Introduction
Osteosarcoma is the most common primary bone tumor
in adolescents and young adults (1). Despite progress in therapeutic
technologies, including surgery, chemotherapy, radiotherapy and
biological therapy, the overall survival (OS) of patients with
osteosarcoma remains unsatisfactory. Approximately 80% of patients
will eventually develop local relapse or metastatic disease
following surgical treatment (2),
and pulmonary metastasis is the major cause of fatal outcome
(3). Like other malignancies, the
development of osteosarcoma is a multistep process with
accumulation of genetic and epigenetic changes. However, to date,
the highly complex molecular mechanisms underlying its initiation
and progression are poorly understood. Therefore, it is necessary
to search for novel markers for osteosarcoma, which can accurately
identify the biological characteristics of tumors, improve
therapeutic strategies and predict clinical outcome.
MicroRNAs (miRNAs) are single-stranded, small
noncoding RNAs of 18–25 nucleotides in length (4). They can negatively regulate gene
expression through base-pairing to the 3′ untranslated region
(3′UTR) of target messenger RNA (mRNA), resulting in translation
inhibition or mRNA degradation (5,6).
Beyond involvement in diverse biological processes, including cell
growth, apoptosis, development, differentiation and endocrine
homeostasis (7), emerging evidence
strongly suggests that the deregulation or dysfunction of miRNAs
contributes to human carcinogenesis and cancer progression
(8–10). miRNAs can function as either
oncogenes or tumor suppressors according to the roles of their
target genes. In terms of osteosarcoma, in vitro functional
assays have shown that miR-126 and miR-133b inhibit the
proliferation, invasion and migration of osteosarcoma cells
(11,12). Clinical analysis has demonstrated
that decreased miR-145 and increased miRNA-214 expression levels in
osteosarcoma are associated with advanced clinical stage and poor
prognosis (13,14). Furthermore, Zhou et al
reported that the upregulation of miR-33a promoted the
chemoresistance of MG63 cells to cisplatin (15). These findings indicate that miRNAs
may act not only as diagnostic and prognostic markers, but also as
potential therapeutic targets of human osteosarcoma.
One of the cancer-related miRNAs is miR-124. It was
first reported to be highly expressed in neuronal cells, where it
regulates neuronal development and neural plasticity (16). Subsequent studies revealed that
miR-124 may modulate the process of tumorigenesis and the behavior
of cancer cells. It has been corroborated to be downregulated and
exert tumor suppressive function in medulloblastoma (17), breast cancer (18), ovarian cancer (19), cervical cancer (20), gastric cancer (21), colorectal cancer (22), hepatocellular carcinoma (23), pancreatic cancer (24) and prostate cancer (25). However, the expression and function
of miR-124 in osteosarcoma is largely unknown. In the current
study, miR-124 expression was investigated in paired osteosarcoma
and adjacent noncancerous bone tissues by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
assay. The correlation of miR-124 levels with clinicopathological
factors and prognosis was also statistically analyzed. Furthermore,
the effects of miR-124 on malignant phenotypes of osteosarcoma
cells were elucidated.
Materials and methods
Patients and tissue samples
This study was approved by the Research Ethics
Committee of General Hospital of PLA, (Beijing, China). Written
informed consent was obtained from all patients. All specimens were
handled and made anonymous according to ethical and legal
standards.
A total of 105 primary osteosarcoma and
corresponding noncancerous bone tissue samples were collected from
the General Hospital of PLA (Beijing, China) for RT-qPCR analysis
between March 2002 and February 2008. No patients had previously
received a blood transfusion, chemotherapy or radiotherapy. All
patients underwent neoadjuvant chemotherapy and wide resection of
the tumor. Tumor biopsies were collected prior to neoadjuvant
chemotherapy and were fresh-frozen and stored at −80°C. The patient
information is summarized in Table
I. Clinical tumor stage was classified according to the
Enneking staging system (26).
Tumor response to pre-operative chemotherapy was assessed using the
Huvos grading system (27), on the
basis of tumor necrosis in the resected specimen. Good response
indicated ≥90% tumor necrosis and poor response indicated <90%
tumor necrosis. All of the patients were followed up periodically.
The OS time was defined as the time from primary surgery to
mortality of the patient or, for living patients, the date of last
follow-up.
 | Table ICorrelation of miR-124 expression with
clinicopathological features of osteosarcoma. |
Table I
Correlation of miR-124 expression with
clinicopathological features of osteosarcoma.
| | miR-124
expression | |
|---|
| |
| |
|---|
| Clinicopathological
features | Number of cases | High n (%) | Low n (%) | P-value |
|---|
| Age |
| <25 years | 45 | 21 (46.7) | 24 (53.3) | 0.322 |
| ≥25 years | 60 | 32 (53.3) | 28 (46.7) | |
| Gender |
| Male | 57 | 26 (45.6) | 31 (54.4) | 0.329 |
| Female | 48 | 27 (56.3) | 21 (43.7) | |
| Tumor size |
| >8 cm | 55 | 23 (41.8) | 32 (58.2) | 0.079 |
| ≤8 cm | 50 | 30 (60.0) | 20 (40.0) | |
| Anatomical
location |
| Tibia/femur | 64 | 29 (45.3) | 35 (54.7) | 0.231 |
| Elsewhere | 41 | 24 (58.5) | 17 (41.5) | |
| Serum level of
lactate dehydrogenase |
| Elevated | 69 | 36 (52.2) | 33 (47.8) | 0.501 |
| Normal | 36 | 17 (47.2) | 19 (52.8) | |
| Serum level of
alkaline phosphatase |
| Elevated | 71 | 37 (52.1) | 34 (47.9) | 0.680 |
| Normal | 34 | 16 (47.1) | 18 (52.9) | |
| Clinical stage |
| IIA | 46 | 39 (84.8) | 7 (15.2) | <0.001 |
| IIB/III | 59 | 14 (23.7) | 45 (76.3) | |
| Distant
metastasis |
| Absent | 74 | 44 (59.5) | 30 (40.5) | 0.005 |
| Present | 31 | 9 (29.0) | 22 (71.0) | |
| Response to
chemotherapy |
| Good | 40 | 31 (77.5) | 9 (22.5) | 0.013 |
| Poor | 65 | 22 (33.8) | 43 (66.2) | |
Cell culture
Four human osteosarcoma cell lines (MG63, U2OS,
Saos-2 and SW1353) and a human normal osteoblastic cell line hFOB
1.19 were purchased from the Institute of Biochemistry and Cell
Biology of the Chinese Academy of Sciences (Shanghai, China). Cells
were cultured in RPMI-1640 medium (Invitrogen Life Technologies,
Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum
(FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin in
humidified air at 37°C with 5% CO2.
RNA extraction and RT-qPCR
Total RNA was isolated using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according
to the manufacturer’s instructions. Reverse transcription reaction
was carried out using 10 ng total RNA, 50 nmol/l stem-loop RT
primer, 1X RT buffer (TIANGEN Biotech Co., Ltd., Beijing, China),
0.25 mmol/l each deoxynucleotide triphosphate (Sigma-Aldrich,
Beijing, China), 3.33 U/μl MultiScribe reverse transcriptase
(Sigma-Aldrich) and 0.25 U/μl RNase inhibitor (Sigma-Aldrich). The
7.5 μl reaction mixture was initially incubated at 16°C for 30 min,
42°C for 30 min and 85°C for 5 min, and then maintained at 4°C.
qPCR was performed using the standard TaqMan MicroRNA assays
protocol on an ABI 7500 Real-Time PCR detection system (Applied
Biosystems by Life Technologies, Foster City, CA, USA), with
cycling conditions of 95°C for 10 min, followed by 40 cycles of
95°C for 15 sec and 60°C for 60 sec. U6 small nuclear RNA was used
as an internal control. The RT primers were 5′-GTC
GTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAT ACGACGGCATTCT-3′ for miR-124
and 5′-TGGTGT CGTGGAGTCG-3′ for U6. The PCR primers for mature
miR-124 or U6 were designed as follows: miR-124 forward,
5′-GATACTCATAAGGCACGCGG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′. U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The threshold cycle (Ct) was defined as
the fractional cycle number at which the fluorescence passed the
fixed threshold. Each sample was measured in triplicate, and the
relative amount of miR-124 to U6 was calculated using the equation
2−ΔCt, where ΔCT = (CTmiR–124 −
CTU6).
Cell transfection
For RNA transfection, the cells were seeded into
each well of 96-well plate and incubated overnight, then
transfected with either miR-124 mimic or negative control (NC)
(TIANGEN Biotech Co., Ltd.) using Lipofectamine 2000 (Invitrogen
Life Technologies) following the manufacturer’s instructions. The
sequences of NC were nonhomologous to any human genome sequences,
and were used to eliminate potential nonsequence-specific effects.
At 48 h after transfection, cells were harvested for further
experiments.
Cell proliferation assay
Cell proliferation capacity was evaluated with an
MTT assay. Cells were seeded into 96-well culture plates at a
density of 2,000 cells in 200 μl/well and incubated at 37°C for 24
h, after transfection. Then, 100 μl MTT solution (0.5 mg/ml;
Sigma-Aldrich, St. Louis, MO, USA) was added to each well, and the
cells were incubated for another 4 h. The medium was then replaced
with 150 μl DMSO. Spectrometric absorbance at 490 nm was measured
using a Multilabel Counter microplate reader (Safire; Tecan Austria
GmbH, Grödig, Austria). Cell proliferation was assessed daily for
four consecutive days, and the MTT assay was repeated three
times.
Detection of apoptosis by flow
cytometry
Apoptosis was detected by flow cytometric analysis.
Briefly, the cells were washed and resuspended in 0.5 ml
phosphate-buffered saline (pH 8.0) at a concentration of
1×106 cells/ml. Then, the cells were stained with
Annexin V and propidium iodide (PI), using the Annexin V Apoptosis
Detection kit (TIANGEN Biotech Co., Ltd.). After incubation at room
temperature in the dark for 15 min, the cell apoptosis was analyzed
on a FACSC LSR II (Becton Dickinson and Co., San Jose, CA,
USA).
Cell migration and invasion assays
The migration and invasion assays were performed
using 24-well Transwell chambers (8 μm; Corning Inc., Corning, NY,
USA). For the migration assay, 1×105 cells suspended in
200 μl serum-free RPMI-1640 medium were seeded into the upper
chamber of the Transwell invasion system, and 500 μl RPMI-1640
medium containing 10% FBS was added to the lower chamber. Following
a 24-h-incubation, cells on the upper surface of the membrane were
scrubbed off, and the migrated cells were fixed with 95% ethanol
and stained with 0.1% crystal violet for 10 min. The number of
migrated cells was determined by counting five random fields on
each membrane. The invasion assay protocol was similar to that of
the migration assay, with the exception that the upper chambers
were first covered with 1 mg/ml Matrigel.
Statistical analysis
SPSS software, version 16.0 for Windows (SPSS Inc.,
Chicago, IL, USA) was used for statistical analysis. Data are shown
as the mean ± standard deviation (SD). The differences between
groups were analyzed using the Student’s t-test or Chi-square test.
Patient survival and their differences were determined by the
Kaplan-Meier method and log-rank test. A Cox’s regression model was
used for univariate and multivariate analysis. P<0.05 was
considered to indicate a statistically significant result.
Results
Decreased expression of miR-124 in
osteosarcoma cell lines and primary tumor samples
The expression levels of miR-124 in osteosarcoma
tissues, corresponding noncancerous bone biopsy samples,
osteosarcoma cell lines and the human normal osteoblastic cell line
hFOB 1.19 were detected by RT-qPCR and normalized to U6 small
nuclear RNA. As shown in Fig. 1A,
the results revealed that miR-124 expression levels were
significantly lower in osteosarcoma tissues (8.3±2.1) than in the
corresponding noncancerous bone tissues (19.6±4.2; P<0.001).
Decreased miR-124 expression was also observed in osteosarcoma cell
lines compared with that in hFOB 1.19 cells (Fig. 1B; P<0.001). The MG63 cell line,
which possessed the lowest levels of miR-124 expression among all
tested osteosarcoma cell lines, was selected for analysis in
further experiments.
miR-124 expression and
clinicopathological features in osteosarcoma
The correlations of miR-124 expression with various
clinicopathological parameters of osteosarcoma tissues are
summarized in Table I. Using the
median miR-124 expression in all 105 osteosarcoma patients as a
cutoff, the patients were divided into a high miR-124 expression
group and a low miR-124 expression group. As shown in Table I, miR-124 was significantly
downregulated in patients with osteosarcoma of advanced Enneking
stage (P<0.001), positive distant metastasis (P=0.005) and poor
response to neoadjuvant chemotherapy (P=0.013). No significant
difference was observed between miR-124 expression levels and
patient age, gender, tumor size, anatomical location, or the serum
levels of lactate dehydrogenase and alkaline phosphatase.
Correlation between miR-124 expression
and prognosis of osteosarcoma patients
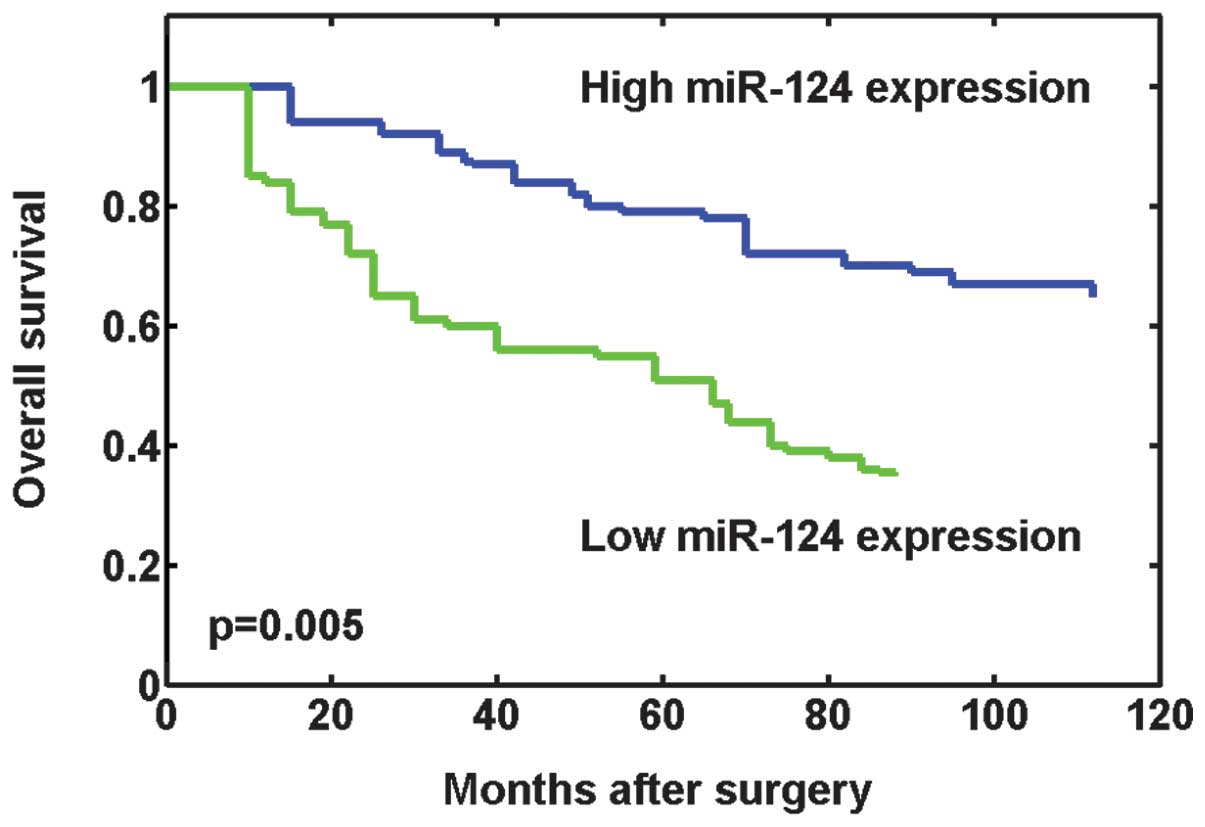
Whether miR-124 expression has prognostic potential
for the OS of osteosarcoma patients was investigated. Using the
Kaplan-Meier method and log-rank test, the OS times of patients
with low miR-124 expression levels were found to be significantly
shorter than those of patients with high miR-124 expression levels
(P=0.005; Fig. 2). In addition,
survival benefits were also found in those with smaller tumor size
(P=0.034), lower Enneking stage (P<0.001), without metastasis
(P=0.011) and a better response to preoperative chemotherapy
(P=0.006). Multivariate Cox regression analysis including the
aforementioned significant parameters revealed that miR-124
expression [relative risk (RR) 6.325; P=0.015], clinical stage (RR
8.973; P=0.008), metastasis status (RR 3.576; P=0.032), and
response to preoperative chemotherapy (RR 4.728; P=0.022) were
independent prognostic markers (Table
II).
 | Table IIUnivariate and multivariate analysis
of overall survival in 105 patients with osteosarcoma. |
Table II
Univariate and multivariate analysis
of overall survival in 105 patients with osteosarcoma.
| Variables | Univariate log-rank
test (P) | Cox multivariable
analysis (P) | Relative risk |
|---|
| MiR-124 expression
(high vs. low) | 0.005 | 0.015 | 6.325 |
| Clinical stage (IIA
vs. IIB/III) | <0.001 | 0.008 | 8.973 |
| Distant metastasis
(absent vs. present) | 0.011 | 0.032 | 3.576 |
| Tumor size (>8
cm vs. ≤8 cm) | 0.034 | - | - |
| Response to
chemotherapy (good vs. poor) | 0.006 | 0.022 | 4.728 |
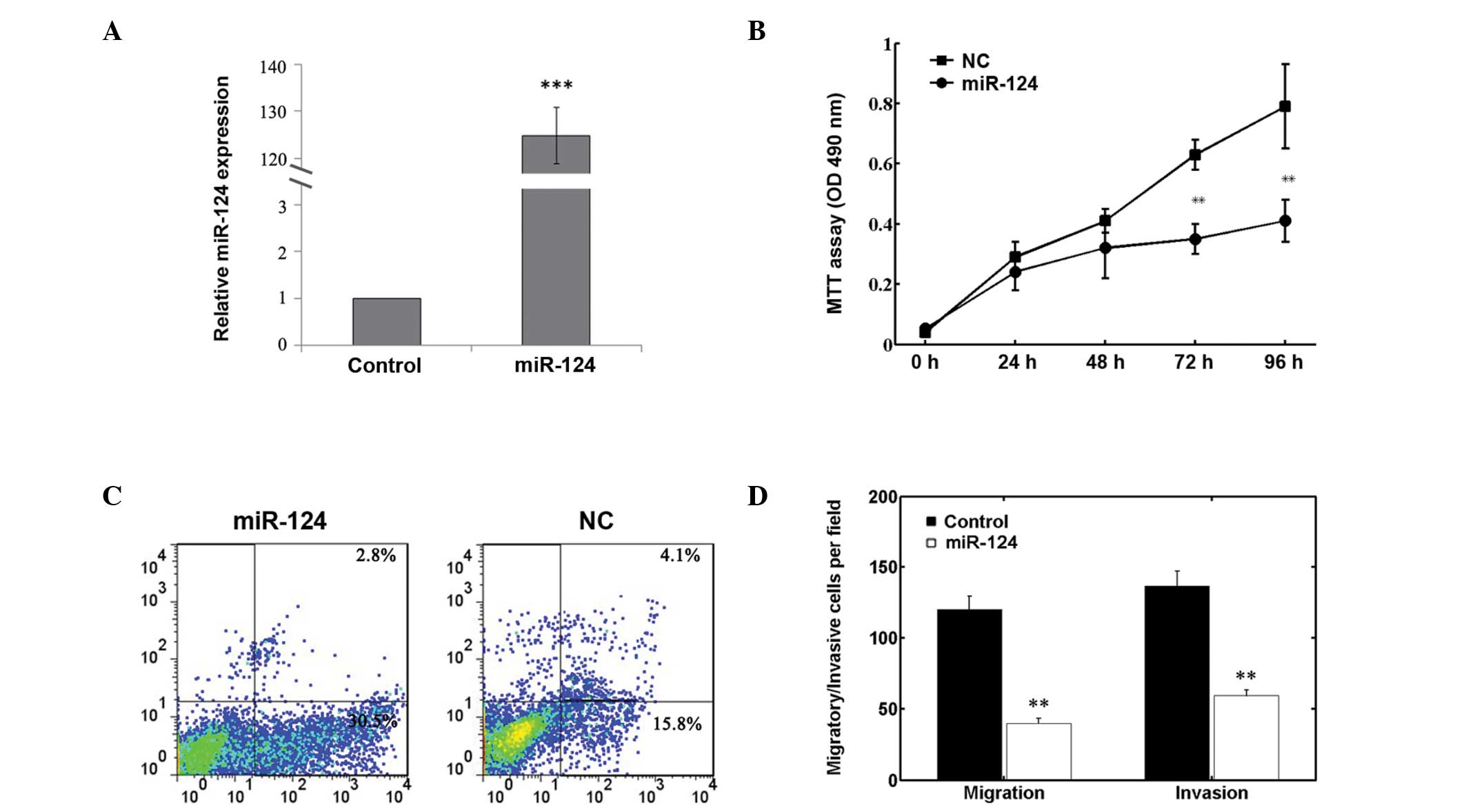
Effects of miR-124 on cell proliferation,
apoptosis, invasion and migration
As shown in Fig.
3A, the expression level of miR-124 in the miR-124
mimic-transfected cells was significantly higher compared with that
in NC-transfected cells (P<0.01). The MTT assay demonstrated
that transfection with miR-124 mimic reduced the proliferation of
MG63 cells (Fig. 3B). In addition,
promotion of cell apoptosis was also observed in the miR-124
mimic-transfected cells (Fig. 3C).
Furthermore, Transwell invasion and migration assays showed a
significant reduction in invaded or migrated MG63 cell numbers
following miR-124 transfection (Fig.
3D). These results indicate that miR-124 is involved in the
negative regulation of osteosarcoma cell growth, invasion and
migration in vitro.
Discussion
Dysregulation of miRNAs has been demonstrated to be
involved in tumorigenesis and progression in various types of
tumor; however, the elucidation of their potential roles in
osteosarcoma remains in the early stage of development. In the
current study, it was first demonstrated that miR-124 was
downregulated in osteosarcoma cell lines and primary tumor samples.
Low levels of miR-124 expression were found to be correlated with
aggressive clinicopathological features and unfavorable to
survival. Furthermore, the transfection of miR-124 mimic into MG63
cells was able to reduce cell proliferation, invasion and
migration, and promote cell apoptosis in vitro. To the best
of the authors’ knowledge, this is the first study regarding the
clinical significance and functional attributes of miR-124 in
osteosarcoma.
Previous research has reported the tumor suppressive
function of miR-124 in numerous human malignancies. In
vitro, ectopic miR-124 expression inhibits cell growth and
induces apoptosis in gastric cancer (21), colorectal cancer (22), pancreatic cancer (24), prostate cancer (28) and cervical cancer (20). The upregulation of miR-124 also
reduces cell invasion and migration in ovarian cancer (19), pancreatic cancer (24), hepatocellular carcinoma (23) and breast cancer (18). In addition, miR-124 radiosensitizes
human glioma cells (29). In
vivo, Zhang et al revealed decreased miR-124 expression
and its association with high tumor grade (Dukes C and D) in
colorectal cancer (22). Liang
et al identified that low miR-124 levels correlated with
poor differentiation of breast cancer (18). Zheng et al observed that
miR-124 downregulation occurred more frequently in hepatocellular
carcinoma patients with large tumor size, multiple tumor nodes and
advanced tumor stage (23).
Moreover, lower expression levels of miR-124 indicated worse
prognosis of patients suffering from pancreatic cancer, colorectal
cancer or hepatocellular carcinoma (23,24).
In xenotransplanted models, miR-124-treated nude mice exhibited
smaller tumor sizes and lower tumor weights in comparison with
those in the control group (21,22,24,28).
These findings suggest that miR-124 might play an important role
not only in tumor initiation and progression but also in the
molecular-targeted therapy of human malignancies.
The mechanism by which miR-124 expression affects
carcinogenesis and cancer development is complex. Some useful
targets have been identified during the past few years, including
SphK1 (19), STAT3 (22), SLC16A1 (17), Rac1 (24), ROCK2 (23), EZH2 (23), Slug (18), the androgen receptor (28) and CDK4 (29). However, there is no ‘one-to-one’
connection between miRNAs and target mRNAs. An average miRNA can
have more than 100 targets (30).
Conversely, several miRNAs can converge on a single transcript
target (31). Thus, the potential
regulatory circuitry afforded by miR-124 may be enormous, and the
identification of the complex molecular network involved in its
function remains an important subject for future investigation.
In conclusion, the results of the present study
revealed that miRNA-124 was downregulated in osteosarcoma cell
lines and clinical samples. Low-level expression of miR-124 was
significantly associated with a more aggressive and poor prognostic
phenotype of patients. Restored miR-124 expression in MG63 cells
exhibited antitumor effects in vitro. These data suggest an
important role of miR-124 in the molecular etiology and gene
therapy of osteosarcoma.
References
|
1
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eppert K, Wunder JS, Aneliunas V, Kandel R
and Andrulis IL: von Willebrand factor expression in osteosarcoma
metastasis. Mod Pathol. 18:388–397. 2005. View Article : Google Scholar
|
|
4
|
Osman A: MicroRNAs in health and disease -
basic science and clinical applications. Clin Lab. 58:393–402.
2012.
|
|
5
|
Zhao G, Cai C, Yang T, et al: MicroRNA-221
induces cell survival and cisplatin resistance through PI3K/Akt
pathway in human osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mendell JT and Olson EN: MicroRNAs in
stress signaling and human disease. Cell. 148:1172–1187. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
9
|
Dieckmann KP, Spiekermann M, Balks T, et
al: MicroRNAs miR-371-3 in serum as diagnostic tools in the
management of testicular germ cell tumours. Br J Cancer.
107:1754–1760. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takahashi M, Cuatrecasas M, Balaguer F, et
al: The clinical significance of MiR-148a as a predictive biomarker
in patients with advanced colorectal cancer. PLoS One.
7:e466842012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Hou C, Zhang H, et al: miR-126
functions as a tumor suppressor in osteosarcoma by targeting Sox2.
Int J Mol Sci. 15:423–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao H, Li M, Li L, et al: MiR-133b is
down-regulated in human osteosarcoma and inhibits osteosarcoma
cells proliferation, migration and invasion, and promotes
apoptosis. PLoS One. 8:e835712013. View Article : Google Scholar
|
|
13
|
Wang Z and Cai H, Lin L, Tang M and Cai H:
Upregulated expression of microRNA-214 is linked to tumor
progression and adverse prognosis in pediatric osteosarcoma.
Pediatr Blood Cancer. 61:206–210. 2014. View Article : Google Scholar
|
|
14
|
Tang M, Lin L, Cai H, Tang J and Zhou Z:
MicroRNA-145 downregulation associates with advanced tumor
progression and poor prognosis in patients suffering osteosarcoma.
Onco Targets Ther. 6:833–838. 2013.PubMed/NCBI
|
|
15
|
Zhou Y, Huang Z, Wu S, et al: miR-33a is
up-regulated in chemoresistant osteosarcoma and promotes
osteosarcoma cell resistance to cisplatin by down-regulating TWIST.
J Exp Clin Cancer Res. 33:122014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chandrasekar V and Dreyer JL: microRNAs
miR-124, let-7d and miR-181a regulate cocaine-induced plasticity.
Mol Cell Neurosci. 42:350–362. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li KK, Pang JC, Ching AK, et al: miR-124
is frequently down-regulated in medulloblastoma and is a negative
regulator of SLC16A1. Hum Pathol. 40:1234–1243. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang YJ, Wang QY, Zhou CX, et al: MiR-124
targets Slug to regulate epithelial-mesenchymal transition and
metastasis of breast cancer. Carcinogenesis. 34:713–722. 2013.
View Article : Google Scholar :
|
|
19
|
Zhang H, Wang Q, Zhao Q and Di W: MiR-124
inhibits the migration and invasion of ovarian cancer cells by
targeting SphK1. J Ovarian Res. 6:842013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wilting SM, van Boerdonk RA, Henken FE, et
al: Methylation-mediated silencing and tumour suppressive function
of hsa-miR-124 in cervical cancer. Mol Cancer. 9:1672010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xia J, Wu Z, Yu C, et al: miR-124 inhibits
cell proliferation in gastric cancer through down-regulation of
SPHK1. J Pathol. 227:470–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang J, Lu Y, Yue X, et al: MiR-124
suppresses growth of human colorectal cancer by inhibiting STAT3.
PLoS One. 8:e703002013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zheng F, Liao YJ, Cai MY, et al: The
putative tumour suppressor microRNA-124 modulates hepatocellular
carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut.
61:278–289. 2012. View Article : Google Scholar
|
|
24
|
Wang P, Chen L, Zhang J, et al:
Methylation-mediated silencing of the miR-124 genes facilitates
pancreatic cancer progression and metastasis by targeting Rac1.
Oncogene. 33:514–524. 2014. View Article : Google Scholar
|
|
25
|
Shi XB, Xue L, Ma AH, et al: Tumor
suppressive miR-124 targets androgen receptor and inhibits
proliferation of prostate cancer cells. Oncogene. 32:4130–4138.
2013. View Article : Google Scholar
|
|
26
|
Enneking WF, Spanier SS and Goodman MA: A
system for the surgical staging of musculoskeletal sarcoma. Clin
Orthop Relat Res. 106–120. 1980.PubMed/NCBI
|
|
27
|
Rosen G, Caparros B, Huvos AG, et al:
Preoperative chemotherapy for osteogenic sarcoma: selection of
postoperative adjuvant chemotherapy based on the response of the
primary tumor to preoperative chemotherapy. Cancer. 49:1221–1230.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Chen J, Gan S, et al: DNA damage
strength modulates a bimodal switch of p53 dynamics for cell-fate
control. BMC Biol. 11:732013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng X, Ma L, Wu M, et al: miR-124
radiosensitizes human glioma cells by targeting CDK4. J Neurooncol.
114:263–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krek A, Grün D, Poy MN, et al:
Combinatorial microRNA target predictions. Nat Genet. 37:495–500.
2005. View
Article : Google Scholar : PubMed/NCBI
|