Introduction
Worldwide, lung cancer has become the cancer with
the highest morbidity and mortality (1). Non-small cell lung cancer (NSCLC)
accounts for 80–85% of all cases of lung cancer (2). The echinoderm microtubule-associated
protein-like 4 (EML4)-anaplastic lymphoma kinase (ALK) fusion gene
was discovered in 2007 as a lung cancer-specific fusion gene in
patients with lung cancer, with a positive rate of 3–8% (3). The presence of the EML4-ALK fusion gene
is mutually exclusive with epidermal growth factor receptor (EGFR)
mutation. The most common EML4-ALK fusion gene variants are
variants 1, 2 and 3 in patients with NSCLC. The positive rates of
variants 1, 2 and 3 are ∼33, 29 and 9%, respectively. These three
variants represent the majority of all variants; other variants are
found in relatively low proportions (4–10).
The thymidylate synthase (TYMS) gene encodes the
rate-limiting enzyme in the de novo synthesis of
deoxythymidine monophosphate, plays an important role in DNA
synthesis, replication and repair (11), leads to DNA breaks and cell death,
and is an effective target for anticancer drugs. Studies have shown
that TYMS activity is significantly higher than that in normal
tissue in a variety of malignant tumors (12), affecting cell cycle by regulating the
expression of p53, and thus affecting tumor cell proliferation
(13). TYMS has been found to be
associated with tumor proliferation (14), and tumor cell populations that
overexpress TYMS have greater growth potential, suggesting that
high TYMS expression correlates with poor prognosis.
For lung cancer patients with low expression levels
of TYMS, the efficacy of the first-line chemotherapy drug
pemetrexed (Alimta) has been demonstrated to be improved (15,16). In
the present multi-center study, the expression levels of the
EML4-ALK fusion gene and TYMS mRNA in 257 patients with stage I–IV
NSCLC were reviewed and the correlation between them was analyzed.
The association of the EML4-ALK fusion gene with the expression of
the TYMS resistance gene in patients with NSCLC was investigated in
order to further explore more effective individualized treatment
plans for patients carrying the EML4-ALK fusion gene.
Materials and methods
Specimens
Paraffin-embedded tissue specimens were collected
from 257 patients from surgeries performed between 2004 and 2013.
There were 103 cases from the General Military Hospital of Beijing
PLA (Beijing, China), 58 cases from the Affiliated Zhongshan
Hospital of Dalian University (Dalian, China) and 96 cases from the
Peoples Hospital of Weifang (Weifang, China). The pathological
diagnosis for the collected specimens was adenocarcinoma without
preoperative chemotherapy, radiotherapy or biological
immunotherapy. The specimens were analyzed for the detection of the
EML4-ALK fusion gene and TYMS mRNA. All protocols were approved by
the Human Clinical and Research Ethics Committees of the General
Military Hospital of Beijing PLA (Beijiang, China), the Affiliated
Zhongshan Hospital of Dalian University (Dalian, China) and the
Peoples Hospital of Weifang (Weifang, China). Written informed
consent was obtained from all patients.
Reagents and instruments
A DNA extraction kit (Qiagen, Hilden, Germany), RNA
extraction kit (Qiagen), EML4-ALK gene expression assay kit (Amoy
Diagnostics Co., Ltd., Xiamen, China) and TYMS Gene Expression
Analysis kit (Amoy Dx Ltd.) were used. In addition, a B-500
spectrophotometer was used to measure nucleic acid protein
concentrations (Shanghai Chong Meng Biotechnology Co. Ltd.,
Shanghai, China), and quantitative polymerase chain reaction (qPCR)
assays were conducted using an ABI 7500 Real-Time PCR system
(Applied Biosystems Life Technologies, Foster City, CA, USA).
Methods
qPCR detection of the EML4-ALK fusion gene
Between four and eight 4-µm paraffin tissue sections
were dewaxed. In accordance with the manufacturer's instructions
provided with the genomic RNA extraction kit, tissue RNA was
extracted and a spectrophotometer was used to detect the purity and
concentration of the extracted RNA. According to the method
provided with the EML4-ALK gene expression assay kit, the gene was
amplified using the ABI 7500 Real-Time PCR instrument. The kit
contained nine fusion mutant primers and probes to amplify the
EML4-ALK gene.
qPCR detection of TYMS mRNA expression in NSCLC
tissues
Tissue sections were dewaxed and tissue RNA was
extracted and spectrophotometrically analyzed as described in the
section above. According to the method provided with the TYMS Gene
Expression Analysis kit, the gene was amplified by qPCR. An
absolute quantitative method was used, with β-actin serving as a
reference gene in the detection of the expression level of TYMS
mRNA. The standard mean ratio of TYMS/β-actin was
4.21×10−3.
Statistical analysis
Data were analyzed using SPSS statistical analysis
software, version 19.0 (SPSS, Inc., Chicago, IL, USA). Results were
analyzed using χ2 and Fisher's exact tests, with a test
level α=0.05. The P-value was set to bilateral distribution, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Correlation between EML4-ALK fusion
gene and TYMS mRNA expression and patient clinical
characteristics
Table I shows the
clinical features of the 257 cases of NSCLC, of which there were 11
cases positive for the EML4-ALK fusion gene (4.28%). The positive
rate of the EML4-ALK fusion gene was higher in non-smoking patients
(11/147, 7.48%; P=0.018), but did not differ significantly
according to patient gender, age, tumor size, lymph node metastasis
or clinical stage. Of the 257 cases of NSCLC, high expression
levels of TYMS mRNA were found in 163 cases (63.42%), and low
expression levels were found in 94 cases (36.58%). The TYMS mRNA
expression level did not differ significantly according to patient
gender, age, smoking status, tumor size, lymph node metastasis or
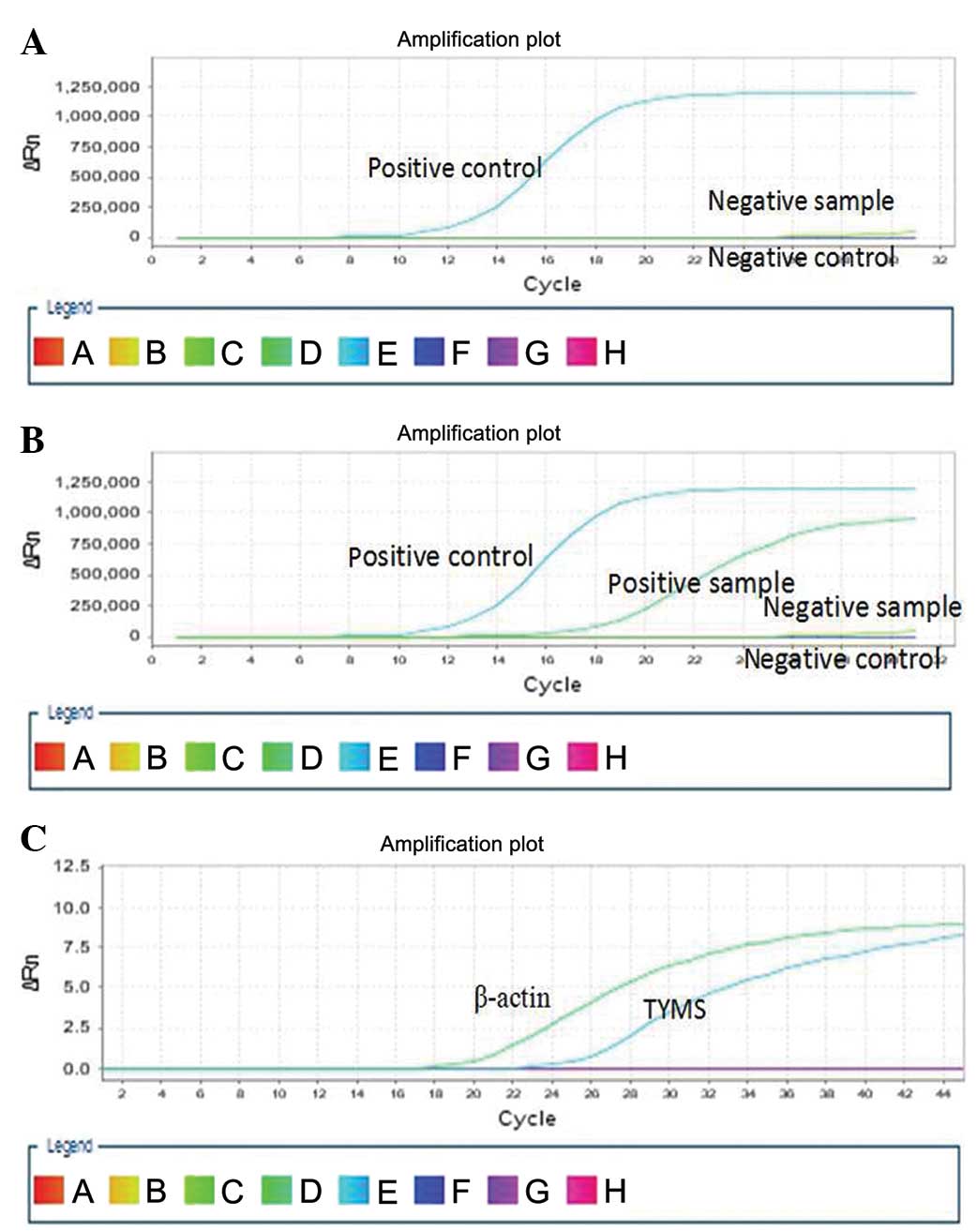
clinical stage, independent of other clinical features (Fig. 1A and B, Table I).
 | Table I.Association between EML4-ALK fusion
gene and TYMS mRNA expression and patient clinical
characteristics. |
Table I.
Association between EML4-ALK fusion
gene and TYMS mRNA expression and patient clinical
characteristics.
| EML4-ALK | | TYMS | |
|---|
|
|
|---|
| Clinical
features | Positive | Negative | P-value | High | Low | P-value |
|---|
| Gender |
|
| 0.137 |
|
| 0.196 |
| Male | 3 | 135 |
| 93 | 45 |
|
|
Female | 8 | 111 |
| 70 | 49 |
|
| Age (years) |
|
| 0.520 |
|
| 0.907 |
| ≥59 | 4 | 126 |
| 82 | 48 |
|
|
<59 | 7 | 120 |
| 81 | 46 |
|
| Smoking history |
|
| 0.018 |
|
| 0.166 |
| Yes | 0 | 99 |
| 68 | 31 |
|
| No | 11 | 147 |
| 95 | 63 |
|
| Tumor
diameter/cm |
|
| 0.637 |
|
| 0.798 |
| ≥5 | 4 | 119 |
| 79 | 44 |
|
|
<5 | 7 | 127 |
| 84 | 50 |
|
| Node metastasis |
|
| 0.751 |
|
| 0.627 |
| Yes | 5 | 100 |
| 70 | 35 |
|
| No | 6 | 146 |
| 93 | 53 |
|
| Clinical stage |
|
| 0.495 |
|
| 0.524 |
| I | 5 | 87 |
| 56 | 36 |
|
|
II+III+IV | 6 | 159 |
| 107 | 58 |
|
Association between the EML4-ALK
fusion gene and the expression level of TYMS mRNA
Table II shows that
11 patients with NSCLC were EML4-ALK fusion gene-positive, 8 of
whom had low levels of TYMS mRNA expression (72.73%). Of the 246
patients with NSCLC that were EML4-ALK fusion gene-negative, low
expression levels of TYMS mRNA were found in 86 cases (34.96%).
Patients with NSCLC that were EML4-ALK fusion gene-positive tended
to have lower expression levels of TYMS mRNA than those who were
negative (72.73 vs. 34.96%, P<0.05; Fig. 1C, Table
II).
 | Table II.Association between the EML4-ALK
fusion gene and the expression level of TYMS mRNA. |
Table II.
Association between the EML4-ALK
fusion gene and the expression level of TYMS mRNA.
| Expression of TYMS
mRNA | |
|---|
|
|---|
| EML4-ALK fusion
gene | High | Low | Total |
|---|
| Positive |
3 | 8 | 11 |
| Negative | 160 | 86 | 246 |
| Total | 163 | 94 | 257 |
Discussion
The EML4-ALK fusion gene is selectively inhibited by
crizotinib (also known as g azole imatinib or Sai Rui™). When
crizotinib was clinically tested in patients with NSCLC, phase I
and II clinical trials obtained good results and no adverse
reactions, and patients that tolerated the treatment entered phase
III clinical trials (17–19). In 2010, Choi et al (20) reported one case of an
EML4-ALK-positive patient with NSCLC who after 5 months of
treatment with crizotinib developed resistance to the drug; two
types of point mutation were identified: C1156Y and L1196M.
Therefore, further exploration of an individualized treatment plan
for certain patients with primary or secondary resistance to
crizotinib is of great practical value and practical
significance.
The results of the present study show that in the
patients with stage I-IV NSCLC, the positive rate of the EML4-ALK
fusion gene was 4.28% (11/257) and was higher in nonsmokers than in
smokers (P<0.05). This positive rate is slightly lower than that
in four other studies, in which the EML4-ALK fusion gene positive
rate was from 4.69%(3/64) to 11.65%(12/103) (7,21–23).
High expression levels of TYMS were exhibited in the NSCLC tissues
at a rate of 63.42% (163/257), and the results are similar to those
reported by Gandara et al (24). The TYMS expression levels were found
not to differ according to patient gender, age, smoking status,
tumor size, lymph node metastasis, pathological stage or other
clinical characteristics.
The present study observed that in NSCLC patients,
the EML4-ALK fusion gene has an association with the expression
level of TYMS (P<0.05), as a majority of the EML4-ALK fusion
gene-positive patients had low expression levels of TYMS. The
development of resistance to chemotherapy is one of the main causes
of tumor treatment failure. Following the short-term application of
pemetrexed chemotherapy, TYMS induction may occur, leading to TYMS
overexpression, increased catalytic activity and the resistance of
tumor cells. High expression levels of TYMS enhance the DNA
synthesis and repair capacity of proliferating cells, making the
lung cancer cells less susceptible to being killed by chemotherapy
drugs and leading to drug resistance; when there is a low
expression level of TYMS, the development of pemetrexed resistance
does not occur so readily, which increases the likelihood of
effective chemotherapy. This may be a theoretical basis for the
mechanism underlying the observation that EML4-ALK fusion
gene-positive NSCLC patients have a higher response rate to
chemotherapy.
In summary, this study initially identified that
patients with NSCLC who are EML4-ALK fusion gene-positive tend to
have low expression levels of TYMS, and it is speculated that
EML4-ALK fusion gene-positive patients will have greater
responsiveness to pemetrexed chemotherapy; however, the inherent
molecular mechanisms underlying this phenomenon require further
study. Our group mainly studies the association between the
chemotherapy drugs cisplatin and gemcitabine, drug resistance genes
and the EML4-ALK fusion gene. In the present study, we examined the
association between the pemetrexed resistance gene TYMS and the
EML4-ALK fusion gene, but did not consider the microtubule drug
resistance gene TUBB3 in first-line chemotherapy. Future studies
will examine the association between the EML4-ALK fusion gene and
TUBB3 to further explore more effective individualized treatment
plans. In particular, research will focus on individualized
treatment plans for patients with primary or secondary resistance
to the EML4-ALK fusion gene-selective inhibitor crizotinib.
Acknowledgements
This study was supported by a grant from the Wu
Jieping Medical Foundation Clinical Research Special Fund (no.
320.6750.1360).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sher T, Dy GK and Adjei AA: Small cell
lung cancer. Mayo Clin Proc. 83:355–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Horn L and Pao W: EML4-ALK: Honing in on a
new target in non-small-cell lung cancer. J Clin Oncol.
27:4232–4235. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Soda M, Choi YL, Enomoto M, Takada S,
Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K,
Hatanaka H, et al: Identification of the transforming EML4-ALK
fusion gene in non-small-cell lung cancer. Nature. 448:561–566.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inamura K, Takeuchi K, Togashi Y, Nomura
K, Ninomiya H, Okui M, Satoh Y, Okumura S, Nakagawa K, Soda M, et
al: EML4-ALK fusion is linked to histological characteristics in a
subset of lung cancers. J Thorac Oncol. 3:13–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perner S, Wagner PL, Demichelis F, Mehra
R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W,
Giordano TJ, et al: EML4-ALK fusion lung cancer: A rare acquired
event. Neoplasia. 10:298–302. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wong DW, Leung EL, So KK, Tam IY, Sihoe
AD, Cheng LC, Ho KK, Au JS, Chung LP and Pik Wong M: University of
Hong Kong Lung Cancer Study Group: The EML4-ALK fusion gene is
involved in various histologic types of lung cancers from
nonsmokers with wild-type EGFR and KRAS. Cancer. 115:1723–1733.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shaw AT, Yeap BY, Mino-Kenudson M,
Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S,
McDermott U, et al: Clinical features and outcome of patients with
non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol.
27:4247–4253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martelli MP, Sozzi G, Hernandez L,
Pettirossi V, Navarro A, Conte D, Gasparini P, Perrone F, Modena P,
Pastorino U, et al: EML4-ALK rearrangement in non-small cell lung
cancer and non-tumor lung tissues. Am J Pathol. 174:661–670. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sasaki T, Rodig SJ, Chirieac LR and Jänne
PA: The biology and treatment of EML4-ALK non-small cell lung
cancer. Eur J Cancer. 46:1773–1780. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diasio RB and Johnson MR: The role of
pharmacogenetics and pharmacogenomics in cancer chemotherapy with
5-fluorouracil. Pharmacology. 61:199–203. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fukushima M, Morita M, Ikeda K and
Nagayama S: Population study of expression of thymidylate synthase
and dihydropyrimidine dehydrogenase in patients with solid tumors.
Int J Mol Med. 12:839–844. 2003.PubMed/NCBI
|
|
13
|
Chu E, Copur SM, Ju J, Chen TM, Khleif S,
Voeller DM, Mizunuma N, Patel M, Maley GF, Maley F and Allegra CJ:
Thymidylate synthase protein and p53 mRNA form an in vivo
ribonucleoprotein complex. Mol Cell Biol. 19:1582–1594.
1999.PubMed/NCBI
|
|
14
|
Kawakami K, Graziano F, Watanabe G, Ruzzo
A, Santini D, Catalano V, Bisonni R, Arduini F, Bearzi I, Cascinu
S, et al: Prognostic role of thymidylate synthase polymorphisms in
gastric cancer patients treated with surgery and adjuvant
chemotherapy. Clin Cancer Res. 11:3778–3783. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Righi L, Papotti MG, Ceppi P, Billè A,
Bacillo E, Molinaro L, Ruffini E, Scagliotti GV and Selvaggi G:
Thymidylate synthase but not excision repair cross-complementation
group 1 tumor expression predicts outcome in patients with
malignant pleural mesothelioma treated with pemetrexed-based
chemotherapy. J Clin Oncol. 28:1534–1539. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gomez HL, Santillana SL, Vallejos CS,
Velarde R, Sanchez J, Wang X, Bauer NL, Hockett RD, Chen VJ,
Niyikiza C and Hanauske AR: A phase II trial of pemetrexed in
advanced breast cancer: Clinical response and association with
molecular target expression. Clin Cancer Res. 12:832–838. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwak EL, Bang YJ, Camidge DR, et al:
Anaplastic lymphoma kinase inhibition in non-small-cell lung
cancer. N Engl J Med. 363:1693–1703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim DW, Ahn MJ, Shi YK, et al: Updated
results of a global phase II study with crizotinib in advanced
ALK-positive non-small cell lung cancer (NSCLC). J Clin Oncol.
30:(Suppl): abstr. 75332012. View Article : Google Scholar
|
|
19
|
Shaw AT, Kim DW, Nakagawa K, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi YL, Soda M, Yamashita Y, Ueno T,
Takashima J, Nakajima T, Yatabe Y, Takeuchi K, Hamada T, Haruta H,
et al: ALK Lung Cancer Study Group: EML4-ALK mutations in lung
cancer that confer resistance to ALK inhibitors. N Engl J Med.
363:1734–1739. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rikova K, Guo A, Zeng Q, Possemato A, Yu
J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al: Global survey
of phosphotyrosine signaling identifies oncogenic kinases in lung
cancer. Cell. 131:1190–1203. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang X, Zhang S, Yang X, Yang J, Zhou Q,
Yin L, An S, Lin J, Chen S, Xie Z, et al: Fusion of EML4 and ALK is
associated with development of lung adenocarcinomas lacking EGFR
and KRAS mutations and is correlated with ALK expression. Mol
Cancer. 9:1882010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun Y, Ren Y, Fang Z, Li C, Fang R, Gao B,
Han X, Tian W, Pao W, Chen H and Ji H: Lung adenocarcinoma from
East Asian never-smokers is a disease largely defined by targetable
oncogenic mutant kinases. J Clin Oncol. 28:4616–4620. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gandara DR, Huang E and Desai S:
Thymidylate synthase (TS) gene expression in patients with ALK
positive(+) non-small cell lung cancer (NSCLC): Implications for
therapy. J Clin Oncol. 30:(Suppl). a75822012.
|