Introduction
Infection of hepatitis B virus (HBV) remains a
global health problem. Patients with chronic hepatitis B (CHB) can
develop progressive liver disease, which can result in cirrhosis
and hepatocellular carcinoma (HCC). These stages of the disease are
associated with an increased risk of morbidity and mortality, and
incur considerable healthcare costs (1). To reduce morbidity and mortality from
chronic HBV infection, antiviral treatment is the only effective
approach (2). Current antiviral
agents for the treatment of CHB include interferon (IFN) and
nucleoside analogues (NUCs). Although treatment with IFN may lead
to a durable response, the unpleasant adverse effects and high cost
limit its use (3). NUCs, including
lamivudine (LAM) and telbivudine (LdT), then become the most common
drugs used for antiviral therapy (4). Currently, LAM, adefovir, entecavir
(ETV), tenofovir and LdT have been licensed for the treatment of
CHB (5,6). NUCs target the HBV reverse
transcriptase (RT), thus inhibiting viral replication and leading
to virological, biochemical and histological improvement in the
majority of patients. Current therapy of CHB does not eradicate HBV
and has limited long-term efficacy, which results in the
requirement for long-term therapy. Emergence of drug-resistant HBV
mutants is an adverse consequence of long-term therapy (7). The high mutational rate of HBV RT
enzyme, due to its lack of proof-reading activity, is the main
cause of HBV mutation occurrence and drug resistance (8). Mutations selected by treatment with
NUCs can be separated into two groups: Those that cause resistance,
sometimes leading to decreased viral fitness; and compensatory
mutations, which partially or fully restore viral fitness (9). LAM is the first safe oral NUC
approved for the treatment of HBV by the Food and Drug
Administration (10). LAM and LdT
belong to L-nucleosides. The main mutation associated with
L-nucleoside resistance is rtM204I/V, a mutation that occurs within
the YMDD motif of the RT region of the polymerase (11).
Previous studies have shown that HBV infection
exists in peripheral mononuclear cells (PBMCs) (12–14),
and HBV viral loads in PBMCs correlated with serum HBV DNA
(15). There is limited knowledge
regarding the association between the quantity of HBV DNA loads in
serum and PBMCs in patients with drug resistance. Although a number
of studies have investigated the characteristics of resistance
mutations of HBV in serum (4,6,16),
few studies focus on the mutational pattern of the polymerase gene
of HBV in PBMCs. In the present study, quantitative polymerase
chain reaction (PCR) was applied for the quantification of total
HBV DNA in serum and PBMCs in patients with LAM or LdT resistance,
and direct sequencing was used to detect the mutation of the
polymerase gene of HBV. Clarification of the association between
serum and PBMC viral loads was attempted, and whether there were
different mutational patterns of the polymerase gene of HBV in
serum and PBMCs of patients with LAM or LdT resistance was
explored.
Materials and methods
Patients and samples
The study was approved by the Ethics Committee of
the Jinan Infectious Disease Hospital, Shandong University (Jinan,
China), and written informed consent was obtained from all the
participants. Between January 2012 and January 2013, 51 patients
with CHB were recruited from the Hepatitis Clinic of the Jinan
Infectious Disease Hospital of Shandong University. All the
patients were on antiviral therapy with LAM (100 mg/day;
GlaxoSmithKline Biological Co., Ltd., Shanghai, China) or LdT (600
mg/day; Novartis Pharma Co., Ltd., Beijing, China) for at least
half a year and experienced a virological breakthrough (HBV DNA
level increased by 1 log10 copies/ml or more than the
treatment nadir). The patients were divided into two groups: LAM
(n=32) and LdT (n=19). All the patients were positive for hepatitis
B surface antigen (HBsAg), had HBV viral loads ≥ 3 log10
copies/ml, and did not have other infectious diseases, including
human immunodeficiency virus, hepatitis C or hepatitis D. The
venous blood samples were collected from the antecubital fossa of
each participant. Serum was separated, divided into aliquots and
maintained frozen at −80°C until testing. PBMCs were isolated using
Lymphocyte Separation medium (Tianjin Haoyang Biological Technology
Co., Ltd., Tianjin, China) and stored at −80°C.
Testing for HBV serological markers and
biochemical parameters
Serological markers for HBV infection were measured
using micro-particle enzyme immunoassay with reagents from Abbott
Laboratories (Abbott Park, IL, USA), and serum biochemical
parameters were also detected (Abbott Laboratories).
Quantitative determination of HBV DNA in
serum and PBMCs
HBV DNA in serum was detected by the fluorescence
quantitative PCR assay (HBV PCR Fluoroscence Diagnostic kit;
Shenzhen PG Biotechnology Co., Ltd., Shenzhen, China). The lower
limit of detection was 500 copies/ml, as according to the
manufacturer’s instructions. PBMCs were washed three times in
phosphate-buffered saline prior to DNA extraction, and the final
cell wash was conserved as a control for contamination with HBV DNA
derived from blood. Total HBV DNA in PBMCs was isolated according
to the manufacturer’s instructions [TIANamp Genomic DNA kit;
Tiangen Biotech (Beijing) Co., Ltd., Beijng, China]. HBV DNA in
PBMCs was also detected by fluorescence quantitative PCR assay.
Detection of HBV genotypes and
mutations
HBV genotypes were detected by using the Genotypes
Detection kit (Yuan Qi Bio-Medicine Co., Ltd., Shanghai, China)
according to the manufacturer’s instructions. The mutations of the
RT region of the polymerase gene of HBV were detected by Shanghai
Shenyou Biotechnology Co., Ltd. (Shanghai, China). Briefly, a pair
of primers was designed (forward, 5′-ATCCCATCATCTTGGGCTTT-3′; and
reverse, 5′-CAA GGTACCCCAACTTCCAAT-3′) for amplification of the HBV
polymerase region. PCR conditions were 95°C for 15 min, followed by
45 cycles consisting of 95°C for 45 sec, 56°C for 45 sec and 72°C
for 45 sec. The PCR products were approximately 300 base pairs. All
the PCR products were purified and directly sequenced. For cycle
sequencing, the following thermal protocol was used: 35 cycles
consisting of 95°C for 20 sec, 50°C for 25 sec and finally 60°C for
2 min. Data were accumulated by direct sequencing and were analyzed
either manually or using the Chromas program (Chromas Lite version
2.1 (2012), Technelysium Pty Ltd, South Brisbane, Queensland,
Australia).
Statistical analysis
The results are expressed as percentages and the
mean ± standard deviation. Differences between categorical
variables were analyzed using the Fisher’s exact or χ2
tests. For continuous variables, the Student’s t-test was used when
the data showed a normal distribution, or the Mann-Whitney U test
was used when the data was not normally distributed. Pearson
correlation was also used for normally distributed variables, and
Spearman correlation for non-normally distributed variables. Values
of P<0.05 (two-tailed) were considered to indicate a
statistically significant difference. All the statistical processes
were carried out by statistical software SPSS 13.0 for Windows
(SPSS, Inc., Chicago, IL, USA).
Results
Characteristics of demographic, clinical
and laboratory data
All the participants were ethnically Chinese and HBV
genotype C positive. The aim was to investigate the mutational
pattern of the polymerase gene of HBV in serum and PBMCs of
patients with LAM or LdT resistance, therefore 20 patients (39.22%,
including 11 patients in the LAM group and nine patients in the LdT
group) were excluded when it was found that there were no drug
resistance mutations in the serum and PBMCs of these patients. The
characteristics of the remaining 31 patients with regard to
demographic, clinical and laboratory data are shown in Table I. No significant differences were
identified between the two groups.
 | Table ICharacteristics of patients in the
different groups. |
Table I
Characteristics of patients in the
different groups.
| Group | | |
|---|
|
| | |
|---|
| Variable | Lamivudine | Telbivudine | t | P-value |
|---|
| Patients, n | 21 | 10 | - | - |
| Male, n (%) | 18 (85.71) | 8 (80.00) | - | 1.00a |
| Ageb, years | 45.76±12.78 | 44.90±7.69 | 0.20 | 0.85 |
| HBsAg positive, n
(%) | 17 (81) | 9 (90) | - | 1.00a |
| Treatment duration,
months | 27.00±17.69 | 20.30±9.32 | 1.12 | 0.27 |
| ALTb, U/l | 173.04±216.13 | 117.80±211.12 | 0.67 | 0.51 |
| ASTb, U/l | 116.61±122.51 | 84.90±128.85 | 0.66 | 0.51 |
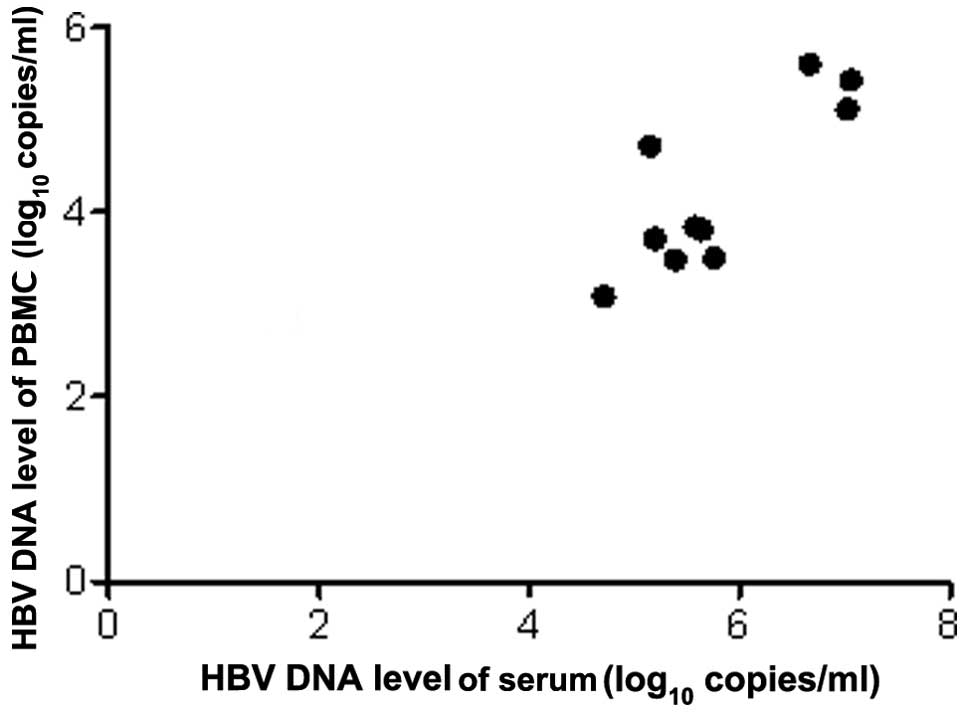
Comparison of HBV DNA levels of serum and
PBMCs
HBV DNA loads of serum and PBMCs in the different
groups are shown in Fig. 1 and
Table II. No significant
differences were observed between the two groups when comparing the
HBV DNA loads in the serum and PBMCs, respectively (serum, t=0.15,
P>0.05; PBMC, t=0.99, P>0.05). While analyzing the HBV DNA
loads of serum and PBMCs, it was found that the HBV DNA level of
serum was significantly higher than that of PBMCs in the two groups
(LAM, t=5.69, P<0.05; LdT, t=9.87, P<0.05).
 | Table IIHBV DNA levels of serum and PBMCs in
the different groups. |
Table II
HBV DNA levels of serum and PBMCs in
the different groups.
| HBV DNA
(log10 copies/ml) |
|---|
|
|
|---|
| Group | Serum | PBMCs |
|---|
| Lamivudine | 5.87±1.04 | 4.63±1.14 |
| Telbivudine | 5.82±0.82 | 4.23±0.90 |
Correlations between serum and PBMCs HBV
DNA loads
The HBV DNA loads of PBMCs were correlated
significantly to that of serum, and there were positive
correlations in the two groups (LAM, r=0.584, P<0.01; LdT,
r=0.829, P<0.01). Scatterplots are shown in Figs. 2 and 3.
Mutational patterns of the polymerase
gene of HBV DNA in serum and PBMCs
Different mutational patterns in the HBV DNA
polymerase gene were distinguished, as are shown in Table III. All the LAM resistant strains
carried the mutation site, rt204, whether in serum or PBMCs. There
were three mutational patterns in the serum of LAM-resistant
patients: Single-site mutation (rtM204I, 3/21, 14.29%), two sites
mutation (rtL180M plus rtM204I, rtM204V or rtM204I/V, 11/21,
52.38%) and three sites mutation (rtL180M plus rtM204I, rtM204V or
rtM204I/V plus another site, 7/21, 33.33%). There were also three
mutational patterns in the PBMCs of LAM-resistant patients.
Analysis of the mutation sites in matched PBMCs and serum from each
LAM-resistant patient revealed that 85.71% (18/21) patients had the
same mutational pattern in the PBMCs and serum. Two mutational
patterns were found in the serum of LdT-resistant patients:
Single-site mutation (rtM204I, 8/10, 80%) and two sites mutation
(rtL180M plus rtM204I, 2/10, 20.00%). Patients 7 and 8 of the LdT
group had wild-type in PBMCs, whereas patients 9 and 10 had
mutational strains in PBMCs, which showed a different mutational
pattern from that in serum. Comparing with mutation sites in
matched PBMCs and serum, 75% (6/8) of patients presented the same
mutational pattern in the LdT group.
 | Table IIIMutational patterns of reverse
transcriptase of the polymerase gene in all the patients. |
Table III
Mutational patterns of reverse
transcriptase of the polymerase gene in all the patients.
| Mutation sites of
reverse transcriptase of the polymerase gene |
|---|
|
|
|---|
| Groups and
patients | Serum | Peripheral blood
mononuclear cells |
|---|
| Lamivudine |
| 1 | rtM204I | rtM204I |
| 2 | rtM204I | rtM204I |
| 3 | rtM204I | rtM204I |
| 4 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 5 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 6 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 7 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 8 | rtL180M + rtM204I +
rtN238S | rtL180M + rtM204I +
rtN238S |
| 9 | rtL180M +
rtM204V | rtL180M +
rtM204V |
| 10 | rtL180M +
rtM204V | rtL180M +
rtM204V |
| 11 | rtL180M +
rtM204V | rtL180M +
rtM204V |
| 12 | rtL180M +
rtM204V | rtL180M +
rtM204V/I |
| 13 | rtL180M + rtM204V +
rtV173L | rtL180M + rtM204V +
rtV173L |
| 14 | rtL180M + rtM204V +
rtM250L | rtL180M + rtM204V +
rtM250L |
| 15 | rtL180M + rtM204V +
rtV207L | rtL180M + rtM204V/I
+ rtV207L |
| 16 | rtL180M + rtM204V +
rtN238S | rtL180M + rtM204V +
rtN238S |
| 17 | rtL180M +
rtM204V/I | rtL180M + rtM204V/I
+ rtT184S |
| 18 | rtL180M + rtM204V/I
+ rtP237H | rtL180M + rtM204V/I
+ rtP237H |
| 19 | rtL180M +
rtM204V/I | rtL180M +
rtM204V/I |
| 20 | rtL180M +
rtM204V/I | rtL180M +
rtM204V/I |
| 21 | rtL180M + rtM204V/I
+ rtV173L | rtL180M + rtM204V/I
+ rtV173L |
| Telbivudine |
| 1 | rtM204I | rtM204I |
| 2 | rtM204I | rtM204I |
| 3 | rtM204I | rtM204I |
| 4 | rtM204I | rtM204I |
| 5 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 6 | rtL180M +
rtM204I | rtL180M +
rtM204I |
| 7 | rtM204I | wild-type |
| 8 | rtM204I | wild-type |
| 9 | rtM204I | rtM204I +
rtM250R |
| 10 | rtM204I | rtL180M + rtM204I +
rtT184S |
Discussion
HBV is not strictly hepatotropic and studies have
established that tissues from the kidney, pancreas and bone marrow,
as well as PBMCs, contain HBV DNA sequences (17–19).
Although hepatocytes are recognized as the main target, HBV has
significant lymphotropic properties. HBV infection of lymphoid
cells is an important mechanism whereby the virus escapes immune
recognition and lymphoid reservoirs, particularly those harboring
drug-resistant HBV, and may be key to the development of antiviral
resistance (20). LAM was approved
for the treatment of CHB in China in 1999. However, LAM is no
longer recommended as a first-line agent for naïve patients with
CHB due to its high incidence of drug resistance (5). A number of patients chronically
infected with HBV remain treated with LAM due to cost reasons and
availability (21). Therefore, the
study of intracellular levels of HBV DNA and mutational patterns of
the polymerase gene of HBV in PBMCs of patients with LAM resistance
are clinically relevant.
Genotypes B and C of HBV have been identified as the
most common strains and account for ~95% of infections among
Chinese patients (22). Compared
with HBV genotype B infections, HBV genotype C infections have been
associated with lower rates of spontaneous clearance of HBsAg in
serum, higher levels of virus replication, more advanced liver
disease (23) and a lower rate of
response to α-interferon therapy (24). All the subjects in the present
study are genotype C, and all the HBV DNA in the PBMCs of the
patients with LAM or LdT resistance was successfully detected. The
HBV DNA loads in PBMCs were significantly lower than those in
serum, whether in the LAM or LdT groups, and there were positive
correlations between them. The present study showed the association
between the quantity of HBV DNA loads in PBMCs and serum in
patients with drug resistance conformed with those in patients
without antiviral therapy (15).
The reasons for the correlation between the viral load in serum and
PBMCs were not clear. Although the HBV DNA loads in PBMCs are
relatively low, it is difficult to eliminate HBV from PBMCs. Ke
et al (14) found that the
negative rate of HBV DNA in serum was 90.48%, but only 59.52% in
PBMCs after 48 weeks of LAM treatment. Coffin et al
(20) found HBV DNA was detected
in 43% (3/7) of plasma, 100% (9/9) of liver and 83% (5/6) of PBMC
samples using sensitive PCR/nucleic acid hybridization assay
despite undetectable plasma HBV DNA by clinical assays following
antiviral therapy. The study also found that PBMCs carried a
drug-resistant virus in patients whose plasma had wild strains
only. HBV can even persist in the serum and PBMCs for years after
clinical and serological recovery from acute viral hepatitis, and
remains transcriptionally active in PBMCs (25–27).
HBV in PBMCs interferes with the immune activity of cells
subsequent to HBV integrating with PBMCs, and decreases the
contents of immunoglobulin, C3, tumor necrosis factor and activity
of natural killer cells, as well as the ratio of cluster of
differentiation 4+ (CD4+)/CD8+
(28). Therefore, the function of
cell-mediated and humoral immunities are reduced and the curative
effects of anti-viral drugs are affected.
As has been described previously, LAM and LdT belong
to the L-nucleosides, therefore the study was also conducted on
patients with LdT resistance. The main mutation associated with
L-nucleoside resistance is rtM204I/V, a mutation that occurs within
the YMDD motif of the RT region of the polymerase (11). The rtL180M mutation is the most
common compensatory mutation, contributing to increasing either
replication efficiency and/or antiviral resistance (29). In the present study, 60.78% (31/51)
of patients who experienced a virological breakthrough during
anti-viral therapy with LAM or LdT appeared to exhibit drug
resistant strains in their serum. All the mutations associated with
LAM or LdT resistance had the mutation site rt204. The majority of
patients associated with LAM resistance had a compensatory mutation
at position rt180 (18/21, 85.71%). Patients 13 and 14 in the LAM
group had three mutation sites that were associated with ETV
resistance despite those two patients never previously being
administered ETV, which offered evidence as to why certain patients
with LAM resistance did not experience beneficial effects when they
changed to ETV for anti-viral therapy. Regarding the mutation
patterns of HBV in PBMCs, 85.71% (18/21) of patients with LAM
resistance and 75% (6/8) of patients with LdT resistance carried
the same mutational pattern as that in the matched serum. In the
LAM group, patients 12 and 15 had different mutational patterns
between the PBMCs and serum despite the same mutation site:
rtM204V/I in the PBMCs and rtM204V in the serum, while patient 17
carried one more mutation site in the PBMCs than in the serum. In
the LdT group, patients 7 and 8 carrying mutants in the serum were
detected to have wild strains in the PBMCs. According to the
present results, it may be inferred that HBV in PBMCs may be
wild-type at first, and gradually mutated following administration
of the antiviral drug. The discrepancy of the mutational patterns
between PBMCs and serum may be partly explained by the differences
in antiviral drug selection pressure in the two compartments. HBV
drug-resistant variants are present in PBMCs, therefore assessing
drug resistance in a single compartment may not be sufficient in
predicting the long-term response to antiviral therapy. Awareness
of resistance patterns in PBMCs may help in antiviral therapy and
predicting clinical outcomes.
In conclusion, the HBV DNA levels in the PBMCs of
patients with LAM or LdT resistance were significantly lower than
those in serum and there were positive correlations between them.
The majority of the mutational patterns of the polymerase gene of
HBV DNA in PBMCs were the same as those in the paired serum. These
finding may help to increase knowledge regarding HBV drug
resistance.
Acknowledgements
The present study was supported by a grant (no.
201221051) from the Science and Technology Development Plan Funds
of Jinan (Shandong, China).
References
|
1
|
Lok AS and McMahon BJ: Chronic hepatitis
B: update 2009. Hepatology. 50:661–662. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment, and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Colacino JM and Staschke KA: The
identification and development of antiviral agents for the
treatment of chronic hepatitis B virus infection. Prog Drug Res.
50:259–322. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li SY, Qin L, Zhang L, et al: Molecular
epidemical characteristics of Lamivudine resistance mutations of
HBV in southern China. Med Sci Monit. 17:PH75–PH80. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
European Association For The Study Of The
Liver. EASL Clinical Practice Guidelines: management of chronic
hepatitis B. J Hepatol. 50:227–242. 2009. View Article : Google Scholar
|
|
6
|
Sayan M, Akhan SC and Senturk O: Frequency
and mutation patterns of resistance in patients with chronic
hepatitis B infection treated with nucleos(t)ide analogs in add-on
and switch strategies. Hepat Mon. 11:835–842. 2011.
|
|
7
|
Nguyen MH and Keeffe EB: Chronic hepatitis
B: early viral suppression and long-term outcomes of therapy with
oral nucleos(t)ides. J Viral Hepat. 16:149–155. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng L and Tang H: Hepatitis B virus drug
resistance to current nucleos(t)ide analogs: Mechanisms and
mutation sites. Hepatol Res. 41:1017–1024. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sheldon J, Rodès B, Zoulim F,
Bartholomeusz A and Soriano V: Mutations affecting the replication
capacity of the hepatitis B virus. J Viral Hepat. 13:427–434. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Josefson D: Oral treatment for hepatitis B
gets approval in the United States. BMJ. 317:10341998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartholomeusz A and Locarnini SA:
Antiviral drug resistance: clinical consequences and molecular
aspects. Semin Liver Dis. 26:162–170. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pontisso P, Poon MC, Tiollais P and
Brechot C: Detection of hepatitis B virus DNA in mononuclear blood
cells. Br Med J (Clin Res Ed). 288:1563–1566. 1984. View Article : Google Scholar
|
|
13
|
Liu MC, Wang GQ, Piao WH, et al: Detection
of hepatitis B virus covalently closed circular DNA in peripheral
blood mononuclear cells from patients with chronic hepatitis B
infection. Zhonghua Gan Zang Bing Za Zhi. 12:249–250. 2004.(In
Chinese). PubMed/NCBI
|
|
14
|
Ke CZ, Chen Y, Gong ZJ, et al: Dynamic
changes of HBV DNA in serum and peripheral blood mononuclear cells
of chronic hepatitis patients after lamivudine treatment. World J
Gastroenterol. 12:4061–4063. 2006.PubMed/NCBI
|
|
15
|
Lu L, Zhang HY, Yueng YH, et al:
Intracellular levels of hepatitis B virus DNA and pregenomic RNA in
peripheral blood mononuclear cells of chronically infected
patients. J Viral Hepat. 16:104–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamidi-Fard M, Makvandi M, Samarbaf-Zadeh
A, et al: Mutation analysis of hepatitis B virus reverse
transcriptase region among untreated chronically infected patients
in Ahvaz city (South-West of Iran). Indian J Med Microbiol.
31:360–365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Murakami Y, Minami M, Daimon Y and Okanoue
T: Hepatitis B virus DNA in liver, serum, and peripheral blood
mononuclear cells after the clearance of serum hepatitis B virus
surface antigen. J Med Virol. 72:203–214. 2004. View Article : Google Scholar
|
|
18
|
Mason A, Wick M, White H and Perrillo R:
Hepatitis B virus replication in diverse cell types during chronic
hepatitis B virus infection. Hepatology. 18:781–789. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pontisso P, Morsica G, Ruvoletto MG, et
al: Hepatitis B virus binds to peripheral blood mononuclear cells
via the pre S1 protein. J Hepatol. 12:203–206. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Coffin CS, Mulrooney-Cousins PM, Peters
MG, et al: Molecular characterization of intrahepatic and
extrahepatic hepatitis B virus (HBV) reservoirs in patients on
suppressive antiviral therapy. J Viral Hepat. 18:415–423. 2011.
View Article : Google Scholar
|
|
21
|
Neumann-Fraune M, Beggel B, et al: High
frequency of complex mutational patterns in lamivudine resistant
hepatitis B virus isolates. J Med Virol. 85:775–779. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Huang Y, Wen S, Zhou B and Hou J:
Hepatitis B virus genotypes and subgenotypes in China. Hepatol Res.
37:S36–S41. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pujol FH, Navas MC, Hainaut P and Chemin
I: Worldwide genetic diversity of HBV genotypes and risk of
hepatocellular carcinoma. Cancer Lett. 286:80–88. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kao JH, Chen PJ, Lai MY and Chen DS:
Hepatitis B genotypes correlate with clinical outcomes in patients
with chronic hepatitis B. Gastroenterology. 118:554–559. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Michalak TI, Pasquinelli C, Guilhot S and
Chisari FV: Hepatitis B virus persistence after recovery from acute
viral hepatitis. J Clin Invest. 94:9071994. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rehermann B, Ferrari C, Pasquinelli C and
Chisari FV: The hepatitis B virus persists for decades after
patients’ recovery from acute viral hepatitis despite active
maintenance of a cytotoxic T-lymphocyte response. Nat Med.
2:1104–1108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cabrerizo M, Bartolomé J, Caramelo C,
Barril G and Carreno V: Molecular analysis of hepatitis B virus DNA
in serum and peripheral blood mononuclear cells from hepatitis B
surface antigen-negative cases. Hepatology. 32:116–123. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pallier C, Castéra L, Soulier A, et al:
Dynamics of hepatitis B virus resistance to lamivudine. J Virol.
80:643–653. 2006. View Article : Google Scholar :
|
|
29
|
Xiong X, Yang H, Westland CE, Zou R and
Gibbs CS: In vitro evaluation of hepatitis B virus polymerase
mutations associated with famciclovir resistance. Hepatology.
31:219–224. 2000. View Article : Google Scholar
|