Introduction
microRNAs (miRs) are a class of small (19–24
nucleotides), non-coding RNAs that are involved in
post-transcriptional gene regulation and/or degradation (1,2). miRs
serve a crucial function in the proliferation, differentiation and
metabolism of a wide range of plant and animal cell types (3,4). In
order to prevent translation or contribute to target mRNA
degradation, miRs bind to target mRNAs at the 3′-untranslated
region (UTR) and/or the 5′-UTR (5).
Nasopharyngeal carcinoma (NPC) is a
non-lymphomatous, squamous cell malignancy arising from the
epithelial lining of the nasopharynx, and is particularly common in
Southeast Asia (6). NPC is an
Epstein-Barr virus-associated cancer (7). The incidence rate of NPC is
30–80/100,000 individuals per year, a rate that has remained
consistently high for decades (8).
The early symptoms of NPC are not often evident and the majority of
patients with NPC are not diagnosed until the advanced stages of
the disease. Radiotherapy and chemotherapy are the most common
treatment options for NPC, and although the radiotherapy technology
has improved considerably over the previous decade, the NPC
survival rate remains low (9). A key
reason for this is the resistance of NPC cells to radiation. Thus,
there is a crucial requirement for studies to elucidate the
mechanisms underlying NPC radiation resistance.
A previous study by Zhang et al indicated
associations between the tumor-related genes, c-Myc, SPLUNC1, Brd3
and UBAP1, with miR-141 in NPC cells (10). Furthermore, Xia et al
(11) and Wong et al
(12) indicated that the let-7
family of miRs was able to inhibit the proliferation of NPC cells.
In addition, Shi et al (13)
observed that miR-100 was able to upregulate PLK1 expression, which
resulted in the progression of NPC. To date, few studies have
investigated the association between radiation resistance and
miR-21 in NPC cells.
In the present study, continual radiation was
applied to a CNE-2 NPC cell line in order to acquire radioresistant
(CNE-2-1) NPC cells. A high-throughput miR sequencing assay was
subsequently used to analyze the regulation of miR between the two
cell lines. The aim of the present study was to investigate the
association between radioresistance and miR-21 in NPC cells.
Materials and methods
Cell culture
The CNE-2 NPC cell line (Shanghai Biological
Technology Co., Ltd., Shanghai, China) was cultured in RPMI 1640
medium (Invitrogen Life Technologies, Carlsbad, CA, USA),
supplemented with 10% fetal bovine serum (Invitrogen Life
Technologies), 100 IU/ml penicillin and 100 IU/ml streptomycin
(Invitrogen Life Technologies), at 37°C under 5% CO2 in
a humidified incubator.
Establishing a radioresistant NPC
CNE-2-1 cell line
CNE-2 cells were cultured in a T75 flask (Corning
Incorporated, Corning, New York, NY, USA) and subjected to 2 Gy
irradiation (IR) using a RS 2000 biological irradiator (Rad Source
Technologies, Inc., Suwanee, GA, USA). Following the first
radiation exposure, the cells were cultured and passaged twice. The
surviving cells were subsequently treated with the same assay as
previously described; however, the IR dose was increased to 4, 6, 8
and 10 Gy after each dose had been administered twice. Following
the complete radiation treatment, the surviving cells were cultured
and defined as the radioresistant NPC cell line, named CNE-2-1.
CNE-2 cells that received no radiation exposure were used as a
control cell line.
Cell viability assay
Cell viability was assessed using a Cell Counting
Kit (CCK)-8 assay (Beyotime Institue of Biotechnology, Haimen,
China), according to the manufacturer's instructions. Briefly,
cells that received various treatments (treated with LNA-antimiR-21
or LNA-control) were cultured in triplicate in a 96-well plate for
24 h, after which the cells were subjected to the assigned IR dose.
CCK-8 reagent was added to each well for 2 h prior to the
termination of the experiment. Absorbance values were measured
using a VersaMax Microplate Reader (Molecular Devices, LLC,
Sunnyvale, CA, USA) and expressed as the viability percentages of
the cells compared with the control cells. All experiments were
performed in triplicate and the data are presented as the mean ±
standard deviation.
Locked nucleic acid (LNA)-antimiR-21
transfection assay
CNE-2-1 cells were maintained in RPMI 1640 medium.
For transfection, the LNA-antimiR-21 or LNA-control
oligonucleotides (Exiqon A/S, Vedbaek, Denmark) were administered
at a final concentration of 50 nM using Lipofectamine 2000 reagent
(Invitrogen Life Technologies).
Cell cycle analysis
Briefly, the CNE-2-1 cells transfected with
LNA-antimiR-21 or LNA-control were exposed to 4 Gy IR, cultured for
3 days and then harvested on day 4. After rinsing twice with cold
phosphate-buffered saline, the cells were fixed with 70%
paraformaldehyde at 4°C. Subsequently, the cells were treated with
RNase A (Beyotime) for 30 min, followed by treatment with trypsin
(0.5% w/v; Beyotime) and ethylenediaminetetraacetic acid (0.2% w/v)
for 5 min. Finally, the cells were stained with 50 µg/ml propidium
iodide (Beyotime) and analyzed using a BD FACSCalibur flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA). A total of
30,000 events were analyzed for each sample. All tests were
performed in triplicate and the data are presented as the mean ±
standard deviation.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assay
Total RNA was extracted from the CNE-2-1 cells
transfected with LNA-antimiR-21 or LNA-control using TRIzol reagent
(Invitrogen Life Technologies), according to the manufacturer's
instructions. For analysis of miR-21 expression, the stem-loop RT
primer, qPCR primers and probe were designed as previously
described (14). Initially, the miR
was reverse transcribed into cDNA using Super-Script II reverse
transcriptase (Invitrogen). qPCR was performed using a standard
TaqMan PCR protocol and a LightCycler 480 II PCR system (Roche
Diagnostics, Basel, Switzerland), according to the manufacturer's
instructions. The relative expression levels were calculated using
the 2−ΔΔCt method and were normalized against the
expression levels of U6 RNA. All RT-qPCR assays were performed in
triplicate and the data are presented as the mean ± standard
deviation.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Statistical analysis was performed with the t-test using
SPSS statistical software, version 13.0 (SPSS, Inc., Chicago, IL,
USA) to evaluate the statistical significance of the differences
between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Establishment of the radioresistant
CNE-2-1 cell line
In order to acquire radioresistant NPC cells, CNE-2
cells were subjected to a series of increasing IR doses. After a
total IR dose of 60 Gy, the surviving cells were harvested,
cultured and designated as radioresistant CNE-2-1 cells. To analyze
the radioresistant capacity of the CNE-2-1 cells, CNE-2-1 and CNE-2
cells were exposed to varying doses of IR, and the cell viability
was assessed using a CCK-8 assay. After exposure to 6 Gy IR on day
0, the two cell lines were cultured for 5 days. Cell viability was
assessed every day, and the results indicated that the viability of
the CNE-2-1 cells was significantly enhanced compared with the
CNE-2 cells. The effect was notable at day 3 following radiation
exposure (Fig. 1). In addition, the
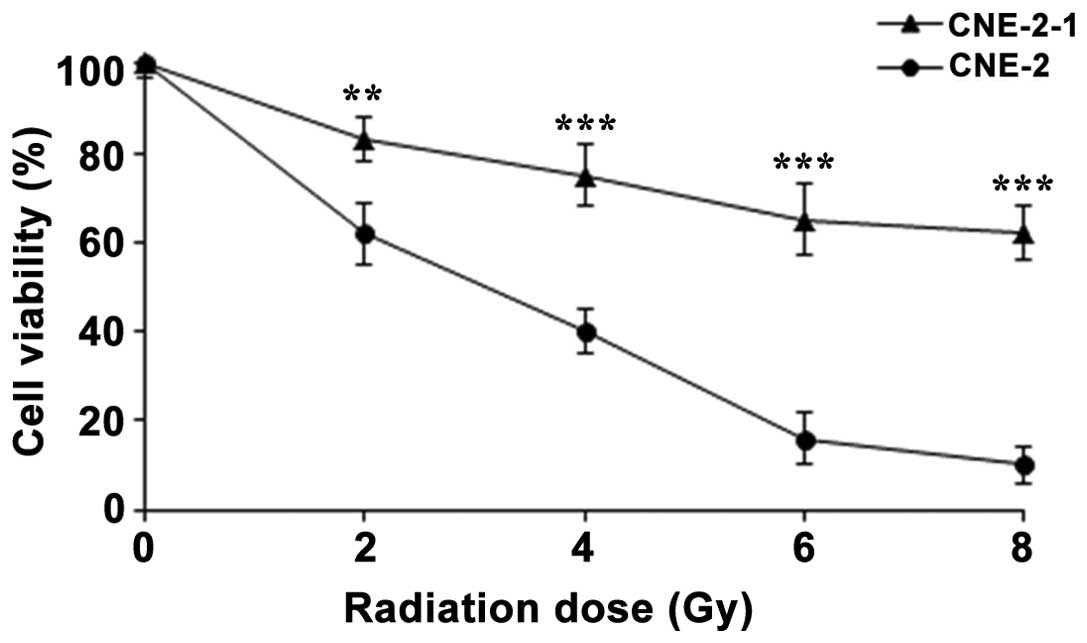
CNE-2-1 and CNE-2 cell lines were exposed to a range of IR doses
(0, 2, 4, 6 or 8 Gy) every day, and the cell viability was assessed
on day 4. The results indicated that the radiation exposure reduced
the viability of the CNE-2-1 and CNE-2 cells in a dose-dependent
manner. Compared with the CNE-2 cells, radiation exposure exhibited
less inhibition on the CNE-2-1 cells, and the difference in cell
viability at each IR concentration between the CNE-2-1 and CNE-2
cell lines was statistically significant (Fig. 2). Therefore, the CNE-2-1 cells
demonstrated a marked radioresistance compared with the CNE-2
cells, and the CNE-2-1 cell model of radioresistant NPC cells was
determined to have been successfully established, while untreated
CNE-2 cells were used as the control.
Regulation of miR in CNE-2-1 and CNE-2
cells
To investigate the difference in miR expression
between the CNE-2-1 and CNE-2 cell lines, cells from each cell line
were cultured in a 6-well plate, harvested and subjected to a
high-throughput miR sequencing assay. The results indicated that 16
miRs were upregulated, while 33 miRs were downregulated in the
CNE-2-1 cell line (data not shown). The altered regulation of
specific miRs has been reported to play a role in tumor
development, including tumor radiation resistance (15,16).
Therefore, it was hypothesized that the regulation of these miRs
may contribute to the radioresistance observed in NPC cells.
Subsequently, the miRs with altered regulation in the CNE-2-1 cells
were selected to determine any association with radioresistance in
NPC. miR-21 was among the three most upregulated miRs detected in
CNE-2-1 cells and to the best of our knowledge there were no
previous studies regarding this area. Therefore, miR-21 was
selected for further investigation.
Quantification of miR-21 expression
levels using RT-qPCR
To confirm the upregulation of miR-21 in CNE-2-1
cells, CNE-2-1 and CNE-2 cells were cultured in a 6-well plate,
harvested and subjected to RT-qPCR. The results indicated that
miR-21 expression was significantly upregulated (∼6 fold) in the
CNE-2-1 cells, as compared with the CNE-2 cells (Fig. 3).
In order to determine whether downregulation of
miR-21 was able to increase the radiosensitivity of the CNE-2-1
cells, an LNA-antimiR-21 transfection assay was performed. CNE-2-1
cells were transfected with LNA-antimiR-21 or LNA-control
oligonucleotides for 48 h, harvested and subjected to RT-qPCR. The
results indicated that the expression levels of miR-21 were
significantly reduced following transfection with the
LNA-antimiR-21 oligonucleotide (Fig.
4). Therefore, downregulation of miR-21 using LNA-antimiR-21
transfection was applied in the further experiments to assess the
function of miR-21 in NPC.
Downregulation of miR-21 increases the
radiosensitivity of CNE-2-1 cells
In order to investigate whether the upregulation of
miR-21 was associated with the radioresistance of CNE-2-1 cells,
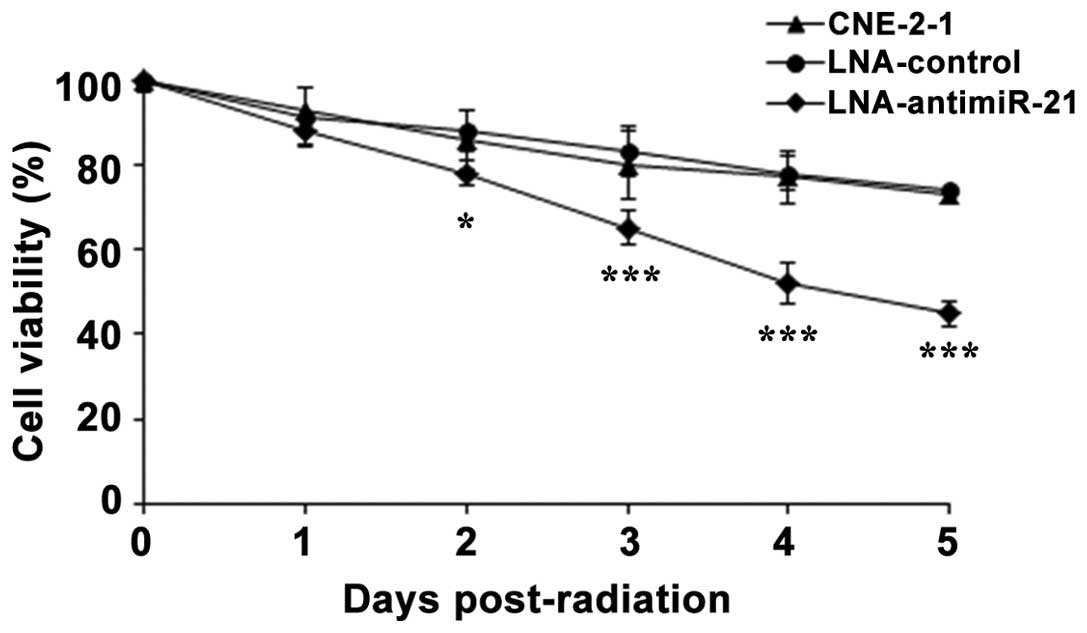
further cell viability assays were performed. CNE-2-1 control
cells, CNE-2-1 cells transfected with LNA-antimiR-21 and
LNA-control cells were exposed to 6 Gy IR on day 0 and cultured for
5 days. The cell viability was assessed every day using a CCK-8
assay. The results indicated that downregulation of miR-21
significantly inhibited the viability of the radiation-exposed
CNE-2-1 cells, as compared with the untreated CNE-2-1 cells, while
no difference was observed in the CNE-2-1 control cells compared
with the untreated CNE-2-1 cells (Fig.
5). Thus, the downregulation of miR-21 was demonstrated to
increase the radiosensitivity of CNE-2-1 cells.
Downregulation of miR-21 affects the
cell cycle of CNE-2-1 cells
Cell cycle assays were performed to determine
whether the inhibition of CNE-2-1 cell viability was associated
with cell cycle regulation. CNE-2-1 control cells and CNE-2-1 cells
transfected with LNA-antimiR-21 or LNA-control were exposed to 4 Gy
IR for 3 days, after which the cells were harvested and subjected
to fluorescence-activated cell sorting (FACS) for cell cycle
analysis. The results indicated that the percentage of cells at the
G1 phase in the LNA-antimiR-21 group was significantly increased,
while the percentage at the G2/M phase was significantly reduced
when compared with the CNE-2-1 control cells (Fig. 6). Thus, the results from the FACS
assay indicated that downregulation of miR-21 inhibited CNE-2-1
cell proliferation by disrupting the G1 phase of the cell
cycle.
Discussion
NPC is a common disease in Southeast Asia,
particularly amongst the Cantonese population of southern China,
including those in the Guangdong and Guangxi provinces. Since the
majority of patients are diagnosed at an advanced stage of the
disease, radiotherapy is the primary therapeutic strategy for
patients (17). Although
developments in radiotherapy technology have led to improved NPC
treatment, the efficacy of radiotherapy remains limited (18). Radioresistant cells are considered to
be the primary reason for this limited efficacy; however, the
mechanisms underlying NPC cell radioresistance are unclear.
miRs are a class of small, non-coding RNA molecules
that function to repress translation or degrade mRNA, subsequently
contributing to the inhibition of gene expression (19,20).
miRs serve crucial functions in a wide range of physiological and
pathological processes, including tumorigenesis (21). Increasing evidence implicates miRs
with various processes associated with cancer progression,
including tumor growth, differentiation, invasion, metastasis and
angiogenesis (22–24). A number of miRs are known to be
dysregulated in NPC cell lines (25). For example, miR-125a-5p has been
demonstrated to regulate and function as a prognostic factor for
gefitinib treatment in NPC (26).
Furthermore, upregulation of miR-324-3p has been shown to inhibit
radioresistance in NPC cells (27).
miR-21 was one of the first miRs to be identified in mammals; and
is highly conserved across mammal species. Previous studies have
demonstrated that miRs are expressed in numerous types of cancer,
including lung, prostate and liver (28). In addition, Deng et al
observed that miR-21 was highly expressed in NPC (29). The present study aimed to investigate
the association between miR-21 and NPC radioresistance.
A CNE-2 NPC cell line was continually exposed to
radiation in order to obtain a radioresistant NPC cell line.
Following the application of a total IR dose of 60 Gy, the
surviving CNE-2 cells were considered to have developed a marked
radiation resistance. The radioresistant CNE-2-1 cells were
subsequently used to investigate the association between miRs and
the radioresistance of NPC cells. A high-throughput miR sequencing
assay identified 16 upregulated and 33 downregulated miRs, and the
expression of miR-21 was observed to be upregulated 6-fold in the
CNE-2-1 cells using a RT-qPCR assay. CCK-8 cell viability assays
indicated that downregulation of miR-21 significantly enhanced the
radiosensitivity of the CNE-2-1 cells. Furthermore, the
downregulation of miR-21 was shown to inhibit CNE-2-1 cell
proliferation at the G1 phase. However, the molecular mechanism
underlying the effects of miR-21 on CNE-2-1 cells remains largely
unclear. Therefore, the results of the present study outline the
novel regulation of miR-21 in CNE-2 radioresistant cells.
In conclusion, the key finding of the present study
is the identification of a potential target of radioresistance in
NPC CNE-2 cells. Therefore, improved understanding of the
functional interaction between miR-21 and radioresistance in NPC
cells may lead to future therapeutic methods.
References
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He H, Jazdzewski K, Li W, et al: The role
of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad
Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Voorhoeve PM, le Sage C, Schrier M, et al:
A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in
testicular germ cell tumors. Cell. 124:1169–1181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu L, Fan J and Belasco JG: MicroRNAs
direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA.
103:4034–4039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei WI and Sham JS: Nasopharyngeal
carcinoma. Lancet. 365:2041–2054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin J-C: Adjuvant chemotherapy in advanced
nasopharyngeal carcinoma based on plasma EBV load. J Radiat Oncol.
1:117–127. 2012. View Article : Google Scholar
|
|
8
|
Li T, Chen JX, Fu XP, et al: microRNA
expression profiling of nasopharyngeal carcinoma. Oncol Rep.
25:1353–1363. 2011.PubMed/NCBI
|
|
9
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2009. CA Cancer J Clin. 59:225–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang L, Deng T, Li X, et al: microRNA-141
is involved in a nasopharyngeal carcinoma-related genes network.
Carcinogenesis. 31:559–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia H, Ng SS, Jiang S, et al:
miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially
inhibits nasopharyngeal carcinoma cell growth, migration and
invasion. Biochem Biophys Res Commun. 391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wong TS, Man OY, Tsang CM, et al: MicroRNA
let-7 suppresses nasopharyngeal carcinoma cells proliferation
through downregulating c-Myc expression. J Cancer Res Clin Oncol.
137:415–422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi W, Alajez NM, Bastianutto C, et al:
Significance of Plk1 regulation by miR-100 in human nasopharyngeal
cancer. Int J Cancer. 126:2036–2048. 2010.PubMed/NCBI
|
|
14
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA, Sevignani C, Dumitru CD, et al:
Human microRNA genes are frequently located at fragile sites and
genomic regions involved in cancers. Proc Natl Acad Sci USA.
101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sevignani C, Calin GA, Nnadi SC, et al:
MicroRNA genes are frequently located near mouse cancer
susceptibility loci. Proc Natl Acad Sci USA. 104:8017–8022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fåhraeus R, Fu HL, Ernberg I, et al:
Expression of Epstein-Barr virus-encoded proteins in nasopharyngeal
carcinoma. Int J Cancer. 42:329–338. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun K and Lai EC: Adult-specific functions
of animal microRNAs. Nat Rev Genet. 14:535–548. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Gene Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen HC, Chen GH, Chen YH, et al: MicroRNA
deregulation and pathway alterations in nasopharyngeal carcinoma.
Br J Cancer. 100:1002–1011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Y, Li Z, Wu L, et al: MiRNA-125a-5p: A
regulator and predictor of gefitinib's effect on nasopharyngeal
carcinoma. Cancer Cell Int. 14:242014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li G, Liu Y, Su Z, et al: MicroRNA-324-3p
regulates nasopharyngeal carcinoma radioresistance by directly
targeting WNT2B. Eur J Cancer. 49:2596–2607. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Deng M, Gu Y, Zheng G, et al: Expression
and clinical significance of miR-21 in nasopharyngeal carcinoma.
Shandong Med J. 52:10–12. 2012.
|