Introduction
Osteosarcoma is one of the most common primary bone
sarcomas in children and adolescents and has a five-year survival
rate of ~70%. Patients with osteosarcoma with metastases at
diagnosis have a poor prognosis, with overall survival rates of
<20% (1). Despite the rapid
development in treatment strategies, the cure rate of patients with
osteosarcoma remains extremely poor. Therefore, there is an urgent
need to develop novel strategies for the diagnosis and treatment of
osteosarcoma.
SUMOylation is an extensively studied modification
that elicits a wide range of effects within the cell (2). A small ubiquitin-like modifier (SUMO)
protein is covalently attached to lysine residues in substrate
proteins in a process similar to ubiquitination (3). SUMO proteins are highly conserved in
a large number of species and have been shown to be indispensable
in numerous cellular processes. In vertebrates, there are four SUMO
family proteins, SUMO-1, -2, -3 and -4. SUMO-2 and SUMO-3 share
~50% identity to SUMO-1, but are highly related to each other,
sharing 96% sequence identity (4).
SUMO-4 is more similar to SUMO-2/3 (5). SUMOylation is a dynamic and
reversible process and six SUMO-specific proteases (SENPs) that
remove SUMO from substrates have been identified, which have
various substrate specificities and subcellular localizations
(6). SENPs are divided into three
subfamilies on the basis of sequence homology, cellular location
and substrate specificity. The first subfamily consists of SENP1
and SENP2, which have broad substrate specificity. The second
subfamily consists of SENP3 and SENP5, which are nucleolar proteins
with preferences for SUMO-2/3. The third subfamily consists of
SENP6 and SENP7, which have an extra loop in their catalytic
domains (7).
DeSUMOylation, induced by SENPs, has been shown to
be crucial in determining protein SUMOylation status and activity
(8). Several SENPs have been shown
to be amplified in a subset of cancer types. Expression of SENP5
has been reported in oral squamous cell carcinoma (OSCC) and the
protease has been associated with the differentiation of OSCC
(9). SENP5 resides primarily
within the nucleoli during interphase, but translocates from the
nucleoli to the mitochondrial surface at the G2/M transition, prior
to nuclear envelope breakdown (10,11).
Knockdown of SENP5 by siRNA results in increased levels of SUMO-1
and SUMO-2/3 conjugates, as well as defects in nuclear and
mitochondrial morphology. These observations have revealed an
essential role for SENP5 in cytokinesis and the maintenance of
mitochondrial function (12–14).
However, the biological role of SENP5 in cancer has yet to be fully
elucidated.
The present study aimed to determine SENP5
expression levels in osteosarcoma cell lines. In addition, the
effect of lentivirus-mediated siRNA of SENP5 on cell growth and
apoptosis in osteosarcoma cells was investigated. Furthermore, the
study aimed to evaluate whether SENP5, as a SUMO-specific protease,
is required for cell growth and apoptosis and may be a promising
drug target for antiosteosarcoma treatment.
Material and methods
Tissue samples
Primary osteosarcoma and distant normal tissues were
collected from routine therapeutic surgeries at the Department of
Orthopedics (Affiliated Jinshan Hospital, Shanghai, China). All
samples were obtained with informed consent and approved by the
Affiliated Jingshan Hospital.
Cell culture
HOS, KHOS, U2OS, Saos-2 and MG-63 cell lines were
purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI
1640 medium (Gibco-BRL, Beijing, China) supplemented with 10% fetal
bovine serum (FBS), 100 IU/ml penicillin and 100 mg/ml streptomycin
(Gibco-BRL).
Quantitative polymerase chain reaction
(PCR)
RNA was extracted using TRIzol reagent (Takara
Biotechnology Co. Ltd., Dalian, China) and reverse transcription
was performed with a Takara RNA PCR kit (Takara Biotechnology Co.
Ltd., Dalian, China), in accordance with the manufacturer’s
instructions. Quantitative PCR was performed using a SYBR Green
Premix Ex Taq kit (Takara Bio, Inc., Shiga, Japan),
according to the manufacturer’s instructions. PCR was performed in
96-well optical plates. The primers used were as follows: The
primers for β-actin 5′-AGAGCTACGAGCTGCCTGAC-3′ and
5′-AGCACTGTGTTGGCGTACAG-3′ and for SENP5 5′-GAGGAAAATTCTATGGAGGA-3′
and 5′-GAGGACAAAGTACTAACATT-3′.
Western blot analysis
Cells were lysed in sample solution. Proteins were
separated on 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) gels, transferred to nitrocellulose
membranes and detected using various antibodies, as indicated. The
membranes were incubated with primary antibodies at 4°C overnight
and horseradish peroxidase-conjugated secondary antibodies for 1 h
at room temperature, prior to detection using the SuperSignal West
Pico Chemiluminescent Substrate kit (Pierce Biotechnology, Inc.,
Rockford, IL, USA). Anti-β-actin and anti-SENP5 antibodies were
purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA)
and anti-cyclin B1 antibodies were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA).
Cell proliferation assay
U2OS and Saos-2 cells, transfected with mock or
SENP5 siRNA, were seeded in 96-well plates and incubated for one to
six days. Subsequently, 20 μl 3-(4,5-dimethylthiazol-2-yl)-2,
5-diphenyltetrazolium bromide solution (5 mg/ml) was added to each
well 3 h prior to the end of incubation. The crystals were
dissolved in 150 μl dimethyl sulfoxide and the absorbance at 570 nm
was measured with a SPECTRAmax 340PC (Molecular Devices, LLC.,
Sunnyvale, CA, USA).
Colony formation assay
U2OS and Saos-2 cells, transfected with mock or
SENP5 siRNA, were seeded in a six-well plate at a density of 500 or
1,000 cells/well. Following incubation at 37°C for 12–21 days, the
colonies were fixed and stained in a dye solution containing 0.1%
crystal violet (Sigma-Aldrich, St. Louis, MO, USA) and 20%
methanol. The number of colonies per well was counted.
Cell cycle analysis
Cells grown in regular growth medium for 24 h were
collected, fixed in 70% cold ethanol overnight and stained with
phosphate-buffered saline (PBS) containing 50 μg/ml propidium
iodide and 100 μg/ml RNase A for 30 min at 37°C. The DNA content of
the labeled cells was measured using the Accuri C6 flow cytometry
system (BD Biosciences, Franklin Lakes, NJ, USA).
Analysis of caspase-3/-7 activity
U2OS and Saos-2 cells, transfected with mock or
SENP5 siRNA, were seeded at a density of 500 cells/well in
triplicate wells in a 384-well plate. Following overnight
incubation, the medium was replaced with Dulbecco’s modified
Eagle’s medium (DMEM) supplemented with 0.2% FBS and incubated for
an additional 48 h. Caspase activity was subsequently measured with
a Caspase-Glo 3/7 Assay System (Promega, Fitchburg, WI, USA),
according to the manufacturer’s instructions. An equal volume of
caspase substrate was added to the cells and the samples were
incubated at room temperature for 1 h. Luminescence was measured
using an EnVision 2103 Multilabel Reader (Perkin Elmer, Inc.,
Waltham, MA, USA). Luminescence of the mock-transfected cells was
set as the standard.
Statistical analysis
The data shown represent the mean ± standard error
(SE) values of three independent experiments. Significance was
analyzed using a Student’s t-test. P<0.05 was considered to
indicate a statistically significant result.
Results
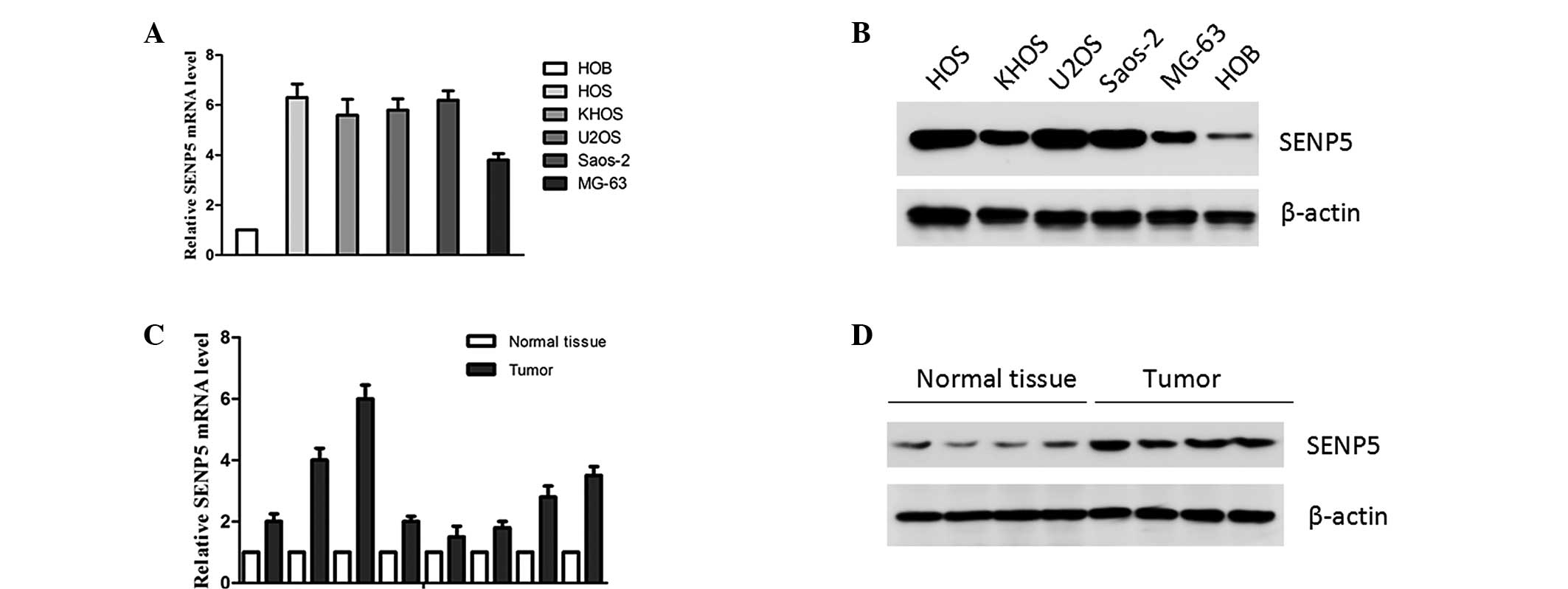
SENP5 is overexpressed in osteosarcoma
cell lines and tissues
To investigate the significance of SENP5 in
osteosarcoma carcinogenesis, the expression levels of SENP5 in
osteosarcoma cell lines (HOS, KHOS, U2OS, Saos-2 and MG-63) and
clinical specimens were analyzed using quantitative PCR and western
blotting. The results showed that SENP5 was significantly
overexpressed in all osteosarcoma cell lines, compared with HOB
cells (human osteoblasts isolated from normal human bone) (Fig. 1A and B). Consistent with these
observations, osteosarcoma clinical specimens expressed high levels
of SENP5 compared with adjacent normal bone tissues (Fig. 1C and D). These results indicated
that increased expression of SENP5 was correlated with osteosarcoma
carcinogenesis.
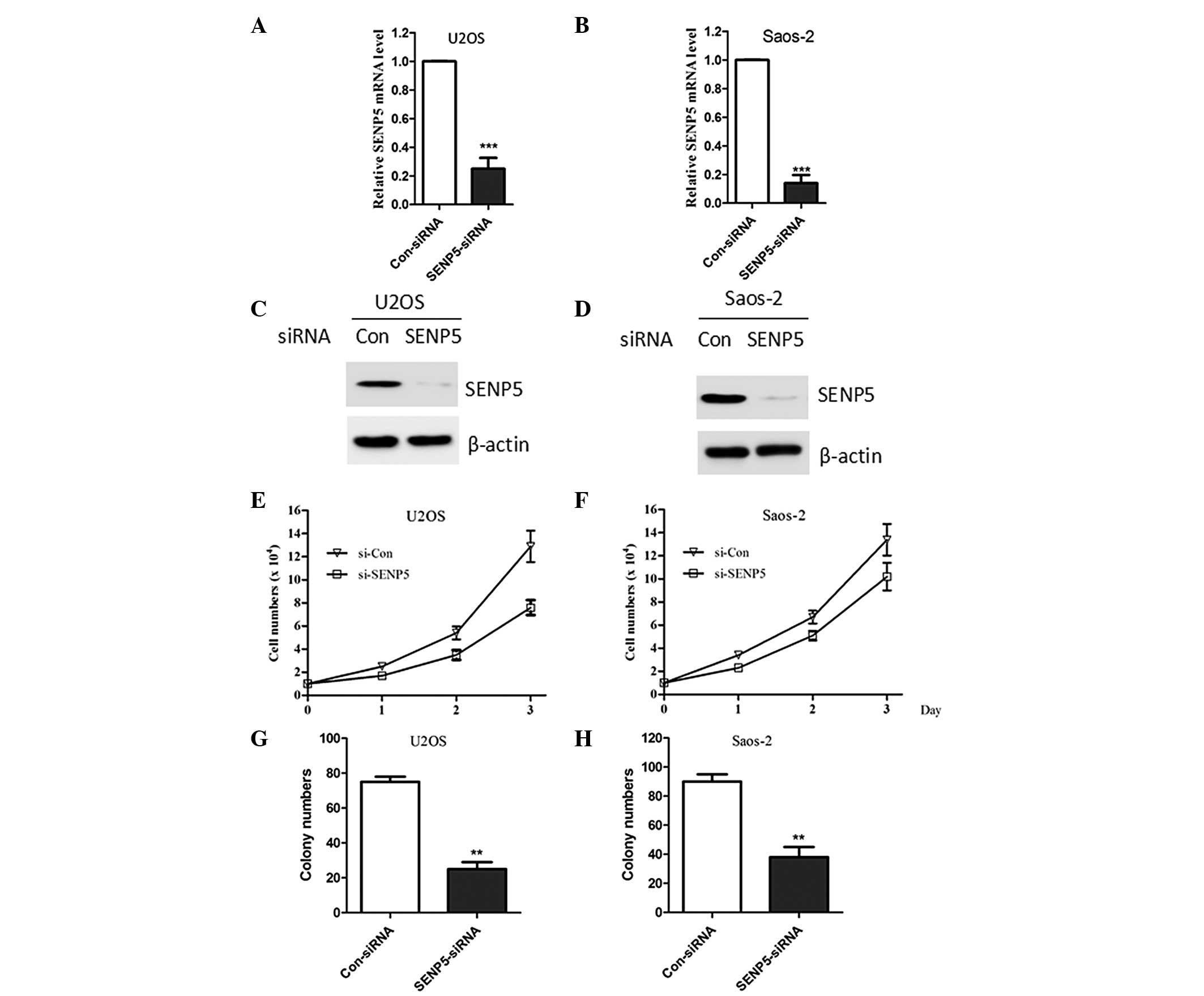
Lentivirus-mediated siRNA of SENP5
significantly inhibits cell growth in osteosarcoma cells
To investigate the biological role of SENP5 in
osteosarcoma, lentivirus-mediated siRNA was utilized to silence the
expression of endogenous SENP5 in osteosarcoma cells. To testify
the silencing effect of lentivirus-mediated siRNA targeting the
SENP5 gene, quantitative PCR and western blot analysis were
performed to detect the expression of SENP5 mRNA and protein in
mock or stably transfected U2OS and Saos-2 cells. As shown in
Fig. 2A–D, mRNA and protein levels
of SENP5 significantly decreased in SENP5-silenced U2OS and Saos-2
cells. These results indicated that the lentivirus-mediated RNAi
system was able to effectively knockdown endogenous SENP5
expression in osteosarcoma cells. Silencing the expression of SENP5
significantly decreased the proliferation of U2OS and Saos-2 cells
(Fig. 2E and F). Consistent with
these observations, silencing the expression of SENP5 resulted in a
marked decrease in the number and size of U2OS and Saos-2 cell
colonies (Fig. 2G and H).
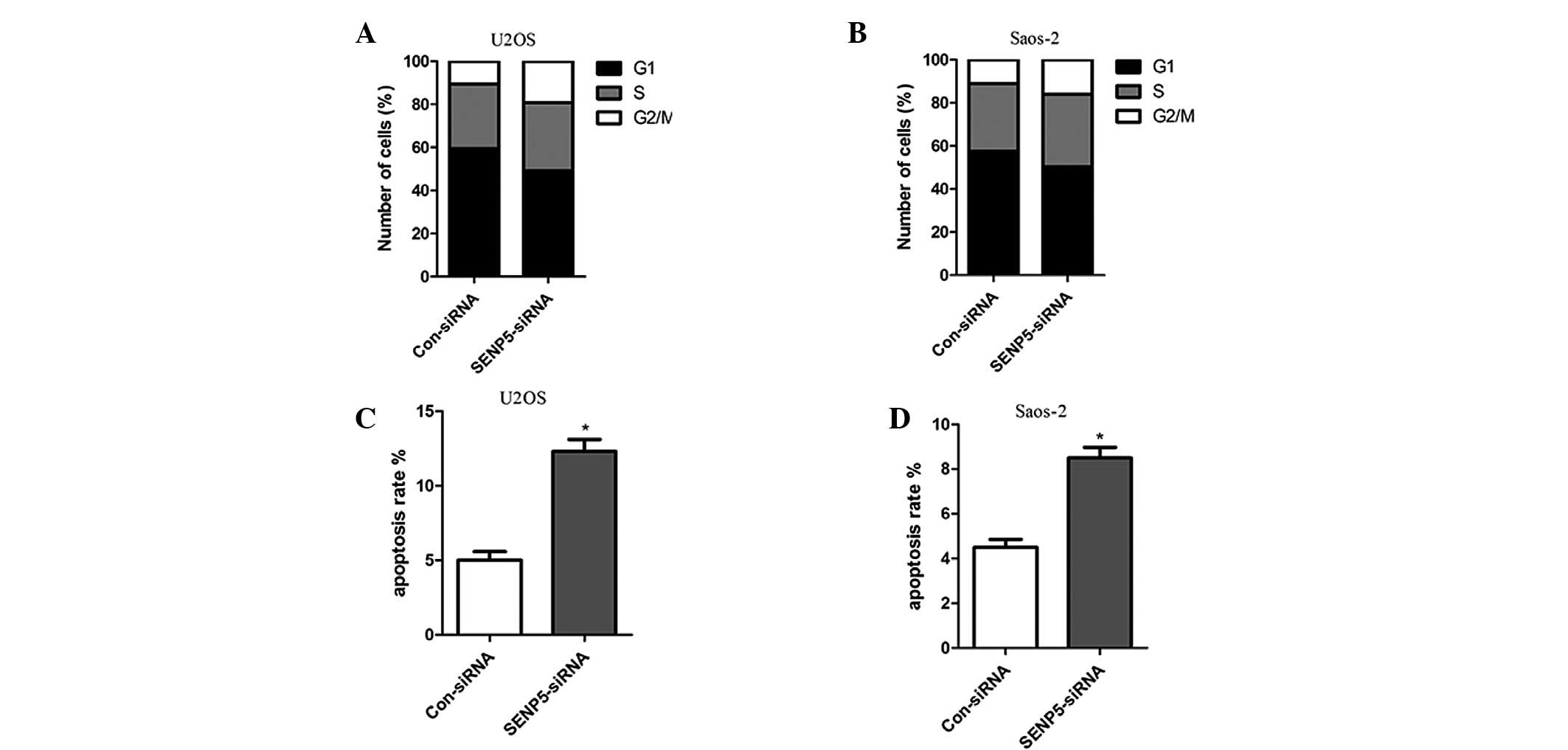
SENP5 inhibition results in G2/M arrest
and apoptosis in U2OS and Saos-2 osteosarcoma cells
The growth inhibition of cells may be caused by a
reduced cell proliferation rate or by increased apoptosis or cell
cycle arrest. The effect of SENP5 on cell cycle distribution was
investigated. Silencing the expression of SENP5 caused a
significant increase in the number of U2OS and Saos-2 cells in the
G2/M phase (Fig. 3A and B), which
was consistent with the previously identified role of SENP5 in cell
division. Moreover, SENP5 inhibition also resulted in spontaneous
osteosarcoma cell apoptosis, when compared with mock inhibition
cells (Fig. 3C and D). In
combination, these results indicated that SENP5 inhibition
decreased osteosarcoma cell proliferation by inducing G2/M arrest
and apoptosis.
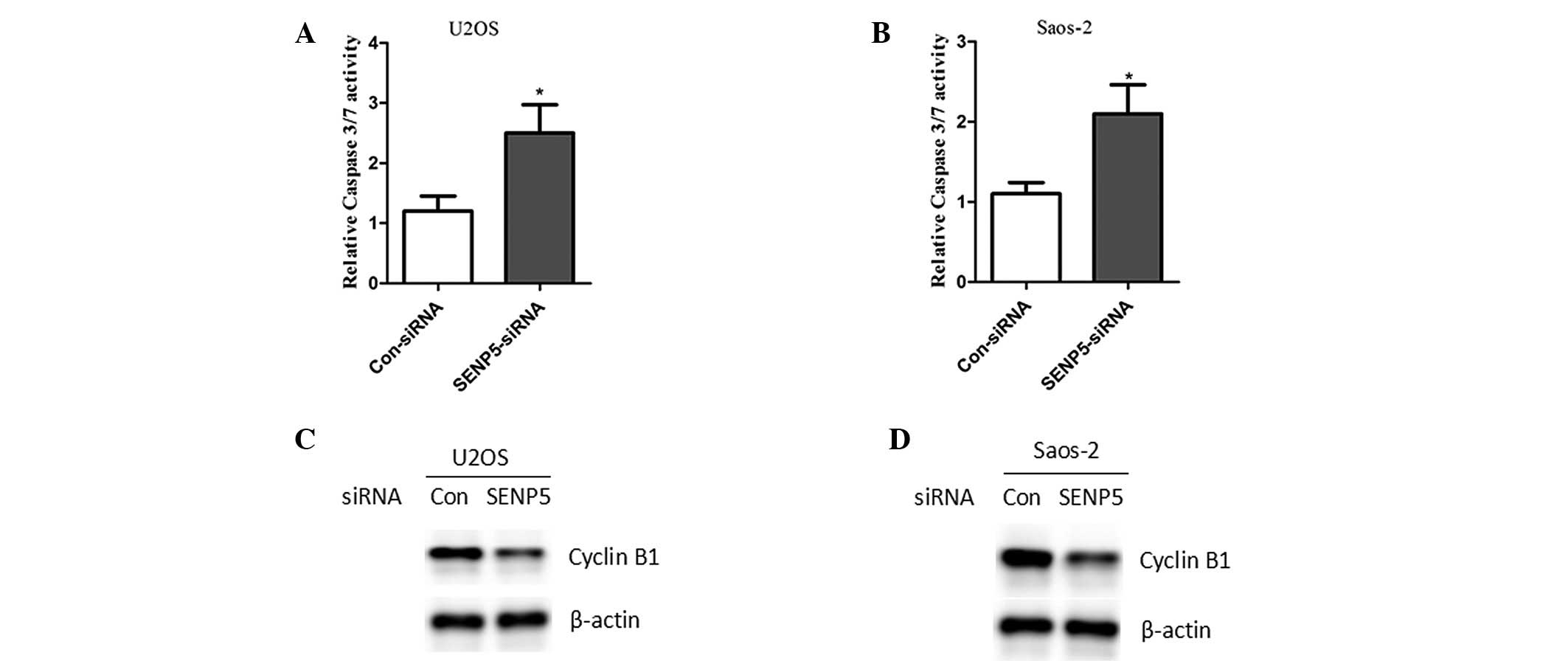
SENP5 inhibition induces caspase-3/-7
activity and inhibits cyclin B1 expression
To investigate the mechanism of SENP5 in the
regulation of osteosarcoma cell apoptosis, it was assessed whether
SENP5 inhibition resulted in the activation of caspases. The
caspase-3/-7 activity of mock- or SENP5-depleted osteosarcoma cells
was evaluated. These cells were serum-starved in 0.2% FBS for two
days. The caspase-3/-7 activity in mock-depleted cells was set as
the standard. SENP5 inhibition resulted in a two-to-three-fold
increase in caspase-3/-7 activity in U2OS and Saos-2 cells
(Fig. 4A and B). The mechanism of
SENP5-depletion-induced G2/M arrest in osteosarcoma cells was also
investigated. It was observed that silencing the expression of
SENP5 significantly decreased the expression of cyclin B1, a key
regulator of G2/M transition in the cell cycle (Fig. 4B and C).
Discussion
SUMOylation has a vital role in tumors (15), with several SENPs identified to be
involved in cancer development. SENP1 has been shown to be crucial
in the development of prostate cancer by modulating the SUMOylation
of the androgen receptor (16).
SENP2 has been shown to be involved in hepatocellular carcinoma
cell growth by modulating the stability of β-catenin (17). SENP3 has been identified to
accumulate in a variety of types of primary human cancer, including
colon adenocarcinoma, by modulating the SUMOylation status of the
tumor suppressor, promyelocytic leukemia protein (PML) (18). SENP6 was previously reported to
induce radiosensitization of hepatocellular carcinoma cells by
blocking radiation-induced NF-κB activation (19).
In the present study, SENP5 was observed to be
overexpressed in osteosarcoma cell lines and tissues. SENP5
primarily resides within the nucleoli during interphase, but
translocates from the nucleoli to the mitochondrial surface at the
G2/M transition, prior to nuclear envelope breakdown (14). Function studies have revealed that
SENP5 is required for cell division and the maintenance of
mitochondrial morphology and function (11,14).
In the present study in osteosarcoma cell lines, it was observed
that silencing the expression of SENP5 significantly decreased cell
proliferation, which was consistent with the function of SENP5 in
cell division. The results indicated that SENP5 regulated
osteosarcoma cell proliferation by inducing G2/M arrest and
apoptosis. Further elucidation of the underlying mechanisms was
also achieved, and it was demonstrated that SENP5 inhibition
increased the activity of caspase-3/-7 and decreased the expression
of cyclin B1. Thus, the present study reported a role of SENP5 in
the regulation of osteosarcoma cell proliferation and
apoptosis.
Although the deSUMOylation activity of SENP5 has
been well-documented, the endogenous SUMOylation substrates of
SENP5 are less well-known. The first substrate of SENP5 identified
was the tumor suppressor, PML, which has an essential role in the
regulation of cell proliferation (10). DRP1, a mitochondrial fission
GTPase, was then identified to be a substrate of SENP5, and SENP5
translocates from the nucleoli to the mitochondria and modulates
DRP1-dependent fission during mitosis (11). However, substrates in osteosarcoma
cells that are responsible for SENP5-regulated G2/M arrest and
apoptosis remain to be identified. The results from the present
study reveal an indispensable role of SENP5 in the regulation of
G2/M arrest and apoptosis in osteosarcoma cells, which supports the
hypothesis that SENP5 may be a promising drug target for
antiosteosarcoma treatment in the future.
Acknowledgements
The authors would like to thank Pubsci for
experimental support during the study.
References
|
1
|
Jaffe N: Osteosarcoma: review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Müller S, Hoege C, Pyrowolakis G and
Jentsch S: SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell
Biol. 2:202–210. 2001.
|
|
4
|
Hannoun Z, Greenhough S, Jaffray E, Hay RT
and Hay DC: Post-translational modification by SUMO. Toxicology.
278:288–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hwang KW, Won TJ, Kim H, Chun HJ, Chun T
and Park Y: Characterization of the regulatory roles of the SUMO.
Diabetes Metab Res Rev. 27:854–861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yeh ET: SUMOylation and De-SUMOylation:
wrestling with life’s processes. J Biol Chem. 284:8223–8227.
2009.PubMed/NCBI
|
|
7
|
Bawa-Khalfe T and Yeh ET: SUMO Losing
balance: SUMO proteases disrupt SUMO homeostasis to facilitate
cancer development and progression. Genes Cancer. 1:748–752. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim JH and Baek SH: Emerging roles of
desumoylating enzymes. Biochim Biophys Acta. 1792:155–162. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding X, Sun J, Wang L, et al:
Overexpression of SENP5 in oral squamous cell carcinoma and its
association with differentiation. Oncol Rep. 20:1041–1045.
2008.PubMed/NCBI
|
|
10
|
Gong L and Yeh ET: Characterization of a
family of nucleolar SUMO-specific proteases with preference for
SUMO-2 or SUMO-3. J Biol Chem. 281:15869–15877. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zunino R, Braschi E, Xu L and McBride HM:
Translocation of SenP5 from the nucleoli to the mitochondria
modulates DRP1-dependent fission during mitosis. J Biol Chem.
284:17783–17795. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Di Bacco A and Gill G: SUMO-specific
proteases and the cell cycle. An essential role for SENP5 in cell
proliferation. Cell Cycle. 5:2310–2313. 2006.PubMed/NCBI
|
|
13
|
Di Bacco A, Ouyang J, Lee HY, Catic A,
Ploegh H and Gill G: The SUMO-specific protease SENP5 is required
for cell division. Mol Cell Biol. 26:4489–4498. 2006.PubMed/NCBI
|
|
14
|
Zunino R, Schauss A, Rippstein P,
Andrade-Navarro M and McBride HM: The SUMO protease SENP5 is
required to maintain mitochondrial morphology and function. J Cell
Sci. 120:1178–1188. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alarcon-Vargas D and Ronai Z: SUMO in
cancer - wrestlers wanted. Cancer Biol Ther. 1:237–242. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cheng J, Wang D, Wang Z and Yeh ET: SENP1
enhances androgen receptor-dependent transcription through
desumoylation of histone deacetylase 1. Mol Cell Biol.
24:6021–6028. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shen HJ, Zhu HY, Yang C and Ji F: SENP2
regulates hepatocellular carcinoma cell growth by modulating the
stability of β-catenin. Asian Pac J Cancer Prev. 13:3583–3587.
2012.PubMed/NCBI
|
|
18
|
Han Y, Huang C, Sun X, et al:
SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic
leukemia is correlated with accelerated cell proliferation under
mild oxidative stress. J Biol Chem. 285:12906–12915. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qian J, Luo Y, Gu X and Wang X: Inhibition
of SENP6-induced radiosensitization of human hepatocellular
carcinoma cells by blocking radiation-induced NF-κB activation.
Cancer Biother Radiopharm. 28:196–200. 2013.PubMed/NCBI
|