Introduction
Sleep apnea-hypopnea syndrome (SAHS) can be divided
into three types: Obstructive, central and mixed. In clinical
practice, obstructive SAHS (OSAHS) is one of the most common forms,
accounting for ~80% of SAHS cases. Previous studies have identified
that the incidence rate of OSAHS in adults is 1–5% (1) or 41% in a subpopulation of patients
with a body mass index (BMI) of >28 kg/m2 (2). The incidence rate of domestic OSAHS
is ~3%. Furthermore, SAHS is an independent risk factor of multiple
systemic diseases (3,4) and seriously affects patient health
and quality of life. The pathogenesis of SAHS remains unclear,
however, previous studies have demonstrated familial aggregation of
OSAHS and genetic predisposition (5). Patients with OSAHS suffer from
recurrent nocturnal intermittent hypoxia, which affects oxidative
phosphorylation in the mitochondrial respiratory chain. Obesity,
hyperlipidemia, hyperglycemia and other pathogenesis factors are
closely associated with OSAHS, and may be associated with
mitochondrial mutations (6,7).
In addition to the nucleus in human cells, the
mitochondrion is an organelle that contains genetic material. Each
mitochondrion contains 210 copies of mitochondrial DNA
(mtDNA) and is able to perform replication, transcription and
translation without relying on nuclear DNA (nDNA). The half-life of
mtDNA is 5–10 times faster than nDNA and the mutation rate of mtDNA
is 10–20 times higher compared with nDNA (8–11).
Thus, mtDNA exhibits greater vulnerability and is damaged more
frequently compared with nDNA; in addition, mtDNA possesses a
greater risk of mutation than nDNA. The displacement loop (D-loop)
is the control region of mtDNA replication and is frequently
mutated. The majority of the regulatory sequences associated with
mtDNA replication, transcription and translation are located within
the D-loop (12). The aim of the
present study was to investigate the correlation between
mitochondrial mutations and OSAHS, as well as the associated
complications, and to investigate whether the genetic
predisposition of OSAHS is associated with mitochondrial
mutations.
Patients and methods
Subjects
In total, 60 male patients with OSAHS and 102
healthy males were recruited from the Sleeping Center of the First
Affiliated Hospital of Wenzhou Medical University (Wenzhou,
China) between June 2007 and December 2007. The subjects were
not related to one another and those presenting with the following
conditions were excluded: Glucose metabolism disorder, lipid
metabolism disorder, endocrine disorder, chronic obstructive
pulmonary disease, heart failure, jaundice or central nervous
system diseases. The age and gender of the two groups were matched.
The study was conducted in accordance with the Declaration of
Helsinki and with approval from the Ethics Committee of Wenzhou
Medical University. Written informed consent was obtained from all
the participants.
Clinical data collection
Clinical symptoms, including height, weight, neck
perimeters, chest and hip complications, were recorded in each
subject.
Biochemical parameter detection
Routine biochemical parameters, including levels of
fasting blood glucose (FBG), triglyceride (TG), total cholesterol
(TC), high-density lipoprotein (HDL) and low-density lipoprotein
(LDL), were measured for each subject.
Genomic DNA extraction
Peripheral blood samples were collected from the
patients and healthy subjects using a K2-EDTA anticoagulant. The
genomic DNA extraction kit (Universal Genomic DNA extraction kit
version 3.0; Takara Biotechnology Co., Ltd., Dalian, China) was
used to extract the peripheral blood genomic DNA, according to the
manufacturer’s instructions.
Polymerase chain reaction (PCR)
PCR reagents and primers, including Taq DNA
polymerase and a DNA fragment purification kit, were purchased from
Takara Biotechnology Co., Ltd.
The D-loop region of the mitochondrial gene was
located at mtDNA np16028-577 and two pairs of primers, with
overlapping product regions, were used to amplify the genes of the
entire D-loop region. The sequences and amplified products of the
two pairs of primers are shown in Table I. In addition, when designing the
two pairs of primers, one universal M13 forward primer,
TGTAAAACGACGGCCAGT and one reverse primer, CAGGAAACAGCTATGACC, were
synthesized at the 5′ end. The PCR conditions were as follows: 35
cycles of predenaturation at 94°C for 5 min, denaturation at 94°C
for 30 sec, annealing at 59°C for 45 sec and extension at 72°C for
1 min, followed by a final extension at 72°C for 5 min. Next, 3 μl
PCR product was mixed with 1 μl 6× loading buffer and the mixture
was added into a gel well. A 2,000 bp DNA marker (3 μl) was then
added and electrophoresis was performed at a voltage of 5 V/cm for
30 min. The gel was photographed using an imager camera and the DNA
fragment purification kit was used for purification, according to
the manufacturer’s instructions.
 | Table IPrimers used to amplify the genes of
the mitochondrial D-loop region. |
Table I
Primers used to amplify the genes of
the mitochondrial D-loop region.
| Primer | Product sequence
(5′-3′) | Location of 3′
(F/R) | Product length
(bp) |
|---|
|
|---|
| F | R |
|---|
| Mit-23 |
TCATTGGACAAGTAGCATCC |
GAGTGGTTAATAGGGTGATAG | 15811/5 | 764 |
| Mit-24 |
CACCATTCTCCGTGAAATCAa |
AGGCTAAGCGTTTTGAGCTG | 16420/775 | 925 |
DNA sequence analysis
Purified PCR products were subjected to DNA sequence
analysis (Shanghai GeneCore BioTechnologies Co., Ltd., Shanghai,
China). The SeqMan function of the DNASTAR software package
(DNASTAR, Inc., Madison, WI, USA) was used for sequence analysis
and the MegAlign function (DNASTAR, Inc.) was used to compare the
products with the reference sequence (human mtDNA revised Cambridge
reference sequence) in order to identify the mutation locus. When
the potential mutation sites were identified, Chromas software
(Technelysium, Brisbane, Australia) was used to observe the
specific peaks, and compare and analyze the sites with the reported
mtDNA mutation sites in the internationally recognized human
mitochondrial database, (www.mitomp.org).
Statistical analysis
SPSS 13.0 statistical analysis software (SPSS, Inc.,
Chicago, IL, USA) was used for analysis and data are expressed as
the mean ± standard deviation. Inter-group comparisons of normally
distributed data were performed using the Student’s t-test, whereas
logistic regression analysis with a bilateral statistical test
level (P<0.05) was used for analyzing the independent variables,
such as the clinical data.
Results
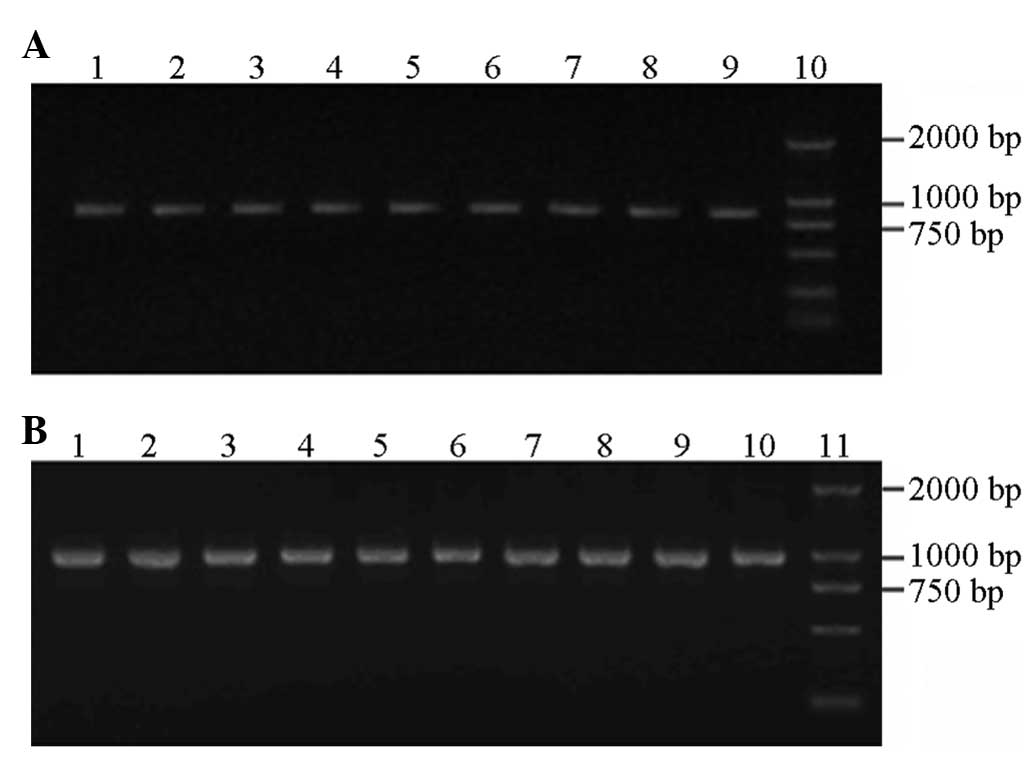
Genomic DNA extraction
Following extraction and purification of the
peripheral blood DNA, a clearly visible band was observed in the
1.0% agarose gel electrophoresis image (Fig. 1).
PCR
Agarose gel (1.2%) electrophoresis was performed
with a 2,000 bp DNA ladder marker that served as the molecular
weight standard to identify the amplification products. The in
vitro amplification of the target genes obtained two fragments
with lengths of 764 and 925 bp (Fig.
1).
Single nucleotide polymorphism
Mutation sites were identified in the mtDNA D-loop
region of each sample (Table II).
A total of 178 mutation sites were found, of which 75 mutation
sites were identified in the 60 OSAHS subjects and 103 mutation
sites were identified in the 102 healthy control subjects. A total
of 115 mutation sites were present in the two groups and the 63
remaining mutation sites were not considered to be common. A total
of 27 mutation sites were present only in the OSAHS group, whereas
36 mutation sites were present only in the healthy control group,
which exhibited a low incidence rate of mutation. The distribution
of the single-mutation sites in the OSAHS and healthy control
groups did not demonstrate a statistically significant difference
(P>0.05). However, for the non-common mutation sites, a
significant difference was identified between the OSAHS group and
the normal control group (P<0.05). No significant differences in
the mutation sites were observed among the mild, moderate and
severe OSAHS subgroups (P>0.05).
 | Table IIMutation sites in the mtDNA D-loop
region of the OSAHS patients and control group. |
Table II
Mutation sites in the mtDNA D-loop
region of the OSAHS patients and control group.
| Mutation sites | Nucleotide
change | OSAHS group (n) | Control group
(n) |
|---|
| 73 | A-G | 57 | 86 |
| 103 | G-A | 2 | 1 |
| 146 | T-A | 4 | 0 |
| 150 | C-T | 8 | 16 |
| 198 | C-T | 2 | 1 |
| 204 | T-C | 2 | 5 |
| 214 | A-G | 2 | 1 |
| 249 delA | 7 | 21 | |
| 263 | A-G | 55 | 89 |
| 298 | C-T | 1 | 1 |
| 310 | T-CTC | 16 | 22 |
| 310 | T-CC | 8 | 11 |
| 16140 | T-C | 7 | 4 |
| 16164 | A-G | 3 | 4 |
| 16316 | A-G | 3 | 1 |
| 16325 | T-C | 3 | 1 |
| 16327 | C-T | 2 | 2 |
| 16357 | T-C | 2 | 1 |
| 16362 | T-C | 11 | 34 |
| 16182 | A-C | 8 | 12 |
| 16183 | A-C | 12 | 23 |
| 16184 | C-T | 7 | 7 |
| 16185 | C-T | 3 | 2 |
| 16189 | T-C | 16 | 30 |
| 16192 | C-T | 2 | 2 |
| 16217 | T-C | 2 | 4 |
| 16223 | C-T | 31 | 46 |
| 16243 | T-C | 4 | 1 |
| 16249 | T-C | 2 | 1 |
| 16260 | C-T | 2 | 7 |
| 16266 | C-T | 1 | 3 |
| 16172 | T-C | 2 | 19 |
| 16319 | G-A | 8 | 12 |
| 16390 | G-A | 2 | 2 |
| 16463 | A-G | 2 | 1 |
| 16519 | T-C | 32 | 47 |
| 16535 | G-A | 1 | 0 |
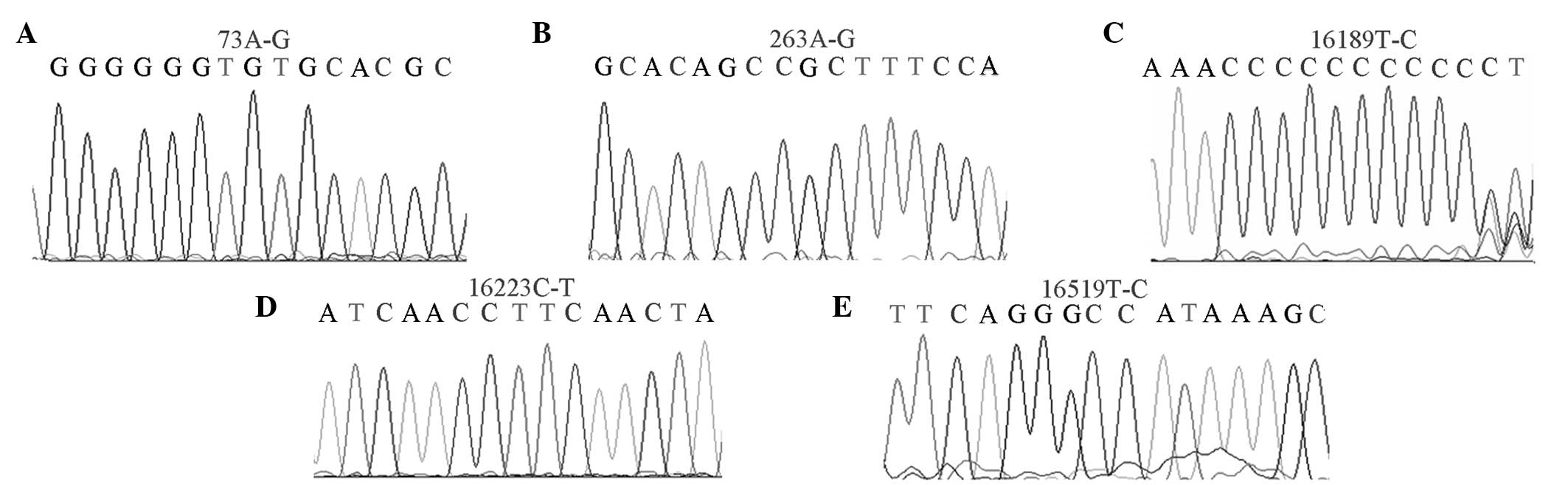
High-mutation sites
In total, 21 high-mutation sites (mutation rate
>10%) were found in the OSAHS group, whereas 24 sites were
identified in the healthy control group (Fig. 2). The np73A-G mutation rates were
93.7 and 84.2%; np263A-G mutation rates were 90.4 and 79.3%;
np16223C-T mutation rates were 50.1 and 47.0%; np16189T-C mutation
rates were 46.7 and 29%; and np16519T-C mutation rates were 53.3
and 46% in the OSAHS and healthy control groups, respectively.
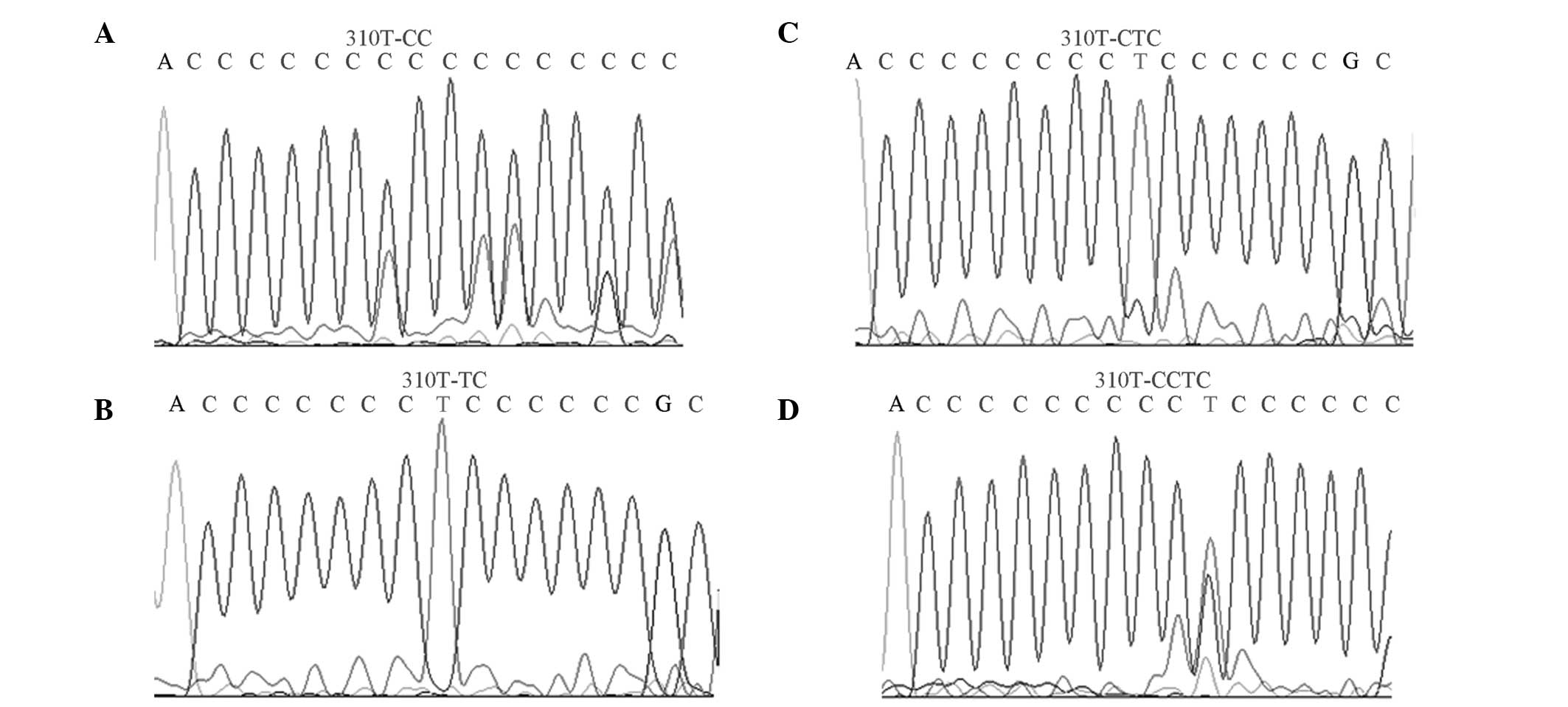
Mutation sites of np303–np315
The mutation site of np303–np315, known as the D310
region, has a high mutation rate. The mutation rate in the D310
region of the OSAHS group was significantly higher compared with
the healthy group (P<0.05; Fig.
3).
No statistically significant difference was
identified in the clinically relevant data (including Rohrer index)
between the mutation and non-mutation groups in the D310 region of
the OSAHS group (P>0.05; Table
III).
 | Table IIIAnalysis of D310 regional mutations
with clinical and biochemical indicators in the OSAHS group. |
Table III
Analysis of D310 regional mutations
with clinical and biochemical indicators in the OSAHS group.
| Indicator | Mutation group
(n=28) | No mutation group
(n=32) | P-value |
|---|
| Age | 39.77±11.35 | 40.47±11.27 | >0.05 |
| BMI | 25.86±2.77 | 26.52±2.28 | >0.05 |
| Rohrer index | 162.90±33.40 | 150.50±17.50 | >0.05 |
| Neck collar | 41.30±2.43 | 34.50±2.20 | >0.05 |
| FBG | 4.95±0.48 | 4.86±0.52 | >0.05 |
| TC | 5.64±0.96 | 5.46±0.96 | >0.05 |
| TG | 3.81±2.44 | 3.39±1.35 | >0.05 |
| HDL-C | 1.13±0.24 | 1.16±0.22 | >0.05 |
| LDL-C | 2.78±0.62 | 2.66±0.54 | >0.05 |
Mutation rates of np16189T-C
Mutation rates of np16189T-C in the OSAHS and
control groups were 46.7 and 29%, respectively. Statistical
significance was observed between the np16189T-C mutation and the
neck perimeter, TG and LDL levels in the OSAHS group (P<0.05;
Table IV).
 | Table IVAnalysis of the np16189T-C mutation
with clinical and biochemical indicators in the OSAHS group. |
Table IV
Analysis of the np16189T-C mutation
with clinical and biochemical indicators in the OSAHS group.
| Indicator | Mutation group
(n=28) | No mutation group
(n=32) | P-value |
|---|
| Age | 38.40±11.65 | 41.44±10.83 | >0.05 |
| BMI | 25.82±2.85 | 26.33±2.40 | >0.05 |
| Rohrer index | 158.90±37.40 | 144.50±18.50 | >0.05 |
| Neck collar | 40.30±2.60 | 38.50±2.40 | <0.05 |
| FBG | 4.90±0.50 | 4.95±0.50 | >0.05 |
| TC | 5.67±0.97 | 5.48±0.95 | >0.05 |
| TG | 3.74±2.73 | 3.73±1.67 | <0.05 |
| HDL-C | 1.09±0.24 | 1.18±0.23 | >0.05 |
| LDL-C | 2.29±0.37 | 3.18±0.52 | <0.05 |
Logistic regression analysis
Logistic regression analysis of the clinical
characteristics of the mtDNA mutations in the OSAHS group indicated
that LDL-cholesterol was associated with a genetic mutation
(Table V).
 | Table VLogistic regression analysis between
clinical features and mutations in the OSAHS group. |
Table V
Logistic regression analysis between
clinical features and mutations in the OSAHS group.
| Characteristic | Score | P-value |
|---|
| BMI | 0.307 | 0.579 |
| Smoking | 0.576 | 0.448 |
| Drinking | 1.603 | 0.206 |
| HBP | 0.066 | 0.798 |
| TC | 1.037 | 0.309 |
| TG | 1.152 | 0.697 |
| HDL-C | 0.152 | 0.697 |
| LDL-C | 4.443 | 0.035 |
Discussion
OSAHS has a number of causes and influencing
factors, including obesity, hyperlipidemia and hyperglycemia, which
are also associated with mitochondrial mutations (13,14).
The D-loop region is the starting point of mtDNA
replication and the site of regulatory functions in mtDNA
transcription and replication for the major non-coding regions of
human mtDNA. Therefore, mutations in the D-loop region lead to
disorders in mitochondrial function. Numerous diseases have been
reported to be associated with mutations in the D-loop region of
mtRNA, and this region is considered to be the common site of DNA
mutations in various gene-associated diseases (15). Therefore, two pairs of primers that
covered the mtDNA D-loop region were selected for amplification and
sequencing, and compared and analyzed with the standard Cambridge
sequence to investigate the correlation between the polymorphisms
in the D-loop region of the mitochondrial genome with the
occurrence, clinical manifestations and complications associated
with OSAHS.
Compared with the Cambridge standard sequence,
mutation sites were identified in the mtDNA D-loop region in each
sample from the OSAHS and healthy control groups. A total of 178
mutation sites were found, of which 75 mutation sites were observed
in the 60 OSAHS cases and 103 mutation sites were found in the 102
healthy control subjects. In total, 115 mutation sites were present
in the two groups; 27 mutation sites were present only in the OSAHS
group and 36 mutation sites were present only in the healthy
control group. Statistical analysis indicated that the non-common
mutation sites found only in the OSAHS group were significantly
greater than those in the control group, indicating that the mtDNA
D-loop region is a high-mutation-incidence region. This observation
is consistent with previous literature (16).
These results indicated that the human mtDNA
standard sequences, when compared between a Zhejiang Han population
and a Western Caucasian population, were not homologous. This
difference may be due to genetic polymorphisms, which may be
determined by the structural characteristics of a particular
population. The D-loop region is the site where mtDNA connects with
the inner mitochondrial membrane, thus, the D-loop region is
increasingly susceptible to peroxide damage. During mtDNA
replication, the D-loop region forms a three-chain structure that
exhibits greater susceptibility to injury (17). The presence of numerous mutations
in the mtDNA D-loop region indicates that mitochondrial oxidative
stress results in mutations (18).
Although the mtDNA D-loop region is the non-coding region of the
mitochondrial genome, the region contains essential elements for
transcription and replication. Thus, mutations in the D-loop region
may affect the transcription and replication of mtDNA. Furthermore,
such mutations can occur in the coding regions through
multiple-site mutations in the D-loop region, which would control
and change the transcription and translation of the coding genes.
Thus, the occurrence and development of diseases may be
affected.
The D310 region consists of 12–18 cytosine residues
and contains the highest number of mitochondrial microsatellite
polymorphisms, thus, is the major microsatellite-unstable region of
mtDNA (19). In addition, the D310
region is responsible for the formation of RNA-DNA complexes that
initiate mtDNA replication (20).
In mitochondria-associated diseases, the repeated deletion and
insertion of a single type of nucleotide (polyC) between np303 and
np310 in the D-loop region has become a major research focus. In
the present study, the mutations in the D310 region in the OSAHS
group were significantly higher compared with the healthy control
group, indicating that the mutations in this region may increase
OSAHS susceptibility. In addition, a greater quantity of mutation
sites with higher frequencies were observed in the OSAHS group.
OSAHS is a chronic disease with multiple causative genes,
therefore, further investigation is required to determine whether
the D-loop region, as the control region, contains numerous
mutation sites that affect the mutation of the entire mitochondrial
genome.
In the OSAHS group, the relevant clinical parameters
included age, BMI, neck perimeter, FBG, TC, TG, HDL and LDL levels.
The clinical data, with or without mitochondrial mutations, were
compared in the OSAHS group and the results revealed statistically
significant differences in the neck perimeter and TG and LDL levels
between the two groups (P<0.05). In addition, logistic
regression analysis was performed on partial clinical
characteristic variables of the patients with mitochondrial
mutations in the OSAHS group, and the results indicated that the
LDL level was associated with gene mutation.
Analysis of the mutation sites revealed 21
high-mutation sites (mutation rate >10%) in the OSAHS group and
24 sites in the healthy control group; the mutation rates of
np16519T-C in the OSAHS and control groups were 53.3 and 46%,
respectively. Poulton et al (21) hypothesized that the mitochondrial
16189 mutation was the candidate gene of the thrifty genome, thus,
a patient with a 16189 mutation experiences an increased risk of
obesity, diabetes and other diseases with increasing age. This
mutation site was considered to promote adult insulin resistance
and contribute significantly to the development of
non-insulin-dependent diabetes (22). Furthermore, Lin et al
(23) indicated that the
np16189T-C mutation reduced antioxidant functions under stress,
thus, this mutation increased the susceptibility to diabetes and
exacerbated the diabetic condition. OSAHS has been demonstrated to
be associated with risk factors of obesity, hypertension, insulin
resistance and high cholesterol levels. In particular, obesity is
the predominant risk factor of OSAHS, and OSAHS and obesity
susceptibility may have a common function through specific common
genes (24). The present study
revealed that the 16189T-C mutation rate of OSAHS patients,
excluding the risk of endocrine disorders, was significantly higher
compared with the healthy group, whereas no statistically
significant difference was identified among the mild, moderate and
severe subgroups of OSAHS. Therefore, the insulin resistance of
OSAHS patients is likely to be greater than the non-OSAHS group,
however, the occurrence rate of insulin resistance is not
associated with the severity of OSAHS. The incidence and associated
complications of OSAHS patients correlate with the 16189T-C
mutation in mtDNA.
In conclusion, the present study demonstrated that
specific sites in the D-loop region are associated with certain
clinical symptoms and complications in patients with OSAHS. The
observations emphasize the importance of early detection,
prevention and treatment, and complication reduction in patients
with OSAHS.
References
|
1
|
Petersen EJ and Reiter ER: Hospital use in
the treatment of sleep apnea. Laryngoscope. 114:460–466. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chmielewska I, Mlak R, Krawczyk P,
Czukiewska E and Milanowski J: Polymorphism of the ACE gene and the
risk of obstructive sleep apnoea. Pneumonol Alergol Pol.
81:207–213. 2013.PubMed/NCBI
|
|
3
|
Ho ML and Brass SD: Obstructive sleep
apnea. Neurol Int. 3:e152011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fusetti M, Fioretti AB, Valenti M, Masedu
F, Lauriello M and Pagliarella M: Cardiovascular and metabolic
comorbidities in patients with obstructive sleep apnoea syndrome.
Acta Otorhinolaryngol Ital. 32:320–325. 2012.PubMed/NCBI
|
|
5
|
Udwadia ZF, Doshi AV, Lonkar SG and Singh
CI: Prevalence of sleep-disordered breathing and sleep apnea in
middle-aged urban Indian men. Am J Respir Crit Care Med.
169:168–173. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel SR, Palmer LJ, Larkin EK, Jenny NS,
White DP and Redline S: Relationship between obstructive sleep
apnea and diurnal leptin rhythms. Sleep. 27:235–239.
2004.PubMed/NCBI
|
|
7
|
Dong JY, Zhang YH and Qin LQ: Obstructive
sleep apnea and cardiovascular risk: meta-analysis of prospective
cohort studies. Atherosclerosis. 229:489–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang D and Hamasaki N: Alterations of
mitochondrial DNA in common diseases and disease states: aging,
neurodegeneration, heart failure, diabetes, and cancer. Curr Med
Chem. 12:429–441. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Abu-Amero KK, Alzahrani AS, Zou M and Shi
Y: Association of mitochondrial DNA transversion mutations with
familial medullary thyroid carcinoma/multiple endocrine neoplasia
type 2 syndrome. Oncogene. 25:677–684. 2006. View Article : Google Scholar
|
|
10
|
Singh KK: Mitochondria damage checkpoint,
aging, and cancer. Ann N Y Acad Sci. 1067:182–190. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chatterjee A, Dasgupta S and Sidransky D:
Mitochondrial subversion in cancer. Cancer Prev Res (Phila).
4:638–654. 2011. View Article : Google Scholar
|
|
12
|
Lièvre A, Chapusot C, Bouvier AM, et al:
Clinical value of mitochondrial mutations in colorectal cancer. J
Clin Oncol. 23:3517–3525. 2005.
|
|
13
|
Mannarino MR, Di Filippo F and Pirro M:
Obstructive sleep apnea syndrome. Eur J Intern Med. 23:586–593.
2012. View Article : Google Scholar
|
|
14
|
Shen GX: Mitochondrial dysfunction,
oxidative stress and diabetic cardiovascular disorders. Cardiovasc
Hematol Disord Drug Targets. 12:106–112. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu M, Wan Y, Zou Q and Xi Y: High
frequency of mitochondrial DNA D-loop mutations in Ewing’s sarcoma.
Biochem Biophys Res Commun. 390:447–450. 2009.PubMed/NCBI
|
|
16
|
Wallace DC and Fan W: Energetics,
epigenetics, mitochondrial genetics. Mitochondrion. 10:12–31. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bianchi NO, Bianchi MS and Richard SM:
Mitochindrial genome instability in human cancers. Mutat Res.
488:9–23. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo W, Yang D, Xu H, et al: Mutations in
the D-loop region and increased copy number of mitochondrial DNA in
human laryngeal squamous cell carcinoma. Mol Biol Rep. 40:13–20.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng Z, Xie C, Wan Q, Zhang L, Li W and Wu
S: Sequence variations of mitochondrial DNA D-loop region are
associated with familial nasopharyngeal carcinoma. Mitochondrion.
11:327–333. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee DY and Clayton DA: Initiation of
mitochondrial DNA replication by transcription and R-LOOP
processing. J Biol Chem. 273:30614–30621. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Poulton J, Marchington DR, Scott-Brown M,
Phillips DI and Hagelberg E: Does a common mitochondrial DNA
polymorphism underlie susceptibility to diabetes and the thrifty
genotype? Trends Genet. 14:387–389. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park KS, Chan JC, Chuang LM, et al; Study
Group of Molecular Diabetology in Asia. A mitochondrial DNA variant
at position 16189 is associated with type 2 diabetes mellitus in
Asians. Diabetologia. 51:602–608. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin TK, Chen SD, Wang PW, et al: Increased
oxidative damage with altered antioxidative status in type 2
diabetic patients harboring the 16189 T to C variant of
mitochondrial DNA. Ann N Y Acad Sci. 1042:64–69. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Palmer LJ, Buxbaum SG, Larkin E, et al: A
whole-genome scan for obstructive sleep apnea and obesity. Am J Hum
Genet. 72:340–350. 2003. View
Article : Google Scholar : PubMed/NCBI
|