Introduction
The steroid-induced osteonecrosis of the femoral
head (SONFH) is a devastating, irreversible and disabling disease
developing following steroid therapy (1). The functions of the hip joint are
markedly impaired when the femoral head collapses. Glucocorticoid
(GC) therapy was the most common cause of ONFH (2). The onset of SONFH is within several
months following the administration of steroids. Regardless of
continuous steroid administration, no expansion of the necrotic
area was found, and recurrence was not identified. Due to ischemia,
patients have no symptoms when SONFH occurs. No pain is noted until
the femoral head collapses. Approximately 5–25% of patients using
GC develop SONFH in the legs. Notably, in China 53.5% of patients
with severe acute respiratory syndrome prescribed GC developed
femoral head osteonecrosis, and the SONFH comprises approximately
half of non-traumatic femoral head necrosis. In total, >50% of
patients with femoral head osteonecrosis admitted to The Third
Affiliated Hospital (Sun Yat-Sen University, Guangzhou, Guangdong,
China) between January 2000 and January 2010 had a history of using
GC, the majority of whom were young males. At present, patients
with SONFH already exceed 10 million in China. However, the
pathogenesis of this disease remains largely unknown, so the
prevention and treatment have yet to be established.
With the progress of systemic biology, proteomic
approaches provides a valuable tool to study the whole protein
content of a biological sample in one set of experiments. One of
the reliable technologies is two-dimensional (2-D) electrophoresis
or 2-D difference gel electrophoresis (DIGE) analysis coupled with
mass spectrometry identification (3,4).
This technology has recently been employed to screen potential
bioactive molecules underlying the pathogenesis of arthritis and
osteopenia, and it is reported that the protein levels in serum,
joint fluid and bone tissue correlate to the severity of rheumatoid
arthritis (5), the autoimmune
response in ankylosing spondylitis in animal models (6), the loss of cartilage integrity in
osteoarthritis (7) and estrogen
loss-induced osteoporosis (8).
However, the changes detected in the serum protein profile of
patients with SONFH have not yet been reported.
The purpose of the present study is to find
potential biomarkers of the SONFH by using proteomic technology to
analyze serum protein profiles in patients with SONFH and the
healthy control group.
Materials and methods
Patients
The study was approved by the Institutional Review
Board of The Third Affiliated Hospital, and informed consent was
obtained from each patient. A total of 12 patients, including five
females and seven males, fulfilled the inclusion and exclusion
criteria. Inclusion criteria included receiving a single
short-course of corticosteroid medication within the three years
prior to presentation, ONFH identified by typical magnetic
resonance imaging (MRI) findings, stage 1 or 2 by the Ficat
classification system and no previous treatment of the femoral head
(9). The exclusion criteria for
sampling were the history or evidence of metabolic bone diseases,
including hyper- or hypoparathyroidism, Paget’s disease, renal
osteodystrophy and the presence of cancers with bone metastasis. In
total, 12 healthy volunteers, including five females and seven
males, were enrolled in the control group. Peripheral venous blood
(5 ml) was drawn from each patient in the Outpatient Department or
in the procedure room prior to general anesthesia for total hip
replacement. The blood samples were processed to collect serum and
stored at −80°C until analysis.
Serum sample preparation
Blood samples (3 ml) from each subject were
collected in a drying tube early in the morning and allowed to clot
for 1 h at room temperature. The samples were centrifuged at 2,500
× g for 15 min at 4°C. The supernatant was dispensed into 0.5 ml
aliquots and stored at −80°C until use. All the serum samples were
processed according to a standard protocol.
Depletion of high abundance proteins and
quantification
All the samples were thawed at room temperature.
High abundance proteins, such as albumin and immunoglobulin G
(IgG), were depleted with the ProteoPrep® Blue Albumin
& IgG Depletion kit (Sigma-Aldrich, Inc., St. Louis, MO, USA)
as per the manufacturer’s instructions. Subsequently, samples were
further cleaned with the Clean-up kit (GE Healthcare, Piscataway,
NJ, USA) according to the manufacturer’s instructions. Proteins
were quantified using 2-D Quant kit (GE Healthcare). The protein
concentrations of samples were adjusted to 8 mg/ml and verified by
SDS-PAGE and Coomassie blue staining.
Fluorescent labeling
To limit experimental variation and ensure accurate
in-gel matching, all the samples were labeled with fluorescence.
The CyDye DIGE Fluor (minimal dye) labeling kit (GE Healthcare) was
used for tagging, following the manufacturer’s instructions.
Cyanine 2 (Cy2) was used for the internal standard sample that was
generated by mixing together an aliquot of samples from the patient
and control groups. Cy3 and Cy5 were used to label samples from the
control and patient groups, respectively. The labeling reaction was
carried out in the dark on ice.
2D-DIGE and in-gel trypsin digestion
All the labeled samples were subjected to 2D-DIGE
with 24 cm immobilized pH gradient (IPG) strips (pH 4–7) (GE
Healthcare) for the first dimensional isoelectric focusing and
subsequently the second dimensional SDS-PAGE with 12%
polyacrylamide gels. The 2D-DIGE was run in triplicate for each
sample to reduce the gel-to-gel variation. The gels were scanned
using Typhoon 9410 Variable Mode Imager (GE Healthcare) at the
excitation/emission wavelengths specific for each CyDyes
immediately. Subsequent to scanning, all the gels were stained with
Deep Purple and stored for subsequent mass spectrometric
identification. Images were then processed with DeCyder
Differential in Gel Analysis V6.0 software (GE Healthcare) to
identify changes in spot fluorescence intensities. Proteins were
considered differentially expressed if the abundance showed
>1.5-fold change between the patient and the control groups with
P<0.05 using one-way analysis of variance. Disparate points were
excised using the Ettan spot handing workstation (GE Healthcare),
then destained and subjected to in-gel trypsin digestion. Peptides
were extracted for subsequent mass spectrometry.
Matrix-assisted laser desorption
ionization time-of-flight mass spectrometry (MALDI-TOF-MS/MS) and
protein identification
Following trypsin digestion, protein identification
was carried out on the Ettan MALDI-TOF mass spectrometer (GE
Healthcare). The trypsin auto-digestion peaks (m/z 842.509 and
2211.104 Da) were used for internal calibration. Each spectrum
corresponded to the sum of 200 acquisitions for each of eight laser
pulses, in which the threshold signal/noise exceeded a set value.
The resulting data were then analyzed with Mascot search engine
(Matrix Science, London, UK) for protein identification and
compared to the National Center for Biotechnology Information
(10) and Swiss-Prot (11) protein databases. The following
keywords were used in the search: Trypsin digestion, Homo
sapiens, 1–100 kDa protein mass, 100 ppm peptide tolerance.
Western blot analysis
Serum protein samples prepared as described above
were diluted 1:25 in Laemmli buffer and resolved by 10% SDS-PAGE
(Invitrogen Life Technologies, Carlsbad, CA, USA). The separated
proteins were transferred to polyvinylidene fluoride. The membranes
were blocked with 5% skimmed dry milk in Tris-buffered saline
containing 0.05% Tween 20 (TBST) for 1 h at room temperature and
incubated with antibodies overnight at 4°C. Subsequent to washing
two times in TBST, the membranes were incubated with corresponding
horseradish peroxidase-conjugated secondary antibody for 1 h at
room temperature. Protein bands were detected using ECL Plus
(Forevergen Bioscience Co., Ltd., Guangzhou, China) and the images
were acquired by the Imaging System (Gel Doc XR System, Bio-Rad,
Hercules, CA, USA).
ELISA
ELISA assays using a microtiter plate assay were
performed individually on the samples in each group chosen randomly
and matched across the groups. Primary and secondary antibodies
were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA,
USA). A standard curve was generated by four-parameter
curve-fitting using SoftmaxPro V 1.11 software, (Molecular Devices
Corp., Sunnyvale, CA, USA).
Statistical analysis
The data analyses were performed by SPSS 15.0 (SPSS,
Inc., Chicago, IL, USA). Statistical significance of the
differences was determined using the Student’s t-test, and the
Tukey method was performed to correct multiple comparisons. All the
values were reported as the mean ± standard deviation, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patients
In total, 12 patients with SONFH (age, 32.3±2.3
years; range, 25–40 years) and 12 healthy volunteers (age, 33±2.3
years; range, 28–41 years) were enrolled. There was no significant
difference in the age (P=0.827) between the SONFH and healthy
volunteer groups. All the patients were diagnosed with ONFH by MRI.
The patients classified as stage 1 or 2 by the Ficat classification
system were enrolled in the study. The medical records show that
all of them had received glucocorticoid therapy due to arthralgia.
The mean steroid dose in equivalent milligrams of prednisone was
850 mg (range, 290–3300 mg). The mean time from administration of
steroids to the development of hip symptoms was 16.6 months (range,
6–33 months), but none of them fulfilled the diagnosis criteria of
rheumatoid arthritis.
2-D DIGE analysis of differential protein
expression
2-D DIGE was performed to analyze differential
protein expressions as previously described (12). Approximately 1,600 protein spots
were detected across all four gels by the DeCyder image analysis
software (GE Healthcare). Fig. 1A
shows the superimposed images in pseudocolor from Cy3- and
Cy5-labelled sera samples and the positions of the spots
corresponding to the proteins revealed by 2-D DIGE analysis.
Fig. 1B presents the differences
in protein expression by the DeCyder 3-D spot simulations. Four
protein spots with >1.4-fold decrease in abundance between the
patient and control groups were identified by computer-assisted
comparative analysis (P<0.05, Student’s t-test; Table I). To further confirm the change of
these four protein spots, two preparative gels loaded with 500 μg
protein from each extract were run in parallel and followed by Deep
Purple staining. The decrease of the protein abundance was
consistent with the result of CyDye labeled images as analyzed by
DeCyder software. Four proteins were revealed to be downregulated
in the sera of patients with SONFH.
 | Table IProteins presenting significant
differences in abundance in the steroid-induced femoral head
osteonecrosis group versus the control group. |
Table I
Proteins presenting significant
differences in abundance in the steroid-induced femoral head
osteonecrosis group versus the control group.
| Position | Master number | T-test | Average ratio | Mw | Identification |
|---|
| 1 | 540 | 0.024 | −1.49 | 89 | ITIH4_HUMAN |
| 2 | 626 | 0.048 | −2.04 | 85 | CO4A_HUMAN or
CO4B_HUMAN |
| 3 | 1092 | 0.032 | −1.58 | 67 | A2MG_HUMAN |
| 4 | 1416 | 0.011 | −2.16 | 48 | CO3_HUMAN |
Protein identification
The above four differential protein spots were
excised and digested with trypsin in gel for MALDI-TOF peptide mass
fingerprinting (PMF) analysis. The product spectra generated by
MALDI-TOF-MS/MS were searched against the Swiss-Prot database for
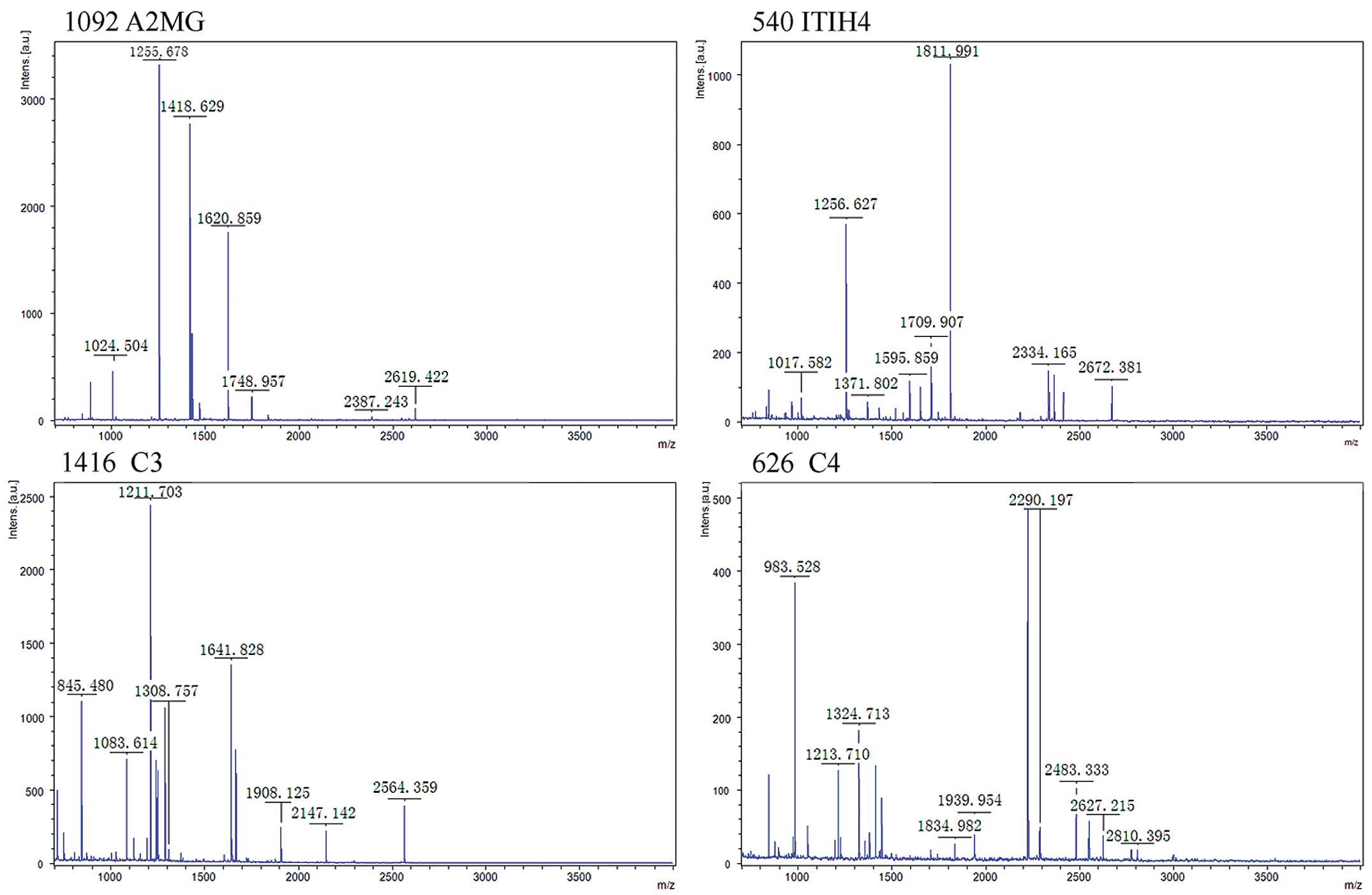
exact matches using the MASCOT by PMF (Fig. 2). The four proteins were
respectively inter-α-trypsin inhibitor heavy chain H4 (ITIH4),
complement component 4 (C4), A2MG and C3 (Table I).
Western blot analysis and ELISA to
confirm the differential expression
To validate the differential expressions of these
four proteins, the eight serum samples used for the DIGE experiment
were assessed by western blot analysis with the specific
antibodies. The levels of C4, ITIH4, A2MG and C3 are significantly
lower in the patient group than in the control group (Fig. 3), which is consistent with the
results from the proteomic study. Subsequently, it was determined
whether proteins express differentially during femoral head tissue
necrosis. To test if it is the case for A2MG, the proteins prepared
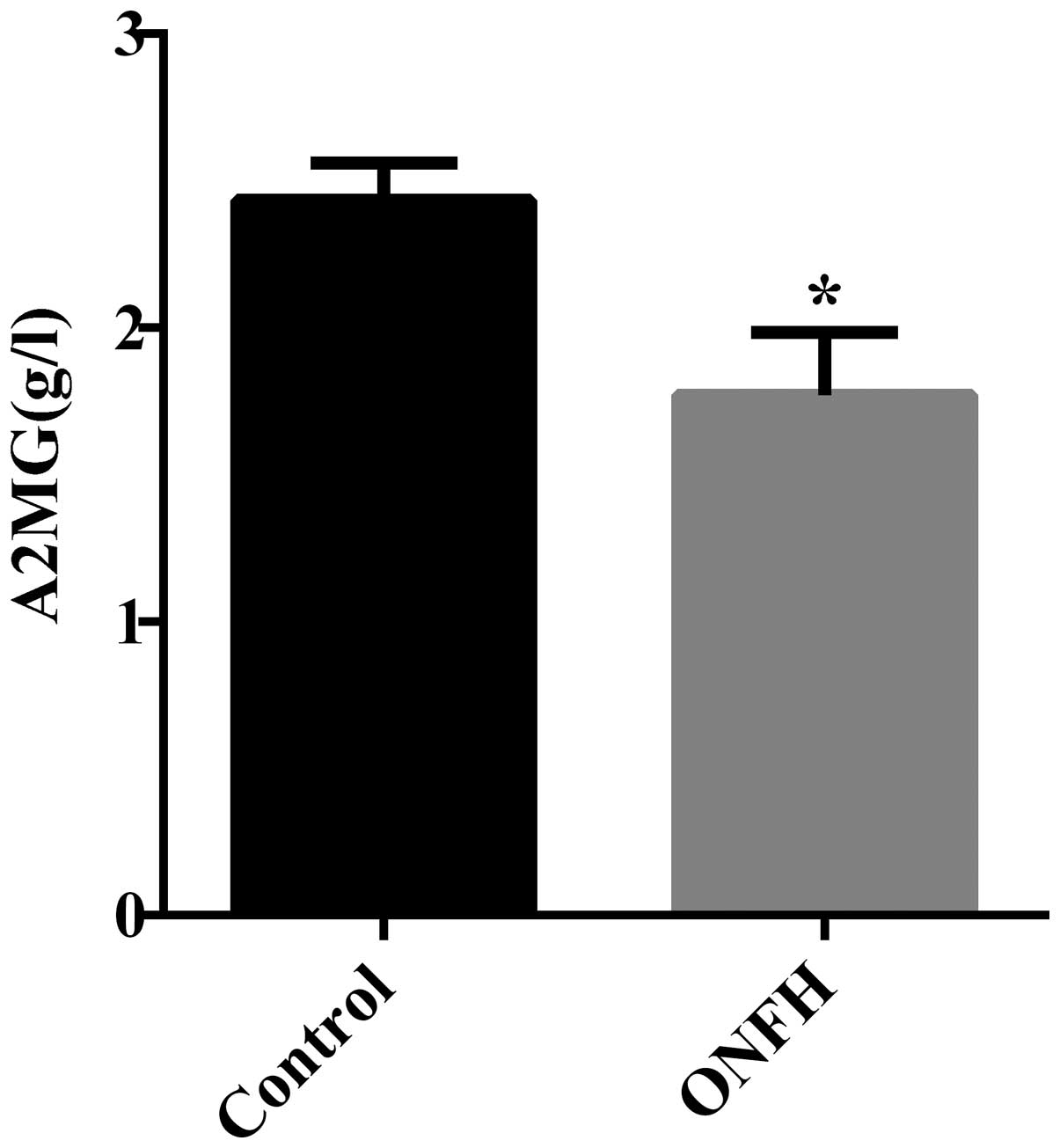
from the necrotic femoral head tissue were analyzed by ELISA. The
expression of A2MG was significantly lower in patient group than in
the control group (Fig. 4), which
is consistent with the result of the serum samples. No change was
observed in the levels of C3, C4 and ITIH4 in necrotic bone tissues
(data not shown).
Discussion
The incidence of SONFH is increasing year by year,
while the diagnosis of this disorder still relies on image
examination, which often fails to detect the lesion at the early
stage. Therefore, a number of patients miss the opportunities for
early treatment. It is important to find the diagnostic biomarkers
for SONFH. Since blood tests are commonly used in clinical
practice, the protein profile in the serum from the patients with
SONFH and healthy volunteers were analyzed using cutting-edge
proteomic technology. The aim was to identify proteins whose level
in the blood was significantly altered in patients with SONFH. To
ensure the reproducibility, accuracy and objectivity of the
experiments, the following steps were implemented to minimize the
experimental errors between samples. First, the sample selection
criteria were strictly followed. Secondly, high abundance proteins
were removed prior to the electrophoresis, which otherwise would
mask the low abundance proteins. Further, fluorescence labeled 2-D
DIGE was employed. An internal standard was used for each protein
spot in 2-D DIGE, and the software designed for 2-D DIGE
automatically corrected the protein amount according to the
internal standard. Thus, these approaches significantly improved
the reproducibility and the sensitivity.
In the present study, four proteins (C3, C4, ITIH4
and A2MG) showed lower expression in the serum of patients with
SONFH than that of the normal subjects. The changes were confirmed
by western blotting. The expressions of these proteins were also
examined in necrotic bone tissues. Unlike serum, necrotic bone
tissue did not have detectable amounts of C4 and ITIH4. In
addition, C3 showed no difference in abundance between the two
groups. Only A2MG was downregulated in the protein levels in
necrotic bone tissue, consistent with the result of the serum.
The pathogenesis of SONFH remains unclear. Several
mechanisms have been proposed, including lipid metabolism
dysfunction, intravascular coagulation, apoptosis and reactive
oxygen imbalances. The present study shows that the expression of
C3, C4, ITIH4 and A2MG were significantly altered in patients with
SONFH. All four proteins are closely associated with apoptosis, and
therefore the present study supports that apoptosis plays a major
role in SONFH.
C3, C4 and their degradation products are cytokines
and acute phase reactive proteins that are produced by macrophages
and hepatocytes. They are key factors in the activation of the
complement system. The activation of the complement cascade can
cause a variety of biological effects, including immune response,
generation of sensitized lymphocytes and altered metabolism of
blood sugar and lipid (13).
However, limited studies have systematically elucidated the
mechanism by which the immune system affects SONFH. Wu et al
(14) showed that the complement
factor C3 precursor is elevated in the serum of patients with ONFH.
This previous study showed that complement factor C3 precursor
plays an important role in the homeostasis of inflammation,
necrosis or apoptosis in ONFH. The present results show that
complement activation is reduced in patients with SONFH. This may
be attributed to the immunosuppressive effect of steroids. Excess
steroids can suppress complement activation and immune complex
formation (15). Familian et
al (16) found that plasma
levels of C3 and C4 increased in the majority of patients with
rheumatoid arthritis prior to therapy, but significantly decreased
following the start of infliximab (an immunosuppressive agent)
treatment. The mechanism of complement inhibition involved in SONFH
requires further study.
ITIH4, when translated, is secreted into the
blood, where it is cleaved by plasma kallikrein into two smaller
forms. ITIH4 mRNA is specifically expressed in the liver.
The gene is part of a cluster of similar genes on chromosome 3. Two
transcription variants encoding different isoforms have been found.
ITIH4 is also an acute phase reactive protein, but its biological
function remains unknown. It was detected in swine, bovine and rat
models with experimentally-induced acute inflammation (17–19).
Pineiro et al (20) showed
that in humans, ITIH4 mRNA and the secreted protein are
highly upregulated by IL-6 in HepG2 hepatoma cells. Bost et
al (21) assumed ITIH4 may
interact with components of the extracellular matrix and modulate
cell migration and proliferation during the development of the
acute-phase response. It is clear that ONFH is accompanied by
inflammation. Aseptic inflammation presents in patients with ONFH
and it is conceivable that persistent consumptive inflammation and
the effects of steroids lead to the decrease of serum ITIH4.
Further study is necessary to address the role of ITIH4 in the
disease.
A2MG is an inhibitor of matrix metalloproteases
(MMP) (22), which is mainly
synthesized by hepatocytes in the liver. Small amounts of A2MG are
also produced by a number of other cells, including lung
fibroblasts, macrophages, astrocytes and tumor cells (23,24).
A2MG functions as a broad irreversible proteinase inhibitor and is
involved in various physiological processes (25,26).
A2MG regulates several key factors of SONFH. The conformational
change can activate A2MG, resulting in exposure of binding sites
for its cell surface receptor, including the low-density
lipoprotein receptor-related protein. Upon binding A2MG-proteinase
complexes from the extracellular matrix are rapidly removed, which
blocks lipid catabolism (27).
A2MG modulates blood coagulation. As reported by Simpson et
al (28), A2MG significantly
enhanced plasmin generation. However, A2MG binds vascular
endothelial growth factor and the resultant A2MG-complex inhibits
heparin activity, leading to elevated coagulation. Human A2MG has
been verified to effectively decrease the release of superoxide
radicals by polynuclear leukocytes following radiation. The
activity of superoxide dismutase in red cells can also be
increased. The free radicals and MMP imbalance exist in the
pathological process of SONFH. Kerachian et al (29) demonstrated that the A2MG
gene is significantly upregulated in avascular necrosis of the rat
femoral head induced with steroids. Along with those findings, the
present study showed that A2MG was significantly lower in the bone
tissue of patients with SONFH. Lower A2MG may affect the process of
SONFH through these aspects. Consistent with the bone tissue, the
serum A2MG level was also decreased.
In conclusion, A2MG is involved in multiple
mechanisms underlying SONFH, including blood coagulation,
hyperlipidemia, free radicals and MMP degradation. This underscores
the critical role of A2GM in the development of SONFH. Therefore,
A2GM may become a novel potential biomarker and a novel therapeutic
target for SONFH.
Acknowledgements
The present study was supported by grants from the
863 Scientific Research of the National Natural Science Foundation
of China (no. 2008AA02Z437), Key Project of Guangdong Provincial
Science and Technology Research (no. 2008A030201026) and the
Sci-tech Research Development Program of Guangzhou (no.
2011Y1-00033). The authors would like to thank Miss Lu Hui-qiong
and Mr Xu Shao-fei (Forevergen Bioscience Co., Ltd.) for providing
biochemical technique support.
Abbreviations:
|
SONFH
|
Steroid-induced osteonecrosis of the
femoral head
|
|
ONFH
|
osteonecrosis of the femoral head
|
|
ITIH4
|
Inter-α-trypsin inhibitor heavy chain
H4
|
|
A2MG
|
α-2-macroglobulin
|
References
|
1
|
Kubo T, Fujioka M and Ishida M: Clinical
condition of steroid-induced osteonecrosis of the femoral head.
Clin Calcium. 17:856–862. 2007.(In Japanese). PubMed/NCBI
|
|
2
|
Kerachian MA, Séguin C and Harvey EJ:
Glucocorticoids in osteonecrosis of the femoral head: a new
understanding of the mechanisms of action. J Steroid Biochem Mol
Biol. 114:121–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiede B and Rudel T: Proteome analysis of
apoptotic cells. Mass Spectrom Rev. 23:333–349. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu W, Zhou XW, Liu S, et al:
Calpain-truncated CRMP-3 and -4 contribute to potassium
deprivation-induced apoptosis of cerebellar granule neurons.
Proteomics. 9:3712–3728. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liao H, Wu J, Kuhn E, et al: Use of mass
spectrometry to identify protein biomarkers of disease severity in
the synovial fluid and serum of patients with rheumatoid arthritis.
Arthritis Rheum. 50:3792–3803. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J, Zhu P, Peng J, et al:
Identification of disease-associated proteins by proteomic approach
in ankylosing spondylitis. Biochem Biophys Res Commun. 357:531–536.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hermansson M, Sawaji Y, Bolton M, et al:
Proteomic analysis of articular cartilage shows increased type II
collagen synthesis in osteoarthritis and expression of inhibin
betaA (activin A), a regulatory molecule for chondrocytes. J Biol
Chem. 279:43514–43521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pastorelli R, Carpi D, Airoldi L, et al:
Proteome analysis for the identification of in vivo
estrogen-regulated proteins in bone. Proteomics. 5:4936–4945. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jawad MU, Haleem AA and Scully SP: In
brief: Ficat classification: avascular necrosis of the femoral
head. Clin Orthop Relat Res. 470:2636–2639. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
National Center for Biotechnology
Information. http://www.ncbi.nlm.nih.gov/uri.
Accessed August 15, 2013
|
|
11
|
Expasy Bioinformatics Resource Portal.
UniProtKB/Swiss-Prot guideline. http://web.expasy.org/docs/swiss-prot_guideline.htmluri.
Accessed August 20, 2013
|
|
12
|
Yu KH, Rustgi AK and Blair IA:
Characterization of proteins in human pancreatic cancer serum using
differential gel electrophoresis and tandem mass spectrometry. J
Proteome Res. 4:1742–1751. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baldo A, Sniderman AD, St-Luce S, et al:
The adipsin-acylation stimulating protein system and regulation of
intracellular triglyceride synthesis. J Clin Invest. 92:1543–1547.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu RW, Wang FS, Ko JY, Wang CJ and Wu SL:
Comparative serum proteome expression of osteonecrosis of the
femoral head in adults. Bone. 43:561–566. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luukkainen R, Hakala M, Sajanti E, Huhtala
H, Yli-Kerttula U and Hameenkorpi R: Predictive value of synovial
fluid analysis in estimating the efficacy of intra-articular
corticosteroid injections in patients with rheumatoid arthritis.
Ann Rheum Dis. 51:874–876. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Familian A, Voskuyl AE, van Mierlo GJ, et
al: Infliximab treatment reduces complement activation in patients
with rheumatoid arthritis. Ann Rheum Dis. 64:1003–1008. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lampreave F, Gonzalez-Ramon N,
Martinez-Ayensa S, et al: Characterization of the acute phase serum
protein response in pigs. Electrophoresis. 15:672–676. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pineiro M, Andres M, Iturralde M, et al:
ITIH4 (inter-alpha-trypsin inhibitor heavy chain 4) is a new
acute-phase protein isolated from cattle during experimental
infection. Infect Immun. 72:3777–3782. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Daveau M, Jean L, Soury E, et al: Hepatic
and extra-hepatic transcription of inter-alpha-inhibitor family
genes under normal or acute inflammatory conditions in rat. Arch
Biochem Biophys. 350:315–323. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pineiro M, Alava MA, Gonzalez-Ramon N, et
al: ITIH4 serum concentration increases during acute-phase
processes in human patients and is up-regulated by interleukin-6 in
hepatocarcinoma HepG2 cells. Biochem Biophys Res Commun.
263:224–229. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bost F, Diarra-Mehrpour M and Martin JP:
Inter-alpha-trypsin inhibitor proteoglycan family-a group of
proteins binding and stabilizing the extracellular matrix. Eur J
Biochem. 252:339–346. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tu G, Xu W, Huang H and Li S: Progress in
the development of matrix metalloproteinase inhibitors. Curr Med
Chem. 15:1388–1395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Borth W: Alpha 2-macroglobulin, a
multifunctional binding protein with targeting characteristics.
FASEB J. 6:3345–3353. 1992.PubMed/NCBI
|
|
24
|
Baker AH, Edwards DR and Murphy G:
Metalloproteinase inhibitors: biological actions and therapeutic
opportunities. J Cell Sci. 115:3719–3727. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zorin NA, Zorina VN and Zorina RM: Role of
alpha-2 macroglobulin in oncologic diseases. Vopr Onkol.
50:515–519. 2004.(In Russian).
|
|
26
|
Mocchegiani E, Costarelli L, Giacconi R,
Cipriano C, Muti E and Malavolta M: Zinc-binding proteins
(metallothionein and alpha-2 macroglobulin) and immunosenescence.
Exp Gerontol. 41:1094–1107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Ge G and Greenspan DS: Inhibition
of bone morphogenetic protein 1 by native and altered forms of
alpha2-macroglobulin. J Biol Chem. 281:39096–39104. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Simpson ML, Goldenberg NA, Jacobson LJ,
Bombardier CG, Hathaway WE and Manco-Johnson MJ: Simultaneous
thrombin and plasmin generation capacities in normal and abnormal
states of coagulation and fibrinolysis in children and adults.
Thromb Res. 127:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kerachian MA, Cournoyer D, Harvey EJ, et
al: New insights into the pathogenesis of glucocorticoid-induced
avascular necrosis: microarray analysis of gene expression in a rat
model. Arthritis Res Ther. 12:R1242010. View Article : Google Scholar : PubMed/NCBI
|