Introduction
Chromium is widely used as an important industrial
material in leather tanning, dyeing, electroplating, pigment
manufacturing and other industries. However, the uncontrolled
release of industrial waste has contaminated soil water systems
(1). As a result of the secondary
pollution and high cost, traditional, physical and chemical
technologies cannot be extensively used to remedy the contaminated
arable land and water. Thus, a number of microbial approaches to
remedy chromium contamination have been investigated in attempts to
overcome the problems (2–4).
Chromium exists in nature as two main oxidation
states, hexavalent [Cr(VI)] and trivalent chromium [Cr(III)].
Cr(VI) is considered to be a more toxic form, while Cr(III) is
relatively innocuous. Microbial bioremediation of chromium
contamination is achieved mainly through two routes (5). One is the efflux of chromate ions
from the cell cytoplasm, and the other is the direct reduction of
Cr(VI) to Cr(III) by the NAD(P)H-dependent flavin mononucleotide
(FMN) reductase (FMN_red) (6).
Various bacteria and associated genes have been identified to
reduce Cr(VI) to Cr(III) (7–9).
Previous studies have demonstrated that the ChrR enzyme from
Pseudomonas putida (10),
the YieF protein from Escherichia coli (E. coli) and the
FMN_red from Pseudomonas aeruginosa (PAO1) have the ability
to reduce Cr(VI) to Cr(III) (11,12),
all of which are members of the FMN_red protein family.
In the present study, a bacterium strain,
Serratia sp. CQMUS2, that possesses high chromate
resistance and rapid chromate reduction ability, was isolated from
chromate-containing waste water generated in our previous research
(13). The full-length DNA of ChrT
from Serratia sp. CQMUS2 was cloned and the deduced amino
acid sequence and three-dimensional (3D) structure were analyzed.
The results may provide a basis for further studies on ChrT gene
expression and protein function.
Materials and methods
Strains, media and plasmid. Serratia
sp
CQMUS2 was subcultured in Luria-Bertani medium
(Sangon Biotech Co., Ltd., Shanghai, China). E. coli DH5α
was used as a host for gene cloning (Tiangen Biotech Co., Ltd.,
Beijing, China) and the pMD19-T plasmid was used as a cloning
vector (Takara Bio, Inc., Otsu, Japan).
Polymerase chain reaction (PCR)
Genomic DNA was isolated according to boiled
template method (14). Following
culture for 3 h, the cells of Serratia sp. CQMUS2 were
harvested by centrifugation at 6,000 × g for 10 min. Cell pellets
were washed three times with 0.01 M phosphate-buffered saline,
suspended in sterile water and boiled for 10 min. Following
centrifugation at 12,000 × g for 10 min, the supernatant was
collected for PCR. The FMN_red gene from Serratia sp. AS13
(GenBank accession no. NC_017573) was selected as the
reference sequence. A pair of primers, Scfmn1-forward (F) and
Scfmn1-reverse (R), were designed and used to amplify the fragment
of the Serratia sp. CQMUS2. The ChrT gene was amplified
using 2xPfu PCR MasterMix (Tiangen Biotech Co., Ltd.), according to
the manufacturer’s instructions. The PCR program was as follows:
One cycle at 94°C for 3 min; 30 cycles at 94°C for 30 sec, 55°C for
30 sec and 72°C for 1 min; and one cycle at 72°C for 5 min. The
primers are shown in Table I
(Sangon Biotech Co., Ltd.).
 | Table IPrimers used in the study. |
Table I
Primers used in the study.
| Procedure | Primers | Primer sequences
(5′-3′) |
|---|
| ChrT gene
fragment | Scfmn1-F |
GATGTGCAACAAGACGAAGGT |
| Scfmn1-R |
GATGACTCCGCCCATAAACTC |
| 5′-hiTAIL-PCR | Scfmn-SP1 |
GATGACTCCGCCCATAAACTCC |
| Scfmn-SP2 |
AGACTCACCAGAGTCGATGGCGTTTTTCAGGC |
| Scfmn-SP3 |
CTATCACAACCCCATCCGCCT |
| Scfmn-AD1 |
ACGCTAGACTCACCTCVNVNNNGGAA |
| Scfmn-AC1 | ACGCTAGACTCACCTC |
| 3′-hiTAIL-PCR | Scfmn-SP1′ |
AGGCGGATGGGGTTGTGATAG |
| Scfmn-SP2′ |
GAACTGCACACGCCTGAAAAACGCCATCGACT |
| Scfmn-SP3′ |
GTGCGCGCTGCCAGTATCAT |
| Scfmn-AD1′ |
ACGATGAACTGCACTGVVNVNNNCCAA |
| Scfmn-AC1′ | ACGATGAACTGCACTG |
| Full-length ChrT
gene | Scfmn-F |
ATCATGTCAGATACCTTGAAAGTGG |
| Scfmn-R |
TGCTTTAACCCGCCGAATATA |
Based on the acquired DNA fragment sequence, a high
efficiency TAIL-PCR (hiTAIL-PCR) method was used to obtain 5′- and
3′-flanking sequences. This method required three consecutive
rounds of PCR. In the first round, the Scfmn-SP1 and Scfmn-AD1
primers were used for rapid amplification of the 5′-DNA end. The
second round of PCR used the product of the first round as the
template, and used Scfmn-SP2 and Scfmn-AC1 as the primers. The
third round of PCR used the product of the second round as the
template, and used Scfmn-SP3 and Scfmn-AC1 as primers. The
amplification of the 3′-end DNA fragment was the same as the
5′-end. The detailed steps and PCR parameters were performed, as
described previously (15). All
amplified products were run in 2% agarose gel and subsequently
underwent Sanger sequencing.
The full-length ChrT gene was obtained using
specific primers (Scfmn-F and Scfmn-R) corresponding to the 5′- and
3′-ends of the ChrT gene. The PCR parameters were as follows: One
cycle at 94°C for 3 min; 30 cycles at 94°C for 30 sec, 56°C for 30
sec and 72°C for 1 min; and one cycle at 72°C for 5 min. The
purified PCR product was cloned into a pMD19-T vector for
sequencing.
Nucleotide sequence accession number
The nucleotide sequence of ChrT was submitted onto
GenBank under the accession number, KF211434.
Sequence analysis
Sequence analysis of the gene and amino acids was
conducted using the Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST/). Open reading
frame (ORF) analysis was performed using ORF Finder (http://www.ncbi.nlm.nih.gov/projects/gorf/). In
addition, the physicochemical characteristics of the ChrT gene were
analyzed using ProtParam software (http://www.expasy.ch/tools/protparam.html), while the
conserved structural domain of ChrT was calculated via the
Conserved Domain Database (CDD) in the National Center for
Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Multiple alignments between the ChrT gene and other bacterial
FMN_red genes were conducted using Clustal X version 2.0 (16). A phylogenetic tree of FMN_red was
constructed using the neighbor-joining method with Molecular
Evolutionary Genetics Analysis (MEGA) software (version 4.0.2)
(17). The secondary and 3D
structures of ChrT were predicted using the NPS@ service
(http://npsa-pbil.ibcp.fr/) and
SWISS-MODEL program (http://swissmodel.expasy.org/).
Results
ChrT gene cloning from Serratia sp.
CQMUS2
To obtain the full-length ChrT gene from
Serratia sp. CQMUS2, a hiTAIL-PCR method was used. Firstly,
a pair of specific primers was synthesized on the basis of the
FMN_red gene sequence of Serratia sp. AS13. When the genomic
DNA from the Serratia sp. CQMUS2 cells was used as a
template, an expected 305-bp fragment of the ChrT gene was
amplified using the Scfmn1-F/R primers, which was subsequently
sequenced (Fig. 1A). Based on this
DNA fragment, the 5′- and 3′-end fragments were amplified by 5′-
and 3′-hiTAIL-PCR. Two DNA fragments were obtained in the third
round of the hiTAIL-PCR. The fragments were 321 and 747 bp in
length, respectively (Fig. 1B and
C). Finally, on the basis of the three DNA fragment assembly, a
full-length ChrT gene of 567 bp was obtained with the specific
Scfmn-F/R primers (Fig. 1D). This
result demonstrated that the ChrT gene was able to be successfully
cloned from Serratia sp. CQMUS2.
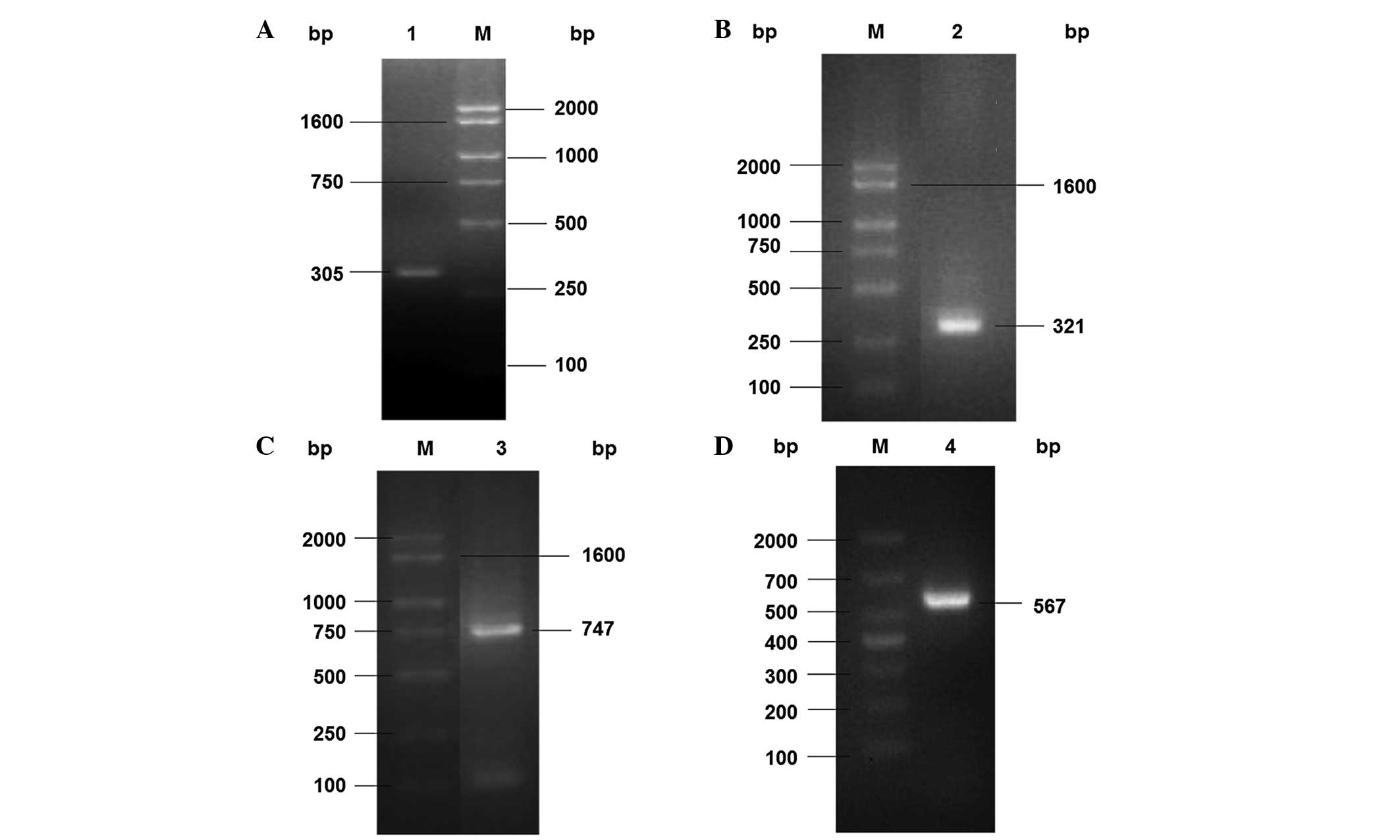 | Figure 1Cloning of Serratia sp. CQMUS2
ChrT gene fragments. (A) PCR amplification of the ChrT gene
fragment: Lane M, DNA marker; lane 1, a 305-bp fragment obtained by
PCR amplification with the Scfmn1-F and Scfmn1-R primers. (B) PCR
amplification of the 5′-end DNA fragment: Lane M, DNA marker; lane
2, a 321-bp fragment obtained by the third round hiTAIL-PCR
amplification with the Scfmn1-SP3 and Scfmn1-AC1 primers. (C) PCR
amplification of the 3′-end DNA fragment: Lane M, DNA marker; lane
3, a 747-bp fragment obtained by the third round hiTAIL-PCR
amplification with the Scfmn1-SP3′ and Scfmn1-AC1′ primers. (D) PCR
amplification of the complete DNA: Lane M, DNA marker; lane 4, the
567-bp ChrT gene obtained by PCR amplification with the Scfmn-F and
Scfmn-R primers. PCR, polymerase chain reaction; F, forward; R,
reverse. |
ChrT protein may be an NADPH-dependent
FMN_red and a member of the flavodoxin-2 superfamily
To understand the characteristics of the ChrT
protein, its nucleotide sequence was investigated. Nucleotide
sequence analysis indicated that the Serratia sp. CQMUS2
ChrT gene contained 567 bp nucleotides with an ORF of 567 bp, which
encoded a 188-amino acid peptide with a theoretical molecular
weight of 20.4 kDa and an isoelectric point of 5.18. The conserved
domain was predicted by CDD in NCBI, and revealed that the putative
polypeptide from Serratia sp. CQMUS2 contained an integrated
conserved region of FMN_red formed by 5–150 amino acids.
Consequently, the result predicted that the deduced protein was an
NADPH-dependent FMN_red and a member of the flavodoxin-2
superfamily; thus, the peptide was named as FMN_red ChrT.
ChrT is closely associated with FMN_red
members in Klebsiella pneumonia (YP_002241302), Raoultella
ornithinolytica (YP_007875969) and Klebsiella oxytoca
(YP_005017294.1)
To compare the sequence differences between ChrT and
other FMN_red members, multiple sequence alignment and phylogenetic
analyses were performed. The deduced amino acid homology alignment
of ChrT with the other nine FMN_red genes from different species
was conducted by Clustal X software (version 2.0). As shown in
Fig. 2, the sequences and GenBank
accession numbers are as follows: Klebsiella pneumonia
(YP_002241302), Raoultella ornithinolytica (YP_007875969),
Enterobacter cloacae (YP_006479989.1), Klebsiella
oxytoca (YP_005017294.1), Enterobacter asburiae
(YP_004830940.1), Citrobacter rodentium (YP_003367456.1),
Enterobacter lignolyticus (YP_003943949.1), E. coli
(WP_001513430.1) and Cronobacter sakazakii (YP_007442551.1).
The similarities between Serratia sp. CQMUS2 and the
aforementioned nine bacteria were 99, 94, 92, 92, 90, 89, 88, 85
and 86%, respectively. A phylogenetic tree of the FMN_red members,
including ChrT, was constructed using the neighbor-joining method
with MEGA 4.0.2 software. According to the phylogenetic tree
(Fig. 3), ChrT was found to be
closely associated with FMN_red members, including Klebsiella
pneumonia (YP_002241302), Raoultella ornithinolytica
(YP_007875969) and Klebsiella oxytoca (YP_005017294.1).
 | Figure 2Multiple sequence alignments of the
predicted protein of ChrT with FMN_red genes from other species.
Ss-ChrT (Serratia sp. CQMUS2, KF211434); Kp-FMN_red
(Klebsiella pneumonia, YP_002241302.1); Ro-FMN_red
(Raoultella ornithinolytica, YP_007875969.1); Ecl-FMN_red
(Enterobacter cloacae, YP_006479989.1); Ko-FMN_red
(Klebsiella oxytoca, YP_005017294.1); Ea-FMN_red
(Enterobacter asburiae, YP_004830940.1); Cr-FMN_red
(Citrobacter rodentium, YP_003367456.1); El-FMN_red
(Enterobacter lignolyticus, YP_003943949.1); Ec-FMN_red
(Escherichia coli, WP_001469560.1); Cs-FMN_red
(Cronobacter sakazakii, YP_007442551.1); FMN_red, flavin
mononucleotide reductase. |
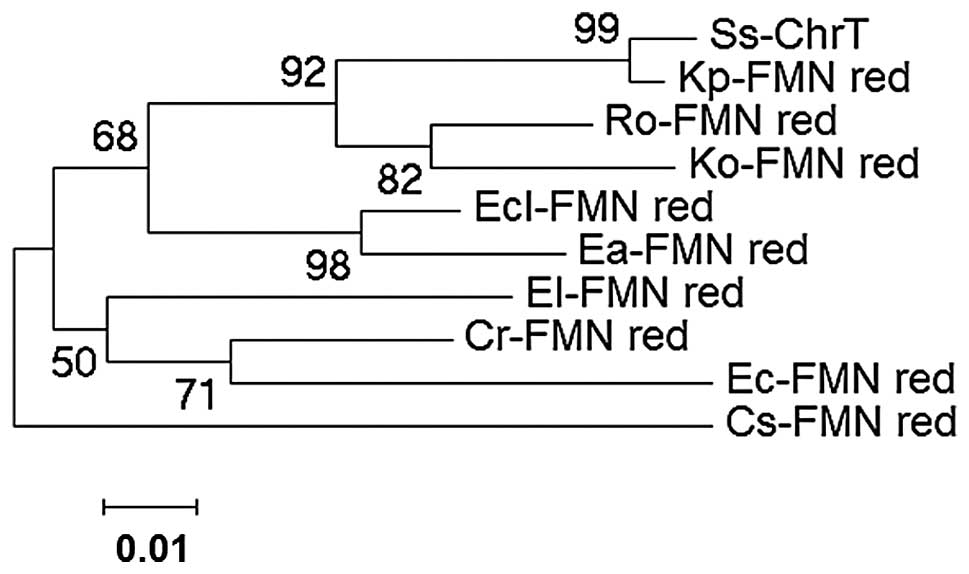 | Figure 3Phylogenetic trees derived from the
amino acid sequences of ChrT and FMN_red genes from other species.
Ss-ChrT (Serratia sp. CQMUS2, KF211434); Kp-FMN_red
(Klebsiella pneumonia, YP_002241302.1); Ro-FMN_red
(Raoultella ornithinolytica, YP_007875969.1); Ecl-FMN_red
(Enterobacter cloacae, YP_006479989.1); Ko-FMN_red
(Klebsiella oxytoca, YP_005017294.1); Ea-FMN_red
(Enterobacter asburiae, YP_004830940.1); Cr-FMN_red
(Citrobacter rodentium, YP_003367456.1); El-FMN_red
(Enterobacter lignolyticus, YP_003943949.1); Ec-FMN_red
(Escherichia coli, WP_001469560.1); Cs-FMN_red
(Cronobacter sakazakii, YP_007442551.1); FMN_red, flavin
mononucleotide reductase. |
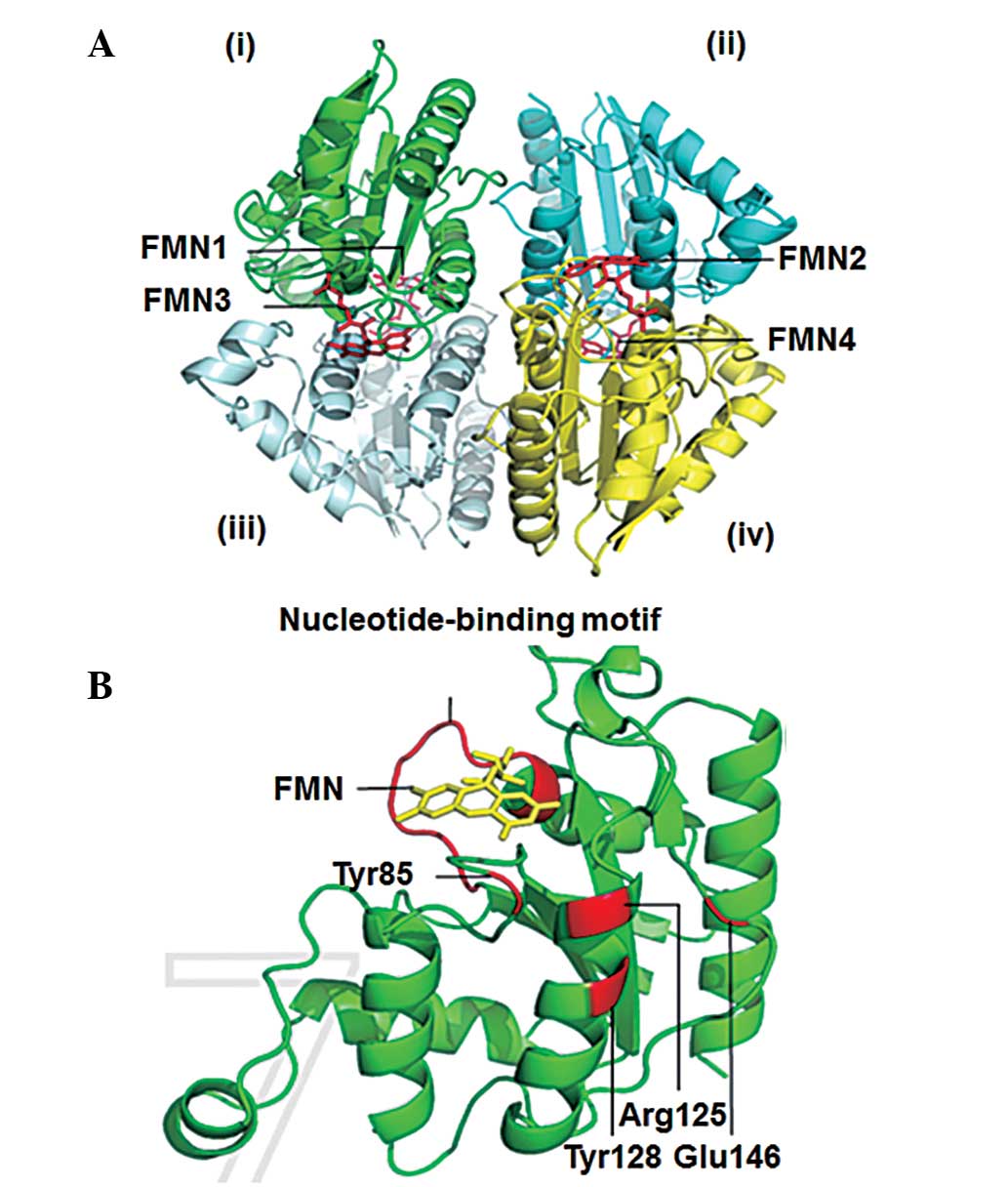
ChrT protein structure confers its
chromate reduction ability
To predict the secondary and 3D structures of ChrT,
the NPS@ service method and SWISS-MODEL program based on a known
crystal structure of the E. coli ChrR enzyme were used,
respectively. The ChrT protein was shown to contain the following
structures: 40.96% α-helix, 11.70% extended strand and 47.34%
random coil, but no π-helix, 310-helix or other
secondary structures. The predicted 3D model and template showed a
high similarity of 85.6%. The structure of the predicted ChrT
protein was a tetramer, formed by two symmetry-related dimers. In
addition, each monomer of the dimers shows an enzyme combined with
FMN that contains the enzyme-active site (Fig. 4A). Furthermore, each monomer was
shown to contain the nucleotide-binding motif, GSLRKGSFN, which
anchors FMN firmly, and four amino acids (Tyr128, Glu146, Arg125
and Tyr85) that are associated with chromate reductase activity
(Fig. 4B). These observations
demonstrated that the ChrT protein structure conferred an ability
for chromate reduction.
Discussion
In the present study, the chromate reductase ChrT
gene was cloned from Serratia sp. CQMUS2 using three
steps of PCR amplification based on different principles. Firstly,
specific primers were designed according to the homologous sequence
from Serratia sp. AS13 for the amplification of the ChrT
gene fragment. Secondly, 5′- and 3′-flanking sequences of ChrT were
obtained by a hiTAIL-PCR method. Thirdly, the full-length gene of
ChrT was obtained using specific primers corresponding to its 5′-
and 3′-ends. In previous studies, rapid amplification of the cDNA
end method has been employed to obtain flanking sequences (18,19),
where RNA is required as the template. The extraction, purification
and reverse transcription of RNA are time-consuming and expensive.
In particular, rapid amplification of 5′-cDNA end technology is
often too difficult to succeed (20). However, the hiTAIL-PCR method uses
DNA as the template; thus, has a number of advantages, including
simple operation, low cost, high specificity and excellent
repetition (21).
FMN exists widely in nature and participates in the
electronic transmission from substrate to electron acceptor as a
coenzyme of flavoenzyme. Therefore, FMN can reduce Cr(VI) to
Cr(III) through electronic transmission from NAD(P)H to Cr(VI)
(22). In the present study, the
FMN_red protein was obtained from Serratia sp.
CQMUS2, and using conserved domain analysis, the protein was
demonstrated to belong to the flavodoxin-2 superfamily. The amino
acid sequence of FMN_red from Serratia sp. CQMUS2 revealed
99 and 94% identity to the enzymes from Klebsiella pneumonia
and Raoultella ornithinolytica, respectively. Moreover, the
predicted 3D model revealed that the protein was a tetramer that
was composed of four enzymes combined with FMN, which had a high
similarity to the ChrR enzyme of E. coli. In a previous
study, the crystal structure of the ChrR enzyme demonstrated that
FMN was anchored by several hydrogen bonds to the
nucleotide-binding motif, GSLRKGSFN, located on each monomer of the
protein (23). In addition, the
amino acids, Tyr128, Glu146, Arg125 and Tyr85, and associated
hydrogen bond networks, were found to play a critical role in
enhancing chromate reductase activity (23). In the present study, the predicted
3D structure of the FMN_red protein was found to contain the
GSLRKGSFN nucleotide-binding motif and the amino acids, Tyr128,
Glu146, Arg125 and Tyr85, in each monomer. Therefore, the predicted
structure theoretically indicates that the proteins share a common
catalytic mechanism for chromate reduction.
In conclusion, ChrT gene cloning and protein
structure prediction demonstrated the ability of the gene for
chromate reduction. Future studies should investigate ChrT
expression in E. coli BL21, the enzyme activity and its
application in removing chromium from waste water.
Acknowledgements
The study was supported by a grant from the Natural
Science Foundation Project of Chongqing Municipal Science and
Technology Commission (no. 2010BB5360).
References
|
1
|
Ren BQ, Wang YL, Zhao LY and Jin Y:
Actuality and trend of chromium containing wastewater treatment
technology. Heilongjiang Kexue. 4:67–69. 2013.(In Chinese).
|
|
2
|
Robins KJ, Hooks DO, Rehm BH and Ackerley
DF: Escherichia coli NemA is an efficient chromate reductase that
can be biologically immobilized to provide a cell free system for
remediation of hexavalent chromium. PLoS One. 8:e592002013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tahri Joutey N, Bahafid W, Sayel H, Ananou
S and El Ghachtouli N: Hexavalent chromium removal by a novel
Serratia proteamaculans isolated from the bank of Sebou River
(Morocco). Environ Sci Pollut Res Int. 21:3060–3072. 2014.
View Article : Google Scholar
|
|
4
|
Christl I, Imseng M, Tatti E, Frommer J,
Viti C, Giovannetti L and Kretzschmar R: Aerobic reduction of
chromium (VI) by Pseudomonas corrugata 28: Influence of metabolism
and fate of reduced chromium. Geomicrobiol J. 29:173–185. 2012.
View Article : Google Scholar
|
|
5
|
Zhitkovich A: Chromium in drinking water:
sources, metabolism, and cancer risks. Chem Res Toxicol.
24:1617–1629. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ramírez-Díaz MI, Díaz-Pérez C, Vargas E,
Riveros-Rosas H, Campos-García J and Cervantes C: Mechanisms of
bacterial resistance to chromium compounds. Biometals. 21:321–332.
2008. View Article : Google Scholar
|
|
7
|
Sandana Mala JG, Sujatha D and Rose C:
Inducible chromate reductase exhibiting extracellular activity in
Bacillus methylotrophicus for chromium bioremediation. Microbiol
Res. pii: S0944-5013(14)00067-6. 2014.PubMed/NCBI
|
|
8
|
Arévalo-Rangel DL, Cárdenas-González JF,
Martínez-Juárez VM and Acosta-Rodríguez I: Hexavalent chromate
reductase activity in cell free extracts of Penicillium sp.
Bioinorg Chem Appl. 2013:9094122013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Belchik SM, Kennedy DW, Dohnalkova AC, et
al: Extracellular reduction of hexavalent chromium by cytochromes
MtrC and OmcA of Shewanella oneidensis MR-1. Appl Environ
Microbiol. 77:4035–4041. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park CH, Keyhan M, Wielinga B, Fendorf S
and Matin A: Purification to homogeneity and characterization of a
novel Pseudomonas putida chromate reductase. Appl Environ
Microbiol. 66:1788–1795. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ackerley DF, Gonzalez CF, Park CH, Blake R
2nd, Keyhan M and Matin A: Chromate-reducing properties of soluble
flavoproteins from Pseudomonas putida and Escherichia coli. Appl
Environ Microbiol. 70:873–882. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Agarwal R, Bonnano JB, Burley SK and
Swaminathan S: Structure determination of an FMN-reductase from
Pseudomonas aeruginosa PAO1 using sulfur anomalous signal. Acta
Crystallogr D Biol Crystallogr. 62:383–391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan XQ, Deng P, Wu Y, et al: In vitro
synthesis and activity identification of chromate reductase T.
Hubei Yixueyuan Xuebao. 7:973–977. 2014.(In Chinese).
|
|
14
|
Sun YB, Mo DJ and Dai TL: Modifying
methods of preparation of PCR template and recovery of DNA. Acta
Academiae Medicinae Hubei. 18:24–25. 1997.(In Chinese).
|
|
15
|
Liu YG and Chen YL: High-efficiency
thermal asymmetric interlaced PCR for amplification of unknown
flanking sequences. Biotechniques. 43:649–650. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Clustal X version 2.0. http://www.ebi.ac.uk/tools/clustalw2uri.
Accessed 27/07/2013
|
|
17
|
Molecular Evolutionary Genetics Analysis
(MEGA) software version 4.0.2. http://www.megasoftware.net/uri.
Accessed 30/09/2013
|
|
18
|
Wang J, Zhang H, Wu M and Tang C: Cloning
and sequence analysis of a novel xylanase gene, Auxyn10A, from
Aspergillus usamii. Biotechnol Lett. 33:1029–1038. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Araya-Garay JM, Feijoo-Siota L,
Veiga-Crespo P and Villa TG: cDNA cloning of a novel gene codifying
for the enzyme lycopene β-cyclase from Ficus carica and its
expression in Escherichia coli. Appl Microbiol Biotechnol.
92:769–777. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pang X, Zhou DS and Yang RF: Summary of
bacteria mRNA extraction. Shengwu Jishu Tongbao. 1:30–34. 2003.(In
Chinese).
|
|
21
|
Tan HQ and Singh J: High-efficiency
thermal asymmetric interlaced (HE-TAIL) PCR for amplification of Ds
transposon insertion sites in Barley. J Plant Mol Biol Biotechnol.
2:9–14. 2011.
|
|
22
|
Jin H, Zhang Y, Buchko GW, et al:
Structure determination and functional analysis of a chromate
reductase from Gluconacetobacter hansenii. PLoS One. 7:e424322012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eswaramoorthy S, Poulain S, Hienerwadel R,
et al: Crystal structure of ChrR - a quinone reductase with the
capacity to reduce chromate. PLoS One. 7:e360172012. View Article : Google Scholar
|