cambridge, UK) for 1 h at room temperature. Antigen-antibody
complexes were visualized using X-ray film following exposure to
enhanced chemiluminescence reagent (Amersham Biosciences,
Fairfield, CT, USA). The gray analysis of western blotting was
completed using ImageJ software (National Institutes of
Health).
 | Table IIPrimary antibodies for western
blotting. |
Table II
Primary antibodies for western
blotting.
| Name | Company | Catalog number | Molecular weight,
kDa |
|---|
| JNK2 | Abcam | 2037-1 | 54 |
| ERK | Abcam | 1171-1 | 44 |
| pSTAT3 | Abcam | 2236-1 | 92 |
| GAPDH | Abcam | ab8245 | 37 |
Data analysis
The qPCR data were analyzed using Bio-Rad iQ5
software. Glyceraldehyde 3-phosphate dehydrogenase was used as an
internal control. Normal PBLs were used as a negative control.
Results were expressed as the mean ± standard error of the mean
using SPSS 16.0 (IBM, SPSS Statistics, Armonk, NY, USA). Values of
P<0.05 and P<0.001 were considered to indicate a significant
difference compared with the control group. The figures were
completed using GraphPad Prism5 (GraphPad Software, Inc., LA Jolla,
CA, USA).
Results
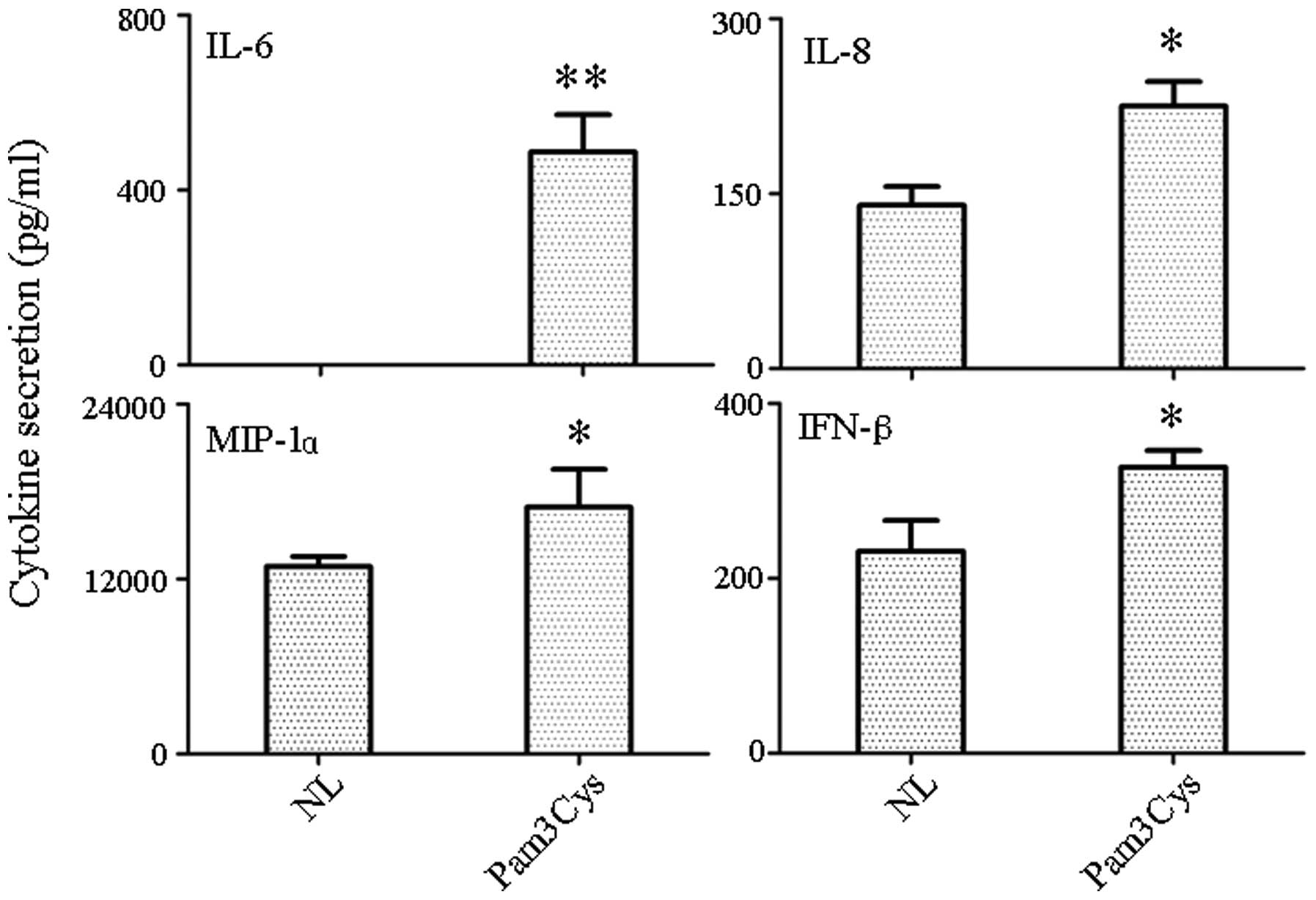
Detection of cytokine/chemokine secretion
in supernatant
Supernatant from TLR1/2 agonist-treated and
untreated PBLs was tested for the presence of secreted chemokines
and cytokines by RayBio antibody-chip assays. A total of 20
molecules were chosen for detection [α-fetoprotein (AFP), albumin,
E-Selectin, intracellular adhesion molecule 1 (ICAM-1), interferon
(IFN)-α and -γ, interleukin (IL)-10, -12, -18, -1β, -4, -5, -6 and
-8, monocyte chemoattractant protein (MCP)-1 and -3, macrophage
inflammatory protein (MIP)-1α, Notch-1, transforming growth factor
β and vascular endothelial growth factor]. The antibody-chip assay
indicated that the TLR1/2 agonist resulted in the increased
secretion of IL-6, IL-8, MIP-1α and IFN-β (Fig. 1). The most significant increase was
in IL-6 (P<0.001), while the other three molecules (IL-8, MIP-1α
and IFN-β) were not increased as significantly as IL-6 (P<0.05).
Other target genes either did not result in detectable protein
levels or had protein levels that remained the same.
Quantification survey of immunomodulatory
genes by qPCR
Cytokine/chemokine expression variations were
analyzed by qPCR in an effort to identify candidate genes
responsible for TLR1/2 agonist-mediated changes in PBLs. Therefore,
changes in the expression of these genes, either due to
microenvironment or pathogen stimulation, could greatly affect the
biological function of PBLs.
Four tumor-related genes were found to be expressed:
Phosphatase and tensin homolog (PTEN), a well-known tumor
suppressor; c-myc, an oncogene; ITGB3, an adhesion molecule; and
IFN-β, a defense factor. Activation of TLR1/2 increased the
expression of all selected genes (P<0.05); the most significant
increase was detected in INF-β (P<0.001; Fig. 2A).
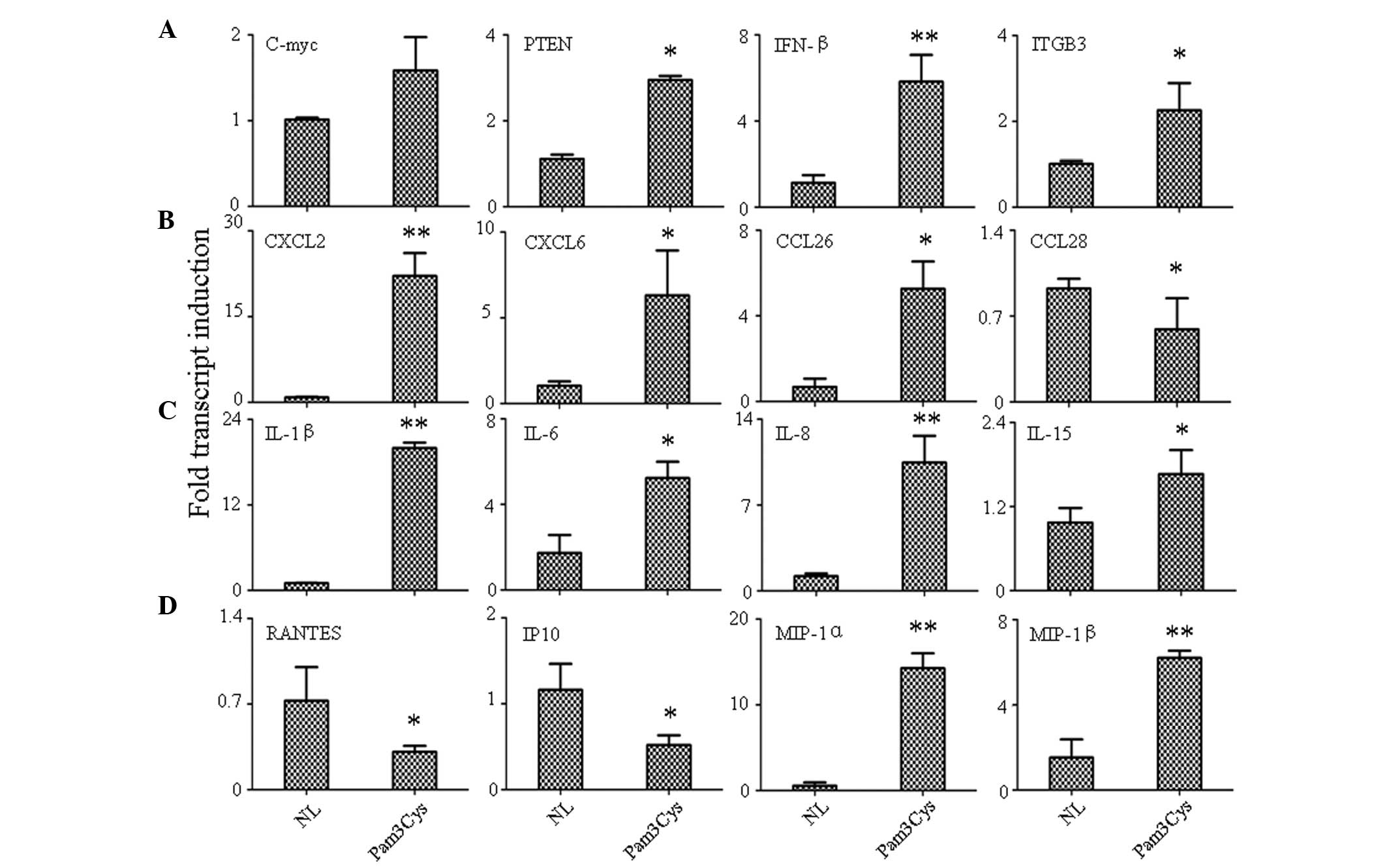 | Figure 2Detection of gene expression variation
by qPCR. (A) Cancer related genes, (B) chemokines, (C) interleukins
and (D) growth factors. **P<0.001 and
*P<0.05 versus control group. qPCR, quantitative
polymerase chain reaction; NL, normal control; Pam3Cys, TLR1
agonist-treated; PTEN, phosphatase and tensin homolog; IFN,
interferon; ITGB3, integrin β3; CXCL, chemokine (C-X-C motif)
ligand; CCL, chemokine (C-C motif) ligand; IL, interleukin; RANTES,
regulated upon activation, normal T cell expressed and secreted;
IP-10, interferon γ-induced protein 10; MIP, macrophage
inflammatory protein. |
In chemokine detection, the result indicated that
the TLR1 agonist could increase the expression of chemokine (C-X-C
motif) ligand 2 (CXCL2; P<0.001), CXCL6 (P<0.05) and
chemokine (C-C motif) ligand 26 (CCL26; P<0.05), while the
expression of CCL28 (P<0.05) was downregulated by Pam3Cys
(Fig. 2B). The expression of other
chemokines, including CCL21, CCL25, CXCL2 and CXCL3 remained the
same along with the normal control (data not shown). In IL
detection, it was found that all tested ILs were either
upregulated, including IL-1β (P<0.001), IL-6 (P<0.05), IL-8
(P<0.001) and IL-15 (P<0.05) (Fig. 2C) or remained unchanged (IL-2, IL7,
IL-9 and IL-18) when stimulated by the TLR1/2 agonist. Finally,
growth factor detection was analyzed and the result showed that the
expression of regulated upon activation, normal T cell expressed
and secreted (RANTES; P<0.05) and interferon γ-induced protein
10 (IP-10; P<0.05) were increased, while MIP-1α (P<0.001) and
MIP-1β (P<0.001) were inhibited by TLR1/2 stimulation (Fig. 2D).
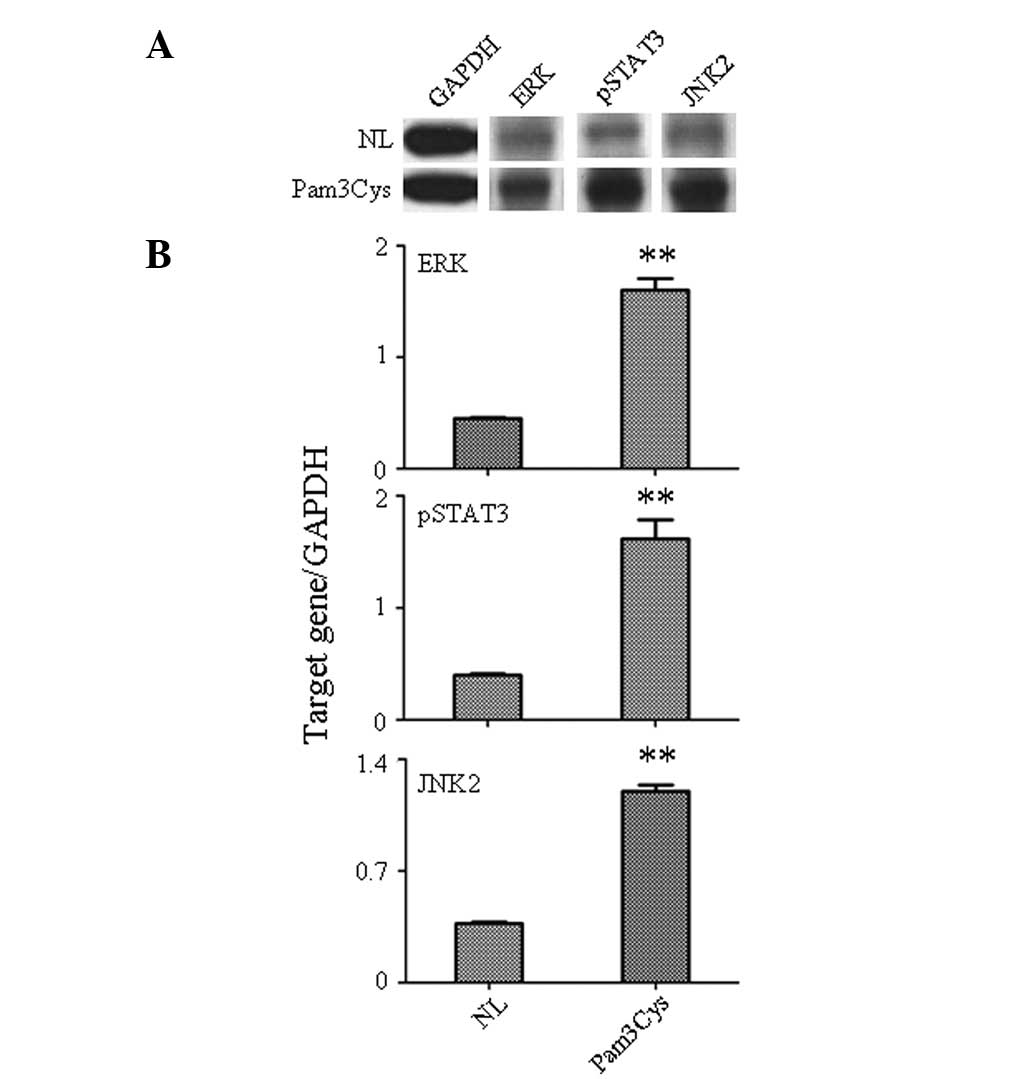
TLR1/2 agonist activates the downstream
signal kinase
Activation of downstream signaling molecules
following Pam3Cys stimulation of PBLs was assessed by western-blot
analysis (Fig. 3). Levels of
phosphorylated signal transducer and activator of transcription 3
(pSTAT3), c-Jun N-terminal kinase 2 (JNK2) and extracellular
signal-related kinase (ERK) were analyzed in the Pam3Cys-treated
PBLs due to their significant role in the control of cell
apoptosis, differentiation, migration and proliferation. The
western-blot analysis indicated the increased expression of pSTAT3,
JNK2 and ERK (Fig. 3A) stimulated
by Pam3Cys. The gray analysis also confirmed that the protein level
of pSTAT3 (P<0.001), JNK2 (P<0.001) and ERK (P<0.001) was
significantly increased in the Pam3Cys-treated PBLs compared with
the untreated group (Fig. 3B).
Since pSTAT3 is mostly activated by IL-6 stimulation (6), this indicates that the TLR1/2
agonist, through increase in the expression of IL-6, plays an
important role in immune response function of PBLs. This result
also confirmed that ERK and JNK were important in the IL-1β-, IL-8-
and MIP-1α-mediated immune response in PBLs caused by Pam3Cys
stimulation (7,8).
Discussion
TLR1 and 2 play an important role in detecting
Gram-positive bacteria, and are involved in the recognition of a
variety of microbial components such as lipoproteins (9). The current study examined the
expressional variation of immunomodulatory molecules of PBL
stimulated by the TLR1/2 agonist. The technique of in vitro
stimulation of human PBLs with TLR agonists followed by
quantification of cytokine expression is not novel. This approach
can be used to identify the PBLs immunological signature and to
understand the TLR signaling pathways. In particular, alterations
in the expression of genes implicated in the following biological
processes were analyzed: i) tumor-related genes, ii) chemokines,
iii) interleukins, iv) growth factors.
Although a former study showed that the TLR1/2
agonist increased the release of IL-8 and TNF-α (10), the present results demonstrated
that activation of TLR1/2 could induce expression of numerous
immunomodulatory factors, including tumor-related genes (c-myc,
PTEN, IFN-β and ITGB3), chemokines (CXCL2, CXCL and CCL26), ILs
(IL-1β, IL-6, IL-8 and IL-15) and growth factors (MIP-1α and
MIP-1β), and only three factors showed decreased expression (CCL28,
RANTES and IP10). However in antibody-chip assays of supernatant,
only four factors showed expressional variation (IL-6, IL-8, MIP-1α
and IFN-β). The explanation for this was either that the increase
in gene expression was not equal with the increase in protein level
or that the treatment time was too short (4 h) to detect the late
expression of numerous factors in the culture supernatant. The
study also uncovered the fact that at least three kinase signal
proteins (pSTAT3, JNK2 and ERK) were significantly induced by the
TLR1/2 agonist, which indicated that there were a number of kinase
signal pathways involved in the immune response that were induced
by activation of the TLR1/2 ligand.
The key difference between the present study and the
previous literature is that the present study surveyed more factors
that were significant in promoting the pro-inflammatory status,
while in the majority of other studies, only a few cytokines were
detected. This difference may miss the complexity of the TLR1/2
response, including the increased expression of c-myc, PTEN and the
CXCLs that was observed in Pam3Cys, which had not been reported
previously.
This study examined a broader range of molecules,
which were significant in immune modulation. The limitation of the
study was that there was only one time-point (4 h) detected; the
treatment time of TLR1/2 should therefore be extended, as certain
later response molecules will fail to be detected. Based on this
study, the enhanced TLR1/2-induced release of pro-inflammatory
conditions by PBLs indicates a possible dysregulation in the innate
immune system. Our further studies will extend the treatment time
of the TLR1/2 agonist and broaden the signal pathway assay.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81300170).
References
|
1
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801. 2006.
View Article : Google Scholar
|
|
2
|
Miggin SM and O’Neill LA: New insights
into the regulation of TLR signaling. J Leukoc Biol. 80:220–226.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith EL, Cools N, Lion E, et al: The
Toll-like receptor 7/8 agonist resiquimod greatly increases the
immunostimulatory capacity of human acute myeloid leukemia cells.
Cancer Immunol Immunother. 59:35–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hornung V, Rothenfusser S, Britsch S, et
al: Quantitative expression of toll-like receptor 1–10 mRNA in
cellular subsets of human peripheral blood mononuclear cells and
sensitivity to CpG oligodeoxynucleotides. J Immunol. 168:4531–4537.
2002.
|
|
5
|
Ozinsky A, Underhill DM, Fontenot JD, et
al: The repertoire for pattern recognition of pathogens by the
innate immune system is defined by cooperation between toll-like
receptors. Proc Natl Acad Sci USA. 97:13766–13771. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Niemand C, Nimmesgern A, Haan S, et al:
Activation of STAT3 by IL-6 and IL-10 in primary human macrophages
is differentially modulated by suppressor of cytokine signaling 3.
J Immunol. 170:3263–3272. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peyssonnaux C and Eychène A: The
Raf/MEK/ERK pathway: new concepts of activation. Biol Cell.
93:53–62. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen YR and Tan TH: The c-Jun N-terminal
kinase pathway and apoptotic signaling (Review). Int J Oncol.
16:651–652. 2000.PubMed/NCBI
|
|
9
|
Alexopoulou L, Thomas V, Schnare M, et al:
Hyporesponsiveness to vaccination with Borrelia burgdorferi
OspA in humans and in TLR1- and TLR2-deficient mice. Nat Med.
8:878–884. 2002.PubMed/NCBI
|
|
10
|
Kwok YH, Hutchinson MR, Gentgall MG and
Rolan PE: Increased responsiveness of peripheral blood mononuclear
cells to in vitro TLR 2, 4 and 7 ligand stimulation in chronic pain
patients. PloS One. 7:e442322012. View Article : Google Scholar : PubMed/NCBI
|