Introduction
Immunoglobulin A (IgA) nephropathy (IgAN), as one of
the most common primary glomerular diseases, accounts for 30%-40%
of primary glomerular diseases, the incidence of which is
increasing year by year (1–3). Relevant follow-up studies have shown
that for 25–30% patients, IgAN will develop into end-stage renal
disease after 20–25 years (4), which
is the primary cause of maintenance hemodialysis in China at
present (5). The clinical
manifestations, pathological patterns and prognoses of IgAN show
diversity, and the pathogenesis is not yet clear. Currently, it is
considered that the pathogenesis is associated with infection,
inflammation, immunological reactions and genetic factors (6,7).
According to previous studies, the prevalence rate of IgAN shows
certain geographical and ethnic differences, and genetic factors
play a very important role in the onset of IgAN (8). Therefore, it is particularly important
to search for genes associated with susceptibility to IgAN in order
to provide a genetic target for therapeutic intervention in IgAN.
There have been many Chinese studies on candidate genes associated
with the severity and complications of IgAN (9–11),
whereas there have been few studies looking into the candidate
genes associated with its pathogenesis. A large-sample study on
IgAN carried out by Li et al in China revealed that
variation of the ST6GALNAC2 gene is associated with genetic
susceptibility to IgAN (12). The
present study analyzed the correlation of polymorphism of a
specific α-2,6 sialyltransferase gene (ST6GALNAC2) and the
susceptibility of the Uyghur population to IgAN, so as to get a
better understanding of the pathogenesis and genetic background of
IgAN in the Uyghur region.
Subjects and methods
Subjects of study
A total of 180 cases of hospital patients and
outpatients of Uyghur ethnicity (86 males and 94 females, average
age, 38.81±11.06 years), diagnosed with IgAN by renal biopsy in the
Nephrology Department of the People's Hospital of Xinjiang Uyghur
Autonomous Region (Urumqi, China) were collected. Renal biopsy
pathological diagnostic criteria established by Zou in 2011
(13) were used as diagnostic
criteria for IgAN. Patients with secondary IgA deposition diseases,
such as systemic lupus erythematosus (SLE), allergic purpura,
chronic liver diseases, ankylosing spondylitic renal damage and
psoriatic renal damage were excluded. All the selected patients
were unrelated, with permanent Uyghur residency, of three different
generations and all lived in Xinjiang. The healthy controls were
180 healthy individuals (84 males and 96 females, average age,
37.53±11.68 years) who went to the aforementioned hospital for
medical examination from July 2008 to January 2013.
All subjects provided informed consent and
participated voluntarily. This study was approved by the Medical
Ethics Committee of The People's Hospital of Xinjiang Uygur
Autonomous Region.
Reagents
The reagents used included the whole blood genomic
DNA extraction kit (Shanghai Sangon Co., Ltd., Shanghai, China),
Taq polymerase, 10X buffer, dNTP (including MgCl2),
ddH2O (Beijing Dingguo Biotechnology Co., Ltd., Beijing,
China) and DNA marker (BBI, SeraCare Life Sciences, Inc., Milford,
MA, USA). The other reagents were conventional molecular biology
reagents.
Design and synthesis of primers
The primer sequences (shown in Table I) of ST6GALNAC2 gene rs3840858 and
rs2304921 were as previously described (12) and were verified using primer5
software (Premier Biosoft, Palo Alto, CA, USA). The primers were
synthesized by Shanghai Sangon Co., Ltd., following the
requirements of the project group.
 | Table I.Primers for rs3840858 and
rs2304921. |
Table I.
Primers for rs3840858 and
rs2304921.
| Locus | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| rs3840858 |
GCTGACAGCCTTAGCTCCCCCACGA |
CACTCCTGCCACTGCGCTCTCTCCA |
| rs2304921 |
AAAGCTTCCAAGGGGTAGGT |
TCATCCTTCTCTGCTGTTGG |
Research methods
Collection of blood samples
A 5 ml sample of venous blood was collected from
each patient on an empty stomach in the morning. EDTA was used for
anticoagulation. The samples were numbered and registered. Whole
blood samples were placed at −80°C for cryopreservation.
DNA extraction
DNA specimens were extracted with the Ezup pillar
blood genomic DNA extraction kit according to the kit instructions
and preserved at −20°C.
Detection of gene polymorphism
i) PCR reaction conditions for SNP rs2304921. The
total volume of the amplification stage of the PCR was 35 µl
(containing 3 µl DNA, 20 µl ddH2O, 5 µl buffer, 2 µl
dNTP, and 1.2 and 2 µl Taq polymerase in the upstream and
downstream directions, respectively). The amplification reaction
conditions of PCR were: denaturation at 95°C for 5 min; main
cycling at 95°C for 45 sec, 61.7°C for 60 sec and 72°C for 45 sec,
38 cycles in total; followed by 72°C for 10 min and preservation at
4°C. The PCR reaction was performed on the GeneAmp® PCR System 9700
Thermal cycler from Applied Biosystems®, Invitrogen Life
Technologies (Foster City, CA, USA).
ii) PCR reaction conditions for SNP rs3840858. The
total volume of the amplification reaction of PCR was 35 µl
(containing 3 µl DNA, 20.6 µl ddH2O, 5 µl buffer, 2 µl
dNTP, 1.5 and 2 µl Taq polymerase in the upstream and downstream
directions, respectively). The amplification reaction conditions of
PCR were: denaturation at 95°C for 5 min; main cycling conditions
at 95°C for 45 sec, 61.0°C for 60 sec and 72°C for 45 sec, 37
cycles in total; followed by 72°C for 10 min and preservation at
4°C.
iii) Genotyping. Every PCR amplification product was
electrophoresed at a voltage of 120 V for 20 min using 1.5% agarose
gel to which ethidium bromide nucleic acid dye had been added in
advance. Imaging was conducted using a UV transmission automatic
image analyzer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All
PCR products for each SNP locus were genotyped by direct sequencing
(conducted by Beijing Dingguo Biotechnology Co., Ltd.).
Statistical analysis
Whether the genotypes of the population were in
Hardy-Weinberg equilibrium was estimated using the χ2
test. Other statistical analyses were performed with SPSS software,
version 17.0 (SPSS, Inc., Chicago, IL, USA). The allele and
genotype frequencies were calculated by χ2 test. A
t-test was applied for comparison of measurement data between
groups. Logistic regression analysis was applied to analyze the
correlation between polymorphism and IgAN. All statistics are
two-sided with a test level a=0.05. P<0.05 was considered to
indicate that a difference was statistically significant.
Results
General patient characteristics
There was no statistically significant difference in
terms of gender and age between the IgAN group and the control
group (P>0.05; Table II).
 | Table II.Gender and age distribution in the
IgAN and control groups. |
Table II.
Gender and age distribution in the
IgAN and control groups.
| Item | IgAN group | Control group | t- or χ2
value | P-value |
|---|
| Age (years) | 38.81±11.06 | 37.53±11.68 | 0.754 | 0.452 |
| Gender (M/F) | 86/94 | 84/96 | 0.045 | 0.833 |
Distributions of genotype and allele
frequencies of rs3840858 in the ST6GALNAC2 gene
Two genotypes, a DD genotype and a DI genotype, were
detected by direct forward sequencing (Fig. 1), and the II genotype was absent. The
distributions of the DD and DI genotypes in the IgAN and control
groups were in accordance with Hardy-Weinberg equilibrium
(P>0.05). As shown in Table
III, the DI genotype ratio (17.8%) in the IgAN group was higher
than that in the control group (5.6%), while the DD genotype ratio
(82.2%) in the IgAN group was lower than that in the control group
(94.4%). A statistically significant difference was observed in the
distributions of each genotype between the IgAN and control groups
(χ2=13.046, P=0.001).
 | Table III.Distribution of genotype and allele
frequencies of rs3840858 polymorphism of the ST6GALNAC2 gene in the
IgAN and control groups. |
Table III.
Distribution of genotype and allele
frequencies of rs3840858 polymorphism of the ST6GALNAC2 gene in the
IgAN and control groups.
| | Genotype frequency, n
(%) | Allele frequency, n
(%) |
|---|
|
|
|---|
| Group | No. of cases | DD | DI | D | I |
|---|
| IgAN | 180 | 148 (82.2) | 32 (17.8) | 328 (91.1) | 32 (8.9) |
| Control | 180 | 170 (94.4) | 10 (5.6) | 350 (97.2) | 10 (2.8) |
|
χ2-value |
| 13.046 |
| 12.238 |
|
| P-value |
| 0.001 |
| 0.001 |
|
In the IgAN group, the I allele frequency (8.9%) was
higher than that in the control group (2.8%), while the D allele
frequency (91.1%) was lower than that in control group (97.2%).
There was a statistically significant difference in the I and D
allele distributions between the IgAN group and the control group
(χ2=12.238, P=0.001).
Univariate logistic regression analysis indicated
that rs3840858 polymorphism may be a risk factor of IgAN (P=0.001).
The risk of developing IgAN in individuals who carried the DI
genotype was 3-fold higher than that in those who carried the DD
genotype [odds ratio (OR)=3.676, 95% confidence interval
(CI)=1.284–10.519], and the risk of developing IgAN in individuals
who carried the I allele was higher than that in those who carried
D allele (OR=3.415, 95% CI=1.223–9.531; Table IV).
 | Table IV.Correlation between polymorphism of
the ST6GALNAC2 gene and the susceptibility to IgAN. |
Table IV.
Correlation between polymorphism of
the ST6GALNAC2 gene and the susceptibility to IgAN.
| Genotype or
allele | Control group, n
(%) | IgAN group, n
(%) | P-value | OR | 95% CI |
|---|
| rs3840858 |
|
|
|
|
|
| Genotype |
|
|
|
|
|
| DD | 170 (94.4) | 148 (82.2) |
| 1 |
|
| DI | 10 (5.6) | 32 (17.8) | 0.001 | 3.676 | 1.284–10.519 |
| Allele |
|
|
|
|
|
| D | 350 (97.2) | 328 (91.1) |
| 1 |
|
| I | 10 (2.8) | 32 (8.9) | 0.001 | 3.415 | 1.223–9.531 |
| rs23840858 |
|
|
|
|
|
| Genotype |
|
|
|
|
|
| AA | 6 (3.3) | 4 (2.2) |
| 1 |
|
| GG | 54 (30.0) | 72 (40.0) | 0.778 | 1.3 | 0.209–8.082 |
| AG | 120 (66.7) | 104 (57.8) | 0.176 | 0.65 | 0.349–1.211 |
| Allele |
|
|
|
|
|
| G | 294 (81.7) | 280 (77.8) |
| 1 |
|
| A | 66 (19.3) | 80 (22.2) | 0.228 | 1.273 | 0.760–2.132 |
Distributions of genotype and allele
frequencies of rs2304921 in the ST6GALNAC2 gene
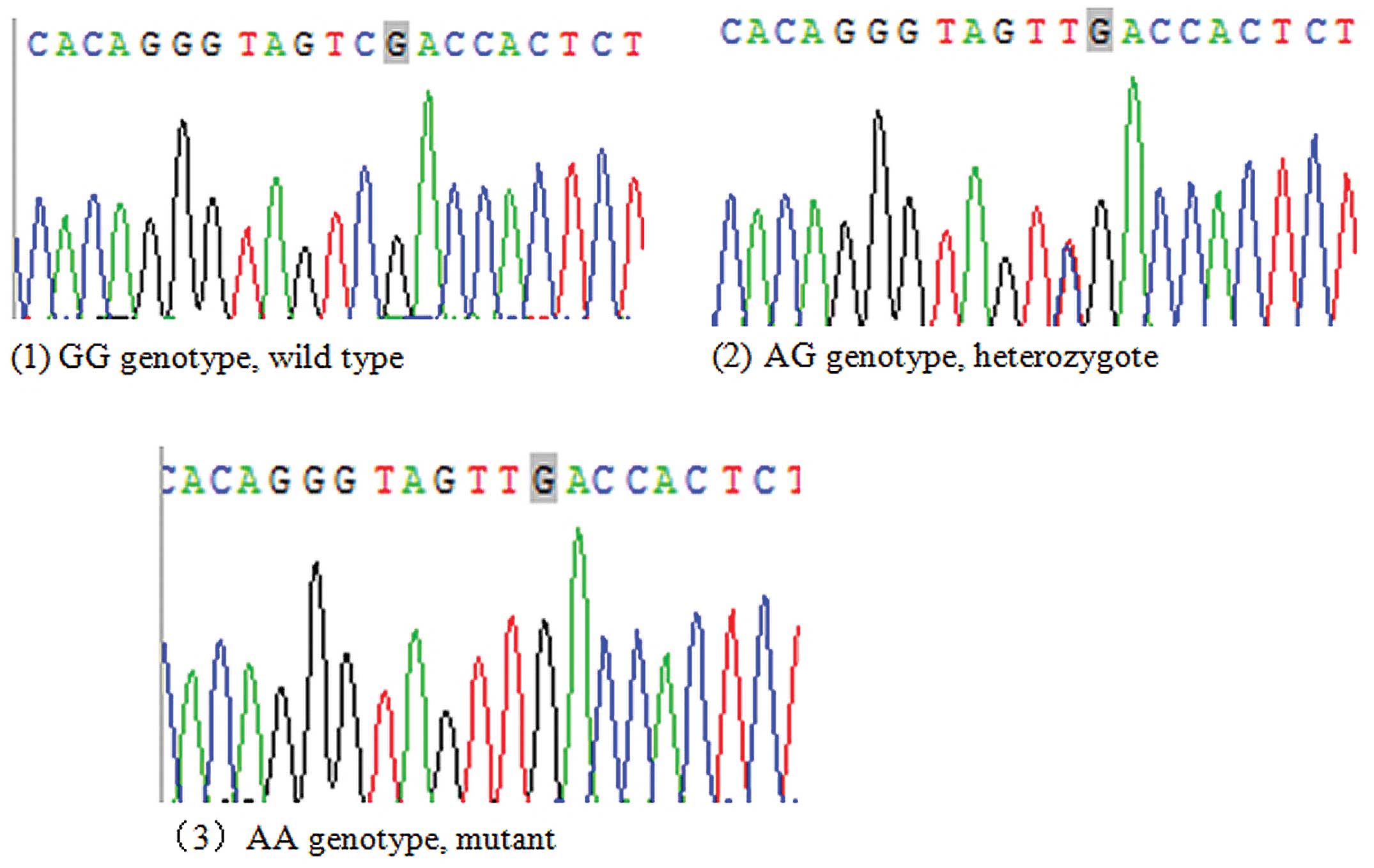
Three genotypes, GG, AG and AA genotypes, were
detected by direct reverse sequencing (Fig. 2). The distributions of the GG, AG and
AA genotypes in the IgAN and control groups were in accordance with
Hardy-Weinberg equilibrium (P>0.05). As demonstrated in Table IV, the AG genotype rate (40.0%) in
the IgAN group was higher than that in control group (30.0%), while
the AA and GG genotype rates (2.2% and 57.8%, respectively) in the
IgAN group were lower than those in the control group (3.3% and
66.7%, respectively). The differences in the distributions of each
genotype of the ST6GALNAC2 gene rs2304921 polymorphism between the
IgAN and control groups were not statistically significant
(χ2=4.114, P=0.128).
The A allele frequency (22.2%) in the IgAN group was
higher than that in the control group (19.3%), while the G allele
frequency (77.8%) in the IgAN group was lower than that in the
control group (81.7%). The difference in A and G allele
distributions between the IgAN group and the control group was not
statistically significant (χ2=1.684, P=0.228).
Univariate logistic regression analysis demonstrated
that the rs2304921 polymorphism is not likely to impact the risk of
developing IgAN (P>0.05; Table
V).
 | Table V.Distribution of genotype and allele
frequencies of rs2304921 polymorphism of the ST6GALNAC2 gene in the
IgAN and control groups. |
Table V.
Distribution of genotype and allele
frequencies of rs2304921 polymorphism of the ST6GALNAC2 gene in the
IgAN and control groups.
| | Genotype frequency, n
(%) | Allele frequency, n
(%) |
|---|
|
|
|---|
| Group | No. of cases | GG | AG | AA | G | A |
|---|
| IgAN | 180 | 104 (57.8) | 72 (40.0) | 4 (2.2) | 280 (77.8) | 80 (22.2) |
| Control | 180 | 120 (66.7) | 54 (30.0) | 6 (3.3) | 294 (81.7) | 66 (19.3) |
|
χ2-value |
|
| 4.114 |
| 1.684 |
|
| P-value |
|
| 0.128 |
| 0.228 |
|
Discussion
The serum IgA levels of the majority of patients
with IgAN increase (14). However,
due to lack of specificity, this cannot be regarded as a criterion
for assisting clinical diagnosis. In recent years, with several
kinds of technologies such as ELISA and mass spectrometry, scholars
from research institutions in Japan, the United States of America
and China all observed that aberrant glycosylation of IgA1
molecules in IgAN patients may be the most important mechanism of
pathogenesis (15–17). It has been widely accepted that
glycosylation defects lead to the formation of immune complexes in
the pathogenesis of IgAN. The study conducted by Gharavi et
al (18) suggests that
glycosylation defects of IgA1 are a genetic characteristic and a
genetic risk factor of IgAN. Thus, aberrant glycosylation provided
a new candidate gene for the study of the genetic susceptibility of
IgAN.
The ST6GLNAC2 gene encodes a specific α-2,6
sialyltransferase enzyme that participates in galactosylation in
the IgA formation process. The study conducted by Patsos et
al (19) indicates that
defective expression of the ST6GLNAC2 gene decreases the activity
of α-2,6 sialyltransferase. According to the literature, α-2,6
sialyltransferase activity on serum IgA1 in patients with focal
proliferative type and sclerotic type IgAN (Haas stage III-IV) is
reduced, which delays the sialylation of GalNAc in the molecular
hinge area of serum IgA1 and affects the galactosylation of
O-linked glycosyl in this area, eventually causing changes to the
molecular charge and spatial structure of IgA1 (20). An increase in the activity of α-2,6
sialyltransferase may cause premature sialylation of GalNAc in the
molecular hinge area of serum IgA1 and then affect the
galactosylation of O-linked glycosyl in this area (21).
Li et al (12)
conducted a large-sample study with sporadic Han IgAN patients as
subjects. The results demonstrated that the frequency of the ADG
haplotype in the promoter region of the ST6GALNAC2 gene was
increased in patients with IgAN, and the ADG haplotype was
associated with a deficiency of α-2,6 sialylation of IgA1. The
authors also analyzed the correlation between variants of C1GALT1
and ST6GALNAC2 genes and the predisposition and severity of IgAN.
The results revealed that the IgA1 O-glycosylation-related genes,
C1GALT1 and ST6GALNAC2, were associated with the disease
predisposition and severity of IgAN.
The analysis of SNP rs3840858 in the ST6GALNAC2 gene
in this study revealed the existence of two genotypes (DD and DI)
as detected by sequencing; the DI genotype rate in the IgAN group
was higher than that in the control group, and the I allele
frequency in the IgAN group was higher than that in the control
group. The differences between the two groups were significant.
Moreover, univariate logistic regression analysis showed that the
risk of developing IgAN in individuals who carried the DI genotype
was 3-fold higher than that in those who carried DD genotype. This
suggests that the rs3840858 polymorphism is associated with the
susceptibility to IgAN. In the study by Li et al (12), three genotypes (DD, DI and II) of SNP
rs3840858 were detected by a restriction enzyme digestion technique
and the differences in the genotype and allele frequencies between
IgAN and control groups was not found to be statistically
significant. The lack of conformity of the results may be due to
geographic and ethnic differences in the gene polymorphism.
The analysis of SNP rs2304921 in the ST6GALNAC2 gene
in this study revealed the existence of three genotypes (GG, AG and
AA) as detected by sequencing. The differences in genotype and
allele frequencies between the IgAN group and control group were
not statistically significant, which suggests that the rs2304921
polymorphism is irrelevant to the susceptibility to IgAN. This
result was in accordance with the study by Li et al
(12).
To conclude, this study indicated that the rs3840858
polymorphism of the ST6GALNAC2 gene might be associated with the
susceptibility to IgAN in an Uyghur population, whereas rs2304921
polymorphism appears to be irrelevant to IgAN in Uyghur. Due to the
limited sample size, the correlation of the ST6GALNAC2 gene with
susceptibility to IgAN requires investigation in a multi-zone and
multi-site study with a larger sample size so as to provide new
evidence for risk prediction as well as prevention and
treatment.
Acknowledgements
This study was supported by the Natural Science
Foundation of Xinjiang Uyghur Autonomous Region (Grant number:
2012211A088) and Science and Technology Support Xinjiang Autonomous
Region Project (Grant number: 2013911114).
References
|
1
|
Zhang YQ: Pathological and clinical
analysis of IgA nephropathy. Zhong Guo Xian Dai Yi Yao Za Zhi.
17:722010.[(In Chinese)].
|
|
2
|
Yang DS, Luo CG, Jiang WM, et al:
Observation of efficacy of combination therapy with Leflunomide and
hormone on IgA nephropathy. Zhong Guo Xian Dai Yi Yao Za Zhi.
17:74–75. 2010.[(In Chinese)].
|
|
3
|
Li LS and Liu ZH: Epidemiologic data of
renal diseases from a single unit in China: analysis based on
13,519 renal biopsies. Kidney Int. 66:920–923. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Barratt J and Feehally J: IgA nephropathy.
J Am Soc Nephrol. 16:2088–2097. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen XM and Xie YS: Attach importance to
basic and clinical research on the postponement of progression of
IgA nephropathy. Chin J Nephrol. 20:235–237. 2004.[(In
Chinese)].
|
|
6
|
Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang
JQ, Sun LD, Sim KS, Li Y, et al: A genome-wide association study in
Han Chinese identifies multiple susceptibility loci for IgA
nephropathy. Nat Genet. 44:178–182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu HH, Chu KH, Yang YH, et al: Genetics
and immunopathogenesis of IgA nephropathy. Clin Rev Allergy
Immunol. 41:198–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kiryluk K, Julian BA, Wyatt RT, et al:
Genetic studies of IgA nephropathy: past, present, and future.
Pediatr Nephrol. 25:2257–2268. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Sui W, Xue W, Wu J, Chen J and Dai
Y: Univariate and multiple linear regression analyses for 23 single
nucleotide polymorphisms in 14 genes predisposing to chronic
glomerular diseases and IgA nephropathy in Han Chinese. Saudi J
Kidney Dis Transpl. 25:992–997. 2014.PubMed/NCBI
|
|
10
|
Xu R, Feng S, Li Z, Fu Y, et al:
Polymorphism of DEFA hinese Han population with IgA nephropathy.
Hum Genet. 133:1299–1309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mao S, Ren X, Huang S and Zhang A:
Association of megsin 2093C/T, 2180C/T and C25663G gene
polymorphism with the risk of IgA nephropathy. Ren Fail.
36:817–822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li GS, Zhu L, Zhang H, et al: Variants of
the ST6GALNAC2 promoter influence transcriptional activity and
contribute to genetic susceptibility to IgA nephropathy. Hum Mutat.
28:950–957. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zou WZ: Guidance on renal biopsy
pathologic diagnostic criteria. Chin J Nephrol. 17:270–275.
2001.[(In Chinese)].
|
|
14
|
Zhu B, Zhu CF, Lin Y, Perkovic V, et al:
Clinical characteristics of IgA nephropathy associated with low
complement 4 levels. Ren Fail. 1–9. Dec 24–2014.(Epub ahead of
print).
|
|
15
|
Shimozato S, Hiki Y, Odani H, Takahashi K,
Yamamoto K and Sugiyama S: Serum under-galactosylated IgA1 is
increased in Japanese patients with IgA nephropathy. Nephrol Dial
Transplant. 23:1931–1939. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gharavi AG, Moldoveanu Z, Wyatt RJ, Barker
CV, Woodford SY, et al: Aberrant IgA1 glycosylation is inherited in
familial and sporadic IgA nephropathy. J Am Soc Nephrol.
19:1008–1014. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin XJ, Ding JX, Zhu L, et al: Aberrant
galactosylation of IgA1 is involved in the genetic susceptibility
of Chinese patients with IgA nephropathy. Nephrol Dial Transplant.
24:3372–3375. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gharavi AG, Moldoveanu Z, Wyatt RJ, et al:
Aberrant IgAl glycosylation is inherited in familial and sporadic
IgA nephropathy. J Am Soc Nephrol. 19:1008–1014. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Patsos G, Hebbe-Viton V, Robbe-Masselot C,
et al: O-glycan inhibitors generate aryl-glycans, induce apoptosis
and lead to growth inhibition in colorectal cancer cell lines.
Glycobiology. 19:382–398. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding JX, Xu LX, Lv JC, et al: Aberrant
sialylation of serum IgA1 was associated with prognosis of patients
with IgA nephropathy. Clin Immunol. 125:268–274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu L, Ding JX, Lv JC, et al: Expression
of ST6GALNAC2 gene in B lymphocytes is influenced in Epstein-Barr
virus transformation. Chinese Journal of Nephrology. 24:130–133.
2008.[(In Chinese)].
|