Introduction
Helicobacter pylori (H. pylori) are
one of the major risk factors for stomach disease. Routine clinical
treatments for clearing H. pylori infection are usually
triple or quadruple antibiotic therapies, but they are often
accompanied with increased incidence of esophageal diseases
(1).
Studies have shown that the incidence of reflux
esophagitis in patients with gastroduodenal ulcer and H.
pylori infection who were treated with antibiotic therapy (26%)
was higher than that in patients who did not receive antibiotic
therapy (13%). It was reported that reflux esophagitis often occurs
following H. pylori eradication therapy (2–6), and
that malignant esophageal adenoma is a complication of reflux
esophagitis. A retrospective examination carried out in patients
with Barrett’s esophagus showed that, following H. pylori
eradication therapy, the columnar epithelium area expanded into the
area of the gastroesophageal junction (7). In 2011, a study in Japan reported the
appearance of lower esophageal malignant adenomas in a patient with
stomach ulcers following H. pylori eradication therapy
(7). These studies suggested that
the use of antibiotic therapy for the eradication of H.
pylori may increase esophageal disease, and H. pylori
may become a potential risk factor for esophageal cancer.
It has been found that simian immunodeficiency virus
infection can cause significant changes in the chimpanzee
intestinal microbiota (8).
Furthermore, in a previous study, the 16S rRNA sequencing of
intestinal microbiota was carried out for individuals with human
immunodeficiency virus (HIV)-negative infection, new HIV-1
infection and old HIV-1 infection. The results showed that HIV
infection was associated with unique changes in the intestinal
microbiota. Antiviral therapy did not allow the microbial
communities to return to the HIV-negative status. HIV-infected
intestines had the characteristics of chronic intestinal enteritis,
but the similarity of the HIV-associated microbiota to the
microbiota of other inflammatory states was limited, which
increased the diversity (9).
As an important part of the esophageal
microenvironment, the microbiota maintains the stability and
balance of the microenvironment through the regulation of various
systems (10). The lower esophagus
is closely connected to the stomach. Changes in the composition of
the lower esophageal microbiota can be caused by the colonization
of H. pylori in the stomach; thus, the association between
these changes and diseases in the lower esophagus warrants
investigation. In the present study, mouse models were used to
analyze the changes in the composition of the lower esophageal
microbiota following H. pylori infection and the eradication
of the H. pylori by antibiotics. In addition, the mechanisms
of diseases in the lower esophagus were further investigated.
Materials and methods
Reagents and equipment
H. pylori standard strain (Hp11637) was
obtained from the Department of Clinical Microbiology, Third
Military Medical Universiry (Chongqing, China), and was stored in
the Laboratory of Pathogen Biology and Immunology (Chongqing,
China). Omeprazole was purchased from Shanxi Jinhua Huixing
Pharmaceutical Co., Ltd. (Yuncheng, China). The antibiotics
ampicillin and clarithromycin were purchased from Zhangjiajie Yuan
Pharmaceutical Co., Ltd. (Zhangjiajie, China) and Xingtai Mingshen
Pharmaceutical Factory (Xingtai, China), respectively. The
bacterial DNA extraction kit was provided by Tiangen Biotech
(Beijing) Co., Ltd. (Beijing, China). Polymerase chain reaction
(PCR) reagents were purchased from Takara Biotech Inc. (Dalian,
China). PCR-denaturing gradient gel electrophoresis (DGGE)
instruments and all associated equipment were purchased from
Bio-Rad (Hercules, CA, USA). The primer sequences specific for the
H. pylori 16S rDNA were as follows: Reverse,
5′-TTTGTTAGAGAAGATAATGACGGTATCTAA-3′; forward,
5′-CATAGGATTTCACACCTGACTGACTATC-3′. The primer sequences for the
prokaryotic rDNA V6 region containing the GC clamp were as follows:
Reverse, 5′-CGGTGTGTACAAGACCC-3′; forward,
5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGCACGGGGGGAACGCGAAGAACCTTAC-3′.
The primer sequences for the prokaryotic rDNA V6 region without the
GC clamp were as follows: Reverse, 5′-CGGTGTGTACAAGACCC-3′;
forward, 5′-AACGCGAAGAACCTTAC-3′. All primers were synthesized by
Shanghai Yingjun Biotechnology Company (Shanghai, China).
Animals
Specific pathogen-free female BALB/c mice (aged 6–8
weeks and weighing 18–20 g), provided by the Experimental Animal
Center of Chongqing Medical University (Chongqing, China), were
randomly divided into three groups of 15 mice. Group A was the
negative control group, which was not infected with H.
pylori, while the mice in group B were infected with H.
pylori (infection group). The mice in group C were treated with
antibiotics subsequent to being infected with H. pylori
(treatment group). Following fasting for 12 h, the mice in groups B
and C were administered 0.5 ml 2×109 CFU/ml fresh H.
pylori solution via gavage, which was repeated every three days
for five times in total. After an interval of four weeks, group C
was fasted for 12 h each day and then orally administered 0.75 ml
solution containing 0.25 ml 0.2 mg/ml omeprazole, 0.25 ml 20 mg/ml
ampicillin and 0.25 ml 50 mg/ml clarithromycin. The mice were
subsequently fasted for a further 3–4 h. These processes were
carried out once a day for seven consecutive days. Similarly, the
mice in group B were treated in the same manner using sterile
saline instead of antibiotics. In group A, sterile saline solution
was used to replace the H. pylori bacteria and antibiotics,
but all other processes were identical. The mice in the three
groups were sacrificed at the same time, once the mice in group C
had been treated with antibiotics for two weeks. All animal
experiments were conducted according to the ethical guidelines of
Chongqing Medical University.
Colonization of H. pylori
Following the sacrifice of the mice by decapitation,
the entire stomach of each mouse was washed and divided into two
parts. One half was tested with a rapid urease test strip, in which
a change from yellow to blue was judged to be a positive result.
The rapid urease test strip was produced according to the methods
of a previous study (11). The
other half was manually ground in 500 μl sterile saline, and more
sterile saline was added to produce a total volume of 1 ml, from
which 100 μl was taken for streaking inoculation on H.
pylori-selective agar plates. The inoculated agar plates were
placed in an anaerobic jar in which a microaerophilic environment
was formed using airbags at 37°C for 74 h.
The remaining homogenate was used for DNA extraction
with a bacterial DNA extraction kit. DNA extractions were performed
according to the manufacturer’s instructions. The primer sequences
specific for the H. pylori 16S rDNA were then used for PCR
amplification of the extracted DNA. The PCR conditions were set as
follows: Initial denaturation at 95°C for 10 min; 30 cycles of 95°C
for 30 sec (denaturation), 60°C for 30 sec (annealing) and 72°C for
30 sec (extension), and a final extension step at 72°C for 10 min.
The extracted H. pylori DNA was used as positive control.
The amplification products were analyzed using agarose gel
electrophoresis. H. pylori colonization was considered to
have occurred if two of the following three tests had positive
results: Isolation culture bacteria, rapid urease test and PCR
analysis. In the treatment group, H. pylori eradication was
considered to have occurred if the results for H. pylori
culture, rapid urease test and PCR analysis were all negative.
Pathohistological observation
The mice were decapitated and the stomachs were
retrieved under sterile conditions for visual inspection of the
control and infected stomachs. The residues inside the stomachs
were washed off by sterile saline. The stomachs were longitudinally
cut open for ulcer observation. According to the clinical criteria
for judging the degree of gastric mucosal lesions (12), the pathological stomach changes in
the infection and treatment groups were classified as follows: No
pathological changes, inflammation, mild ulcers and large ulcers
(ulcer diameter >2 cm).
Hematoxylin-eosin (HE) staining
Tissues of the lower esophagus were fixed with 10%
formaldehyde, dehydrated by a graded ethanol series, embedded by
paraffin and sliced into serial vertical sections. Finally, the
slices were stained by HE for observation under a microscope.
PCR-DGGE
Following the sacrifice of the mice, one-third of
the lower esophagus was removed under sterile conditions, and
further cut longitudinally into two halves. One half was fixed
using 10% formaldehyde solution and was observed under the
microscope subsequent to HE staining. The other half was
homogenized for bacterial DNA extraction using a DNA extraction
kit. The extracted DNA was used as a template, and primers for the
prokaryotic 16S rDNA V6 region containing a GC clamp were used to
amplify the bacterial genomic DNA (13). The amplification products were
analyzed by DGGE using the Bio-Rad gel irrigation system (Quantity
One 1-D Analysis Software version 4.6.2; Bio-Rad Laboratories,
Inc., Hercules, CA, USA), in which the denaturing gradient of
vertical electrophoresis was 0–80%, the temperature was maintained
at 60°C, the voltage was 85 V and the total time of electrophoresis
was 16 h. The gel was rinsed with deionized water and stained with
silver nitrate subsequent to electrophoresis, and the GD7500 gel
imaging analysis system (Bio-Rad) was then used for imaging.
DGGE profiling
DGGE patterns for each group were analyzed. Three
samples were randomly selected from group A, and nine samples were
randomly selected from groups B and C, respectively. PCR-DGGE was
performed for a total of 21 samples on the same piece of gel using
Quantity One® 1-D Analysis Software (Version 4.6.2;
Bio-Rad). Following standard processing, the DGGE staining patterns
were analyzed. The Shannon-Weiner diversity index (14) was calculated using the following
formula: H = −∑ (Ni/N) × ln (Ni/N), in which Ni was the brightness
of the individual bands and N was the brightness of all the bands.
The homogeneity (Pielou) index (14) was calculated using the formula E =
H′/lnS, in which S was the number of the bands, and the abundance
(Margalef) index (14) was
calculated using the formula R = (S−1)/lnN. According to the
criteria provided by Jiang et al (15), the average value of
OD30250 was taken as the cut-off value to analyze the
average number of bands for each experimental group. The Student’s
t-test was used to examine statistically significant differences
between group bands.
Quantity One software was used to analyze the
similarity of profiles between different groups. Data were
processed using the unweighted pair group method with arithmetic
mean, and tree diagrams for similarity between groups were plotted.
The similarity of the microbiota composition in the lower esophagus
was then analyzed on the basis of the tree diagrams.
Sequencing analysis
The recycled amplification products were sent for
sequencing and the DNA sequences were determined using Basic Local
Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
identification (16).
Statistical analysis
All statistical analyses were performed using SPSS
17.0 for Windows (SPSS Inc., Chicago, IL, USA). Results are
expressed as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
H. pylori colonization leads to severe
ulcers in the mouse stomach, which are alleviated by antibiotic
treatment
To test the effect of antibiotic treatment on H.
pylori infection, H. pylori colonization in the mouse
stomach was measured and the stomachs were visually inspected. The
results showed no H. pylori colonization in the negative
control group, while the percentage of colonization in the
infection group and treatment group was 100%, respectively; the
H. pylori eradication rate was 93% in the treatment group
(Table I). Visual inspection
showed that 93% of the mice in the infection group had gastric
lesions, the majority of which were classified as large ulcers
(Fig. 1 and Table II). Following treatment, the
percentage of mice with gastric lesions did not change, but the
ulcers were mitigated (Table II).
These data indicated that H. pylori infection had a serious
effect on the stomachs of mice, and that treatment with antibiotics
reduced the degree of gastric ulceration.
 | Table IHelicobacter pylori
colonization in different groups of mice. |
Table I
Helicobacter pylori
colonization in different groups of mice.
| Group | No. of mice | Positive result in
urease test, n (%) | Positive result in
culture, n (%) | Positive result in
PCR, n (%) | Colonization n
(%) | Eradication n
(%) |
|---|
| A | 15 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | - |
| B | 15 | 14 (93) | 15 (100) | 14 (93) | 15 (100) | - |
| C | 15 | 1 (7) | 1 (7) | 1 (7) | - | 14 (93) |
 | Table IIDegree of gastric mucosal lesions in
mice. |
Table II
Degree of gastric mucosal lesions in
mice.
| Group | No. of mice | Mice without
pathological changes, n (%) | Mice with
inflammation, n (%) | Mice with mild
ulcers, n (%) | Mice with large
ulcers, n (%) |
|---|
| Infection | 15 | 1 (7) | 1 (7) | 4 (27) | 9 (60) |
| Treatment | 15 | 1 (7) | 3 (20) | 8 (53) | 3 (20) |
Antibiotic treatment reduces gastric
lesions induced by H. pylori infection according to histological
examination results
To visualize how antibiotic treatment affected the
H. pylori-infected mouse stomach, HE staining and microscopy
were performed. The results showed mucosal integrity, no damage and
few inflammatory cells in the negative control group (Fig. 2A). By contrast, mucosal injury,
increased number of glands and significantly increased inflammatory
cell infiltration were observed in the infection group (Fig. 2B). In the treatment group, severe
mucosal injury and glandular atrophy were visualized, but the
number of inflammatory cells was significantly decreased (Fig. 2C). These results suggested that
antibiotic treatment attenuated the gastric lesions induced by
H. pylori infection.
PCR amplifies the bacterial 16S rDNA V6
region
To analyze the PCR amplification products, agarose
gel electrophoresis was performed. The data showed that the
bacterial DNA extracted from the lower esophagus was specifically
amplified by PCR, with the amplified fragment length of ~450 bp.
This observation was consistent with the expected amplified
fragment length (Fig. 3), which
suggested that the 16S rDNA V6 region was correctly amplified.
Increased numbers of bacterial species
are observed in the stomachs of the infection group compared with
the stomachs of the control and treatment group mice
To determine the number of bacterial species present
in the microbiota in the three groups of mice, DGGE profiling was
performed. The DGGE profiles in the lower esophagus showed that the
numbers of bands in groups A, B, and C were 8.7±0.6, 16.4±1.8, and
4.9±0.9, respectively (Fig. 4).
These data indicated that more species were present in the
infection group than in the control and treatment groups.
Bacterial species in the lower esophagus
vary among the three groups of mice, but the dominant species and
the relative content are similar
To analyze the diversity of the bacterial species in
the lower esophageal microbiota of mice, diversity index
calculation methods were used to investigate the number of bands
and gray scales for each of the groups in the DGGE fingerprints.
The analysis showed that there were significant differences in the
diversity and abundance indices of the DGGE profiles among the
three groups (P<0.05); however, the homogeneity index showed no
significant differences among the groups (P>0.05) (Table III). The abundance index
reflected the number of bacterial species present, while the
diversity index indicated the heterogeneity among the groups and
the homogeneity index reflected the dominant species present in the
microbiota and the relative content (Fig. 5). These results suggested that the
bacterial species in the lower esophageal microbiota varied among
the three groups of mice, but the dominant species and the relative
content were similar.
 | Figure 5DGGE profiles of the lower esophageal
bacterial 16S rDNA in selected mice. The amplification products of
the bacterial 16S rDNA V6 were analyzed by DGGE using the Bio-Rad
(Hercules, CA, USA) gel irrigation system, in which the denaturing
gradient of vertical electrophoresis was 0–80%, the temperature was
maintained at 60°C, the voltage was 85 V and the total time of
electrophoresis was 16 h. The gel was rinsed with deionized water
and stained with silver nitrate subsequent to electrophoresis, and
the GD7500 gel imaging analysis system (Bio-Rad) was then used for
imaging. Each lane represents a mouse, each band represents a type
of bacteria, and the number of bands in the same lane corresponds
to the number of bacterial species. DGGE, denaturing gradient gel
electrophoresis; A, negative control group; B, infection group; C,
treatment group. |
 | Table IIIAnalysis of V6-denaturing gradient
gel electrophoresis fingerprinting diversity. |
Table III
Analysis of V6-denaturing gradient
gel electrophoresis fingerprinting diversity.
| Group | Diversity index
(mean ± SD) | Margalef index
(mean ± SD) | Pielou index (mean
± SD) |
|---|
| A | 8.7±0.6 | 2.8±0.2 | 1.3±0.1 |
| B | 16.4±1.8 | 5.6±0.5 | 2.0±0.2 |
| C | 4.9±0.9 | 1.8±0.2 | 1.1±0.2 |
H. pylori infection and antibiotic
treatment can change the composition of the stable microbial
community in the lower esophagus
To analyze the similarity of the profiles in the
different groups, tree diagrams were plotted and used for the
comparison of the lower esophageal microbiota composition among the
groups. According to the similarity coefficient, groups with
samples showing a high similarity were classified into one class so
that classes I, II and III were obtained. In class I, A2 mice
exhibited relatively high similarity to A3 mice (0.46), but
relatively low similarity to B3 mice (0.40). In class II, the
groups with the highest similarity were C3 and C4, with a
similarity coefficient of 0.88. Groups C5 and C6/C7 had a
similarity coefficient of 0.82, and the similarity coefficient for
groups C1 and C2 was 0.63. In class III, the highest similarity
occurred between B1 and B2, with a coefficient of 0.62, followed by
B5 and B9, with a similarity coefficient of 0.58. The lowest
similarity coefficient was 0.56 for B6 and B7 (Fig. 6). These results showed that, with
the exception of B3, bacteria in different mice could be separated
on the evolutionary tree, suggesting that stable microbial
community compositions existed in the lower esophagus, but with
different compositions among the groups.
 | Figure 6Analysis of PCR-DGGE patterns of
bacteria in each group. Three samples were randomly selected from
group A, and nine samples were randomly selected from groups B and
C, respectively. PCR-DGGE was performed for a total of 21 samples
on the same piece of gel. Quantity One® software
(Bio-Rad, Hercules, CA, USA) was used to analyze the similarity of
profiles between different groups. Data were processed using the
unweighted pair group method with arithmetic mean, and tree
diagrams for similarity between groups were plotted. The similarity
of the microbiota composition in the lower esophagus was then
analyzed on the basis of the tree diagrams. PCR-DGGE, polymerase
chain reaction-denaturing gradient gel electrophoresis; A, negative
control group; B, infection group; C, treatment group. |
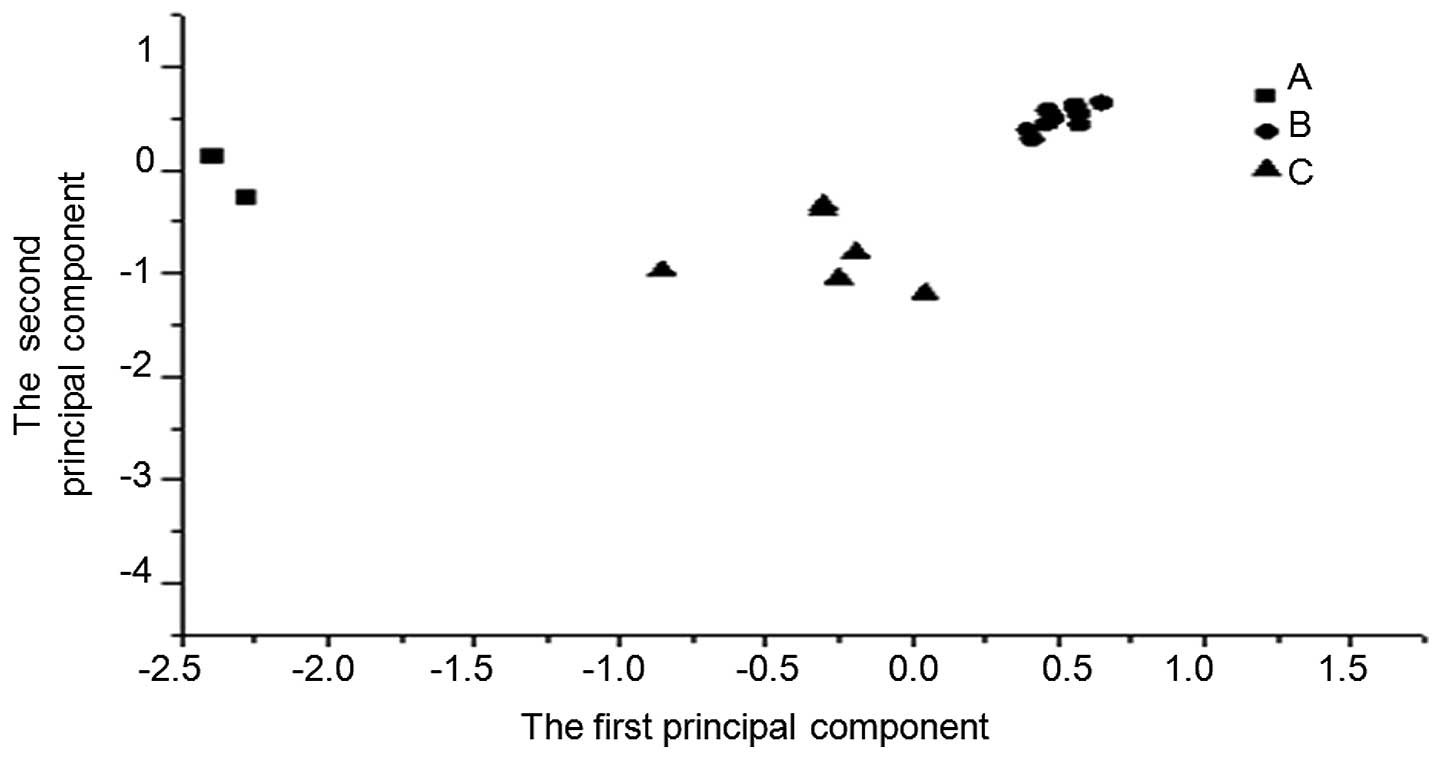
Variations in the dominant flora species
of the control, infection and treatment groups
To analyze the DGGE patterns in the different groups
of mice, principal component analysis was performed. The data
showed that the bacteria in the different groups gathered in
different locations. Although the composition of flora species in
the lower espohagus varied, the dominant species, and their
relative contents, were similar in the control, infection and
treatment groups (Fig. 7).
Certain types of bacteria are found in
the lower esophagus of all three groups, but certain bacteria are
specific solely to the infection or control group
To analyze the bacterial species in the three
groups, the mean OD30250 value was used as the cut-off value. Two
unique bands (a1 and a2) were observed in group A and six (b1–b6)
in group B; however, no unique bands were observed in group C. The
three experimental groups shared three common bands (Fig. 6 and Table IV). The unique bands and common
bands were amplified and BLAST analysis was performed to compare
the sequences to determine the species of bacteria. These results
suggested that certain types of bacteria were found in the lower
esophagus of all three groups, but certain bacteria were specific
solely to the infection or control group.
 | Table IVAnalysis of bacteria species. |
Table IV
Analysis of bacteria species.
| Band
properties | Band no. | Species of
bacteria | Similarity (%) |
|---|
| Specific for Group
A | a1 |
Lactobacillus | 98 |
| a2 |
Bacteroides | 97 |
| Specific for Group
B | b1 |
Staphylococcus | 99 |
| b2 |
Acinetobacter | 97 |
| b3 |
Bacteridium | 99 |
| b4 | Uncultured
bacterium | 99 |
| b5 | Uncultured
bacterium | 99 |
| b6 | Uncultured
bacterium | 99 |
| Common for Groups
A, B and C | 1 |
Enterobacter | 99 |
| 1 |
Klebsiella | 99 |
| 1 | Pseudomonas
aeruginosa | 99 |
Discussion
It has been reported that >200 types of bacteria
can colonize in the lower esophagus (17). In the present study, DGGE
fingerprint spectrum abundance confirmed that the lower esophagi of
the studied mice each contained a microbiota composed of a large
number of bacteria. The dominant types of bacteria included
Lactobacillus, Bacteroides, Staphylococcus,
Escherichia coli, Klebsiella and Pseudomonas
aeruginosa.
In the present study, H. pylori-infected
mouse models were constructed to analyze the DGGE profiles of the
prokaryotic 16S rDNA V6 region in the lower esophagus of the
infected and uninfected mice. It was found that both the number and
species of the dominant bacteria in the lower esophagus increased
significantly subsequent to the H. pylori infection. In
addition, more complex types of bacteria, Acinetobacter,
Klebsiella, Enterobacter, and various unknown species appeared.
Previous studies have reported that H. pylori infection can
change the microenvironment of the stomach, and inhibit or promote
the growth of certain types of bacteria (18,19).
Since the lower esophagus is closely connected to the stomach,
changes in the microenvironment of the stomach may affect the
microenvironment of the lower esophagus. Inflammation and mucosal
injury of the lower esophagus in the infection and treatment groups
indicated that H. pylori infection and antibiotic treatment
could alter the microenvironment of the lower esophagus, causing
changes in the composition of the lower esophageal microbiota.
Regarding the eradication of H. pylori
infection using antibiotics, DGGE profile analysis of the 16S rDNA
V6 region showed that the number of species in the lower esophageal
microbiota was significantly reduced in the treatment group
compared with the infection group, and no specific bacterial
colonization was observed. This could be due to the antibiotics
killing other bacteria as well as H. pylori or due to the
antibiotic treatment changing the microenvironment in the lower
esophagus to such an extent that the bacteria could not
colonize.
Previous studies have suggested that lower
esophageal sphincter pressure and lower esophageal acid reflux are
the most important factors for changes in the pathology of the
lower esophagus (20–23); however, decreased lower esophageal
sphincter pressure may be due to the microbes in the lower
esophagus. A study in mice showed that certain microbes in the
lower esophagus are present from birth, while other microbial
populations appear subsequent to colonization (20). Gram-negative bacteria have been
reported to be the dominant bacteria causing acid reflux in the
stomach; these bacteria cause relaxation of the lower esophageal
sphincter predominantly through the induction of nitric oxide and
enzymes (24).
In this study, following the eradication of H.
pylori infection by antibiotics, the number of Gram-negative
bacteria was not increased, but colonization with
Staphylococcus, Acinetobacter and non-spore Bacillus
was observed. The association between the changes in the lower
esophageal microbiota and esophageal diseases therefore warrants
further investigation.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 31070445).
References
|
1
|
Labenz J, Blum AL, Bayerdöffer E, et al:
Curing Helicobacter pylori infection in patients with duodenal
ulcer may provoke reflux esophagitis. Gastroenterology.
112:1442–1447. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hamada H, Haruma K, Mihara M, et al: High
incidence of reflux oesophagitis after eradication therapy for
Helicobacter pylori: impacts of hiatal hernia and corpus gastritis.
Aliment Pharmacol Ther. 14:729–735. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Inoue H, Imoto I, Taguchi Y, et al: Reflux
esophagitis after eradication of Helicobacter pylori is associated
with the degree of hiatal hernia. Scand J Gastroenterol.
39:1061–1065. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kawanishi M: Development of reflux
esophagitis following Helicobacter pylori eradication. J
Gastroenterol. 40:1024–1028. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Take S, Mizuno M, Ishiki K, et al:
Helicobacter pylori eradication may induce de novo, but transient
and mild, reflux esophagitis: Prospective endoscopic evaluation. J
Gastroenterol Hepatol. 24:107–113. 2009. View Article : Google Scholar
|
|
6
|
Nam SY, Choi IJ, Ryu KH, et al: Effect of
Helicobacter pylori infection and its eradication on reflux
esophagitis and reflux symptoms. Am J Gastroenterol. 105:2153–2162.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abe Y, Koike T, Iijima K, et al:
Esophageal adenocarcinoma developing after eradication of
Helicobacter pylori. Case Rep Gastroenterol. 5:355–360. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moeller AH and Ochman H: Factors that
drive variation among gut microbial communities. Gut Microbes.
4:403–408. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lozupone CA, Li M, Campbell TB, et al:
Alterations in the gut microbiota associated with HIV-1 infection.
Cell Host Microbe. 14:329–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang L, Lu X, Nossa CW, et al:
Inflammation and intestinal metaplasia of the distal esophagus are
associated with alterations in the microbiome. Gastroenterology.
137:588–597. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu ZQ and Qing SD: Study on the self-made
rapid urease test strip for detecting Helicobacter pylori.
Chongqing Yi Ke Da Xue Xue Bao. 17:313–315. 1992.
|
|
12
|
Sezikli M, Çetinkaya ZA, Güzelbulut F, et
al: Effects of alpha tocopherol and ascorbic acid on Helicobacter
pylori colonization and the severity of gastric inflammation.
Helicobacter. 17:127–132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huws SA, Edwards JE, Kim EJ and Scollan
ND: Specificity and sensitivity of eubacterial primers utilized for
molecular profiling of bacteria within complex microbial
ecosystems. J Microbiol Methods. 70:565–569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
von Rosenvinge EC, Song Y, White JR, et
al: Immune status, antibiotic medication and pH are associated with
changes in the stomach fluid microbiota. ISME J. 7:1354–1366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Gao F, Xu X, et al: Changes in
the composition of the bacterial flora on tray-packaged pork during
chilled storage analyzed by PCR-DGGE and real-time PCR. J Food Sci.
76:M27–M33. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Devillard E, Burton JP and Reid G:
Complexity of vaginal microflora as analyzed by PCR denaturing
gradient gel electrophoresis in a patient with recurrent bacterial
vaginosis. Infect Dis Obstet Gynecol. 13:25–31. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chao A: Nonparametric estimation of the
number of classes in a population. Scand J Statist. 11:265–270.
1984.
|
|
18
|
Reid G and Burton J: Use of Lactobacillus
to prevent infection by pathogenic bacteria. Microbes Infect.
4:319–324. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo G, Tong W, Zou Q, et al: The role of
gene HP0318 in adaptative colonization of Helicobacter pylori. Di
San Jun Yi Da Xue Xue Bao. 29:1006–1009. 2007.(In Chinese).
|
|
20
|
Ley RE, Bäckhed F, Turnbaugh P, et al:
Obesity alters gut microbial ecology. Proc Natl Acad Sci USA.
102:11070–11075. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iwakiri K, Hayashi Y, Kotoyori M, et al:
The minimum pressure of the lower esophageal sphincter, determined
by the rapid pull-through method, is an index of severe reflux
esophagitis. J Gastroenterol. 39:616–620. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sano H, Iwakiri K, Kawami N, Tanaka Y and
Sakamoto C: Mechanisms of Acid reflux and how refluxed Acid extends
proximally in patients with non-erosive reflux disease. Digestion.
90:108–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gatenby P and Soon Y: Barrett’s
oesophagus: Evidence from the current meta-analyses. World J
Gastrointest Pathophysiol. 5:178–187. 2014.PubMed/NCBI
|
|
24
|
Fan YP, Chakder S, Gao F and Rattan S:
Inducible and neuronal nitric oxide synthase involvement In
lipopolysaccharide-induced sphincteric dysfunction. Am J Physiol
Gastrointest Liver Physiol. 280:G32–G42. 2001.
|