Introduction
Capparis spinosa L. is a common Uyghur folk
medicine and is a member of the Capparidaceae family, which
is used in phytomedicine worldwide for its reported anti-oxidative
(1–4), anti-fungal (5), anti-bacterial (6), anti-hepatotoxic (7), anti-inflammatory (8,9),
anti-diabetic (10–12), anti-proliferative (13) and anti-tumor (14) effects.
Gastric cancer is the fifth most prevalent type of
cancer and the third leading cause of cancer-associated mortality
worldwide (15). In 2012, ~950,000
cases of gastic cancer were diagnosed, and ~723,000 cases of
gastric cancer-related mortality were recorded. Typical treatments
for gastric cancer include surgical tumor excision, chemotherapy,
radiotherapy and targeted therapy (16). Various combinations of drugs may be
used in the treatment of gastric cancer, including flurouracil,
capecitabine, carmustine, semustine and doxorubicin, in addition to
mitomycin C, cisplatin and docetaxel. However, gastric cancer
frequently exhibits low sensitivity to these treatments, which are
typically used to palliatively reduce the tumor size, relieve
symptoms and increase survival time. Therefore, there is a
requirement for novel treatment options for the treatment and
prevention of gastric cancer.
A preliminary study indicated that in vitro
anti-tumor activity exerted by the n-butanol extract of C.
spinosa L. (CSBE) (17);
however, the underlying molecular mechanism of CSBE-induced tumor
cell apoptosis is currently poorly understood. The present study
aimed to elucidate the mechanisms underlying CSBE-induced apoptosis
of the SGC-7901 human gastric cancer cell line by investigating
alterations to the expression levels and localization of
initiators, effectors, and markers of the mitochondrial apoptosis
pathway, following CSBE exposure.
Materials and methods
Materials
CSBE was provided by the Institute of Materia Medica
at the Harbin University of Commerce (Harbin, China). The MTT cell
proliferation assay, propidium iodide (PI), and Rhodamine 123 dye
were purchased from Sigma-Aldrich (St. Louis, MO, USA). RPMI-1640
medium was purchased from Gibco Life Technologies (Carlsbad, CA,
USA) and fetal calf serum (FCS) was purchased from GE Healthcare
Life Sciences (Logan, UT, USA). Caspase-9 and caspase-3 test kits
were purchased from Biyuntian Co. (Shanghai, China). TRIzol®
reagent was purchased from Invitrogen Life Technologies (Carlsbad,
CA, USA), and monoclonal antibodies against β-actin (sc-47778),
cytochrome c, B-cell lymphoma-2 (BCL-2; sc-7382) and
BCL-2-associated protein X (BAX; sc-7480) were purchased from Santa
Cruz Biotechnology, Inc. (Dallas, TX, USA). Horse anti-mouse
alkaline phosphatase (AP)-conjugated (ZB-2310) and goat anti-mouse
fluorescein-conjugated antibodies (ZF-0312) were purchased (ZSBio;
OriGene Technologies, Inc., Beijing, China).
Cell culture
The SGC-7901 human gastric carcinoma cell line
(Institute of Materia Medica, Chinese Academy of Medical Sciences,
Peking, China) was cultured in RPMI-1640 medium supplemented with
10% FCS at 37°C, in a 5% CO2 incubator.
MTT proliferation assay
The MTT assay is a colorimetric assay for assessing
cell viability (18). Logarithmic
phase cells (2×105/ml) were grown in 96-well plates.
Following 24 h of growth, various concentrations of 100 µl CSBE,
fresh medium and hydroxycamptothecin (HCPT; 20090430; Harbin
Pharmaceutical Group Co., Ltd.), were added to each well. HCPT has
apparent anti-tumor activities and is an alkaloid, while CSBE also
has an alkaloid component. So HCPT was used for positive control.
Six parallel wells were set up for each group: CSBE (1, 5, 25, 50,
75 and 100 µg/ml); HCPT (0.01, 0.1, 1 and 10 µg/ml); and control
(RPMI-1640). After 72 h, cell viability was assayed via the
addition of 0.5 mg/ml MTT dye. After 4 h incubation at 37°C, the
medium was removed, formazan crystals were dissolved in 200 µl
dimethyl sulfoxide/well, and the 96-well plates were read in a
microplate reader (Wellscan MK3; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) at 570 nm. The rate of inhibition of CSBE on
SGC-7901 cell proliferation was calculated as follows: Inhibition
rate = (optical density (OD) of control group - OD of CSBE group) ×
(100%/OD of control group).
Apoptotic morphology assay
SGC-7901 cells were inoculated into 6-well plates
(3×108 cells/ml), in which each well contained a
coverslip. At 24 h, CSBE (15, 30 or 60 µg/ml) was added to each
well. The final concentration of HCPT (positive control group) was
0.2 µg/ml, whereas the negative control group received the same
volume of culture medium. After 40 h the supernatant was discarded,
and the SGC-7901 cells were washed in phosphate-buffered saline
(PBS), and fixed in buffered solution (volume ratio of methyl
alcohol:glacial acetic acid was 3:1) at 4°C for 10 min.
Subsequently, 5 mg/l of Hoechst 33258 (861405; Sigma-Aldrich), the
fluorescent probe, was added into each well. Following 30 min in
the dark at 37°C, the cells were washed in PBS and the coverslip in
each well was removed under a CKX-41 fluorescence microscope, and
the stained cells were visualized using a digital camera (both from
Olympus Corporation, Tokyo, Japan).
Apoptotic rate assay
PI single-staining of SGC-7901 cells was performed
according to a method outlined in previous studies (19,20).
Briefly, SGC-7901 cells were plated into a 6-well plate
(3×105 cells/ml) and CSBE (15, 30 or 60 µg/ml) was added
at 24 h. After 48 h, the cells were collected and fixed in 70%
ethanol and stored at 4°C overnight. Following removal of the
ethanol, the SGC-7901 cells were suspended in PI dye (50 µg/ml) at
37°C for 30 min. Subsequently, the cells were analyzed using flow
cytometry (EPICS XL-MCL; Beckman Coulter, Inc., Brea, CA, USA), in
which the excitation wavelength was 488 nm and the emission
wavelength was 630 nm.
Measurement of mitochondrial membrane
potential
The SGC-7901 cells were inoculated into a 6-well
plate (2×105 cells/ml), after which CSBE (15, 30 or 60
µg/ml) was added after 24 h. The cells were collected at 48 h and
suspended in Rhodamine 123 dye (10 µg/ml) for 30 min. Subsequently,
the cells were centrifuged (340 × g, 10 min) and washed with PBS.
Fluorescence intensity was measured via flow cytometry, in which
the excitation wavelength was 488 nm and the emission wavelength
was 525 nm.
Measurement of caspase-9 and caspase-3
activity
The SGC-7901 cells were inoculated into a 6-well
plate (1×106 cells/ml, cells inoculated into a 100-ml
culture bottle) and CSBE (15, 30 or 60 µg/ml) was added at 24 h.
Subsequently, the cells were incubated (100 µl/2×106
cells) with Ac-DEVD-pNA lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China) in an ice bath for 15 min,
following suspension of the precipitation. Following centrifugation
at 4°C for 10 min at 21,000 × g, the culture supernatant was
transferred to a cooling centrifuge tube, and 90 µl detecting
buffer, and 10 µl Ac-DEVD-pNA colorimetric substrate (2 mM), were
added to the control group. Subsequently, 60 µl detecting buffer,
10 µl Ac-DEVD-pNA (2 mM) and 10 µl sample were added to the sample
group. All of the samples were incubated at 37°C for 60 min. The
Ac-DEVD-pNA (10 mM) was diluted to 0, 10, 20, 50, 100 and 200
µmol/l, which were used to generate a standard curve. Absorbance at
405 nm was determined using an enzyme-labeled instrument (Wellscan
MK3), when the color was significantly altered.
Western blot analysis
The SGC-7901 cells were inoculated into a 6-well
plate (1×106 cells/ml, cells were inoculated into a
100-ml culture bottle), and CSBE (15, 30 or 60 µg/ml) was added
following 24 h. Total protein extraction occurred at 48 h, and
protein content was measured using Bradford Protein Assays
(20100216; Beyotime). After 24 h of administration, cells were
washed twice with PBS, then scraped. The lysate was added, and the
samples were chilled on ice for 30 min. The cells were
centrifugated at 4°C for 10 min (21,000 × g), while the supernatant
fluid was separated from the total protein. Equivalent amounts of
protein (3 µg/ml) were separated using 15% SDS-PAGE. The proteins
were transferred to a nitrocellulose membrane (10401196; Whatman;
GE Healthcare Life Sciences, Shanghai, China), which was blocked
using 5% skimmed milk, washed using Tris-buffered saline containing
Tween 20 (TBST) and stored at 4°C for 2 h. Subsequently, the
nitrocellulose membrane was incubated with anti-BCL-2, anti-BAX,
anti-cytocrome c and anti-β-actin polyclonal antibodies
(1:200) overnight at 4°C. Following washing with TBST, the
nitrocellulose membrane was incubated with AP-conjugated secondary
antibodies (1:500) at room temperature for 2 h, after which the
membrane was incubated with the AP-NBT/BCIP substrate solution
(ZLI-9041; ZSBio; OriGene Technologies, Inc.). Bound antibodies
were detected using the Tanon Gel Imaging system (GIS-2019; Tanon
Science & Technology Co., Ltd., Shanghai, China), and were
analyzed quantitatively using the Gel-Pro Analyzer 3.1 Density
Analysis software (Media Cybernetics, Inc., Rockville, MD,
USA).
Isolation of total RNA from SGC-7901
cells and reverse transcription-polymerase chain reaction
(RT-PCR)
SGC-7901 cells were inoculated into a 6-well plate
(1×106 cells/ml, cells inoculated into a 75-ml culture
bottle), after which CSBE (15, 30 or 60 µg/ml) was added at 24 h.
Total RNA was extracted using TRIzol® reagent after 24 h.
Qualitative analysis of the total RNA was performed using gel
electrophoresis to evaluate the purity and degradation of the RNA.
The RT reaction was performed in accordance with the manufacturer's
instructions (Takara Biotechnology Co., Ltd., Dalian, China).
Primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai,
China). The primer sequences were as follows: BCL-2 (458 bp),
forward 5′-GGT GCC ACC TGT GGT CCA CCT-3′, reverse 5′-CTT CAC TTG
TGG CCC AGA TAG G-3′; BAX (382 bp), forward 5′-CGT CCA CCA AGA AGC
TGA GCG −3′, reverse 5′-AGC ACT CCC GCC ACA AAG ATG-3′; β-actin
(540 bp), forward 5′-GTG GGG CGC CCC AGG CAC CA-3′, reverse 5′-CTT
CCT TAA TGT CAC GCA CGA TTT C-3′. The reaction conditions of the
PCR were: 94°C for 3 min; 94°C for 30 sec; 72°C for 1 min; and 72°C
for 8 min. BCL-2 underwent 32 cycles and BAX underwent 30 cycles.
β-actin underwent 94°C for 2 min, 94°C for 30 sec, 58°C for 30 sec,
and 72°C for 60 sec. The products of the PCR were detected by 2%
sepharose gel electrophoresis, and quantitative analysis of data
was performed using the Tanon gel imaging system.
Statistical analysis
PASW Statistics, version 18 (SPSS, Inc., Chicago,
IL, USA) was used to analyze the results. Data are expressed as the
mean ± standard deviation. Differences between groups were examined
for statistical significance using one-way analysis of variance. In
all cases, P<0.05 was considered to indicate a statistically
significant difference.
Results
Cytotoxic effects of CSBE on SGC-7901
cells
The cytotoxic effects of CSBE on SGC-7901 cells were
measured using MTT assays. The half maximal inhibitory
concentration (IC50) of CSBE on SGC-7901 cells was
31.542 µg/ml, whereas the IC50 of HCPT was 0.175 µg/ml.
These results led to the use of the following doses of CSBE in
subsequent experiments: 15, 30 and 60 µg/ml.
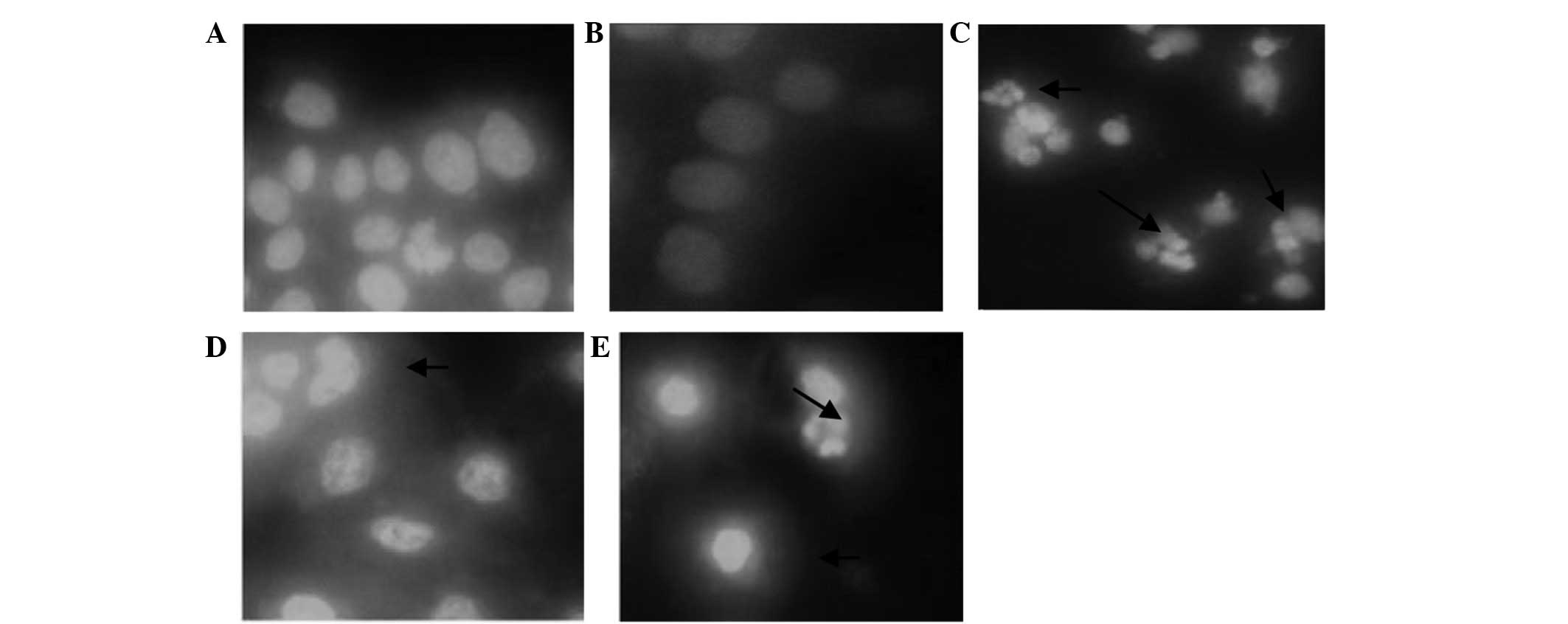
Effect of CSBE on the morphology of
SGC-7901 cells
CSBE administration was associated with significant
apoptosis-associated morphological alterations to SGC-7901 cells,
as compared with the control group, 24 h following CSBE treatment.
Apoptotic cells were distinguished by the typical nuclear
morphology of apoptosis, including profound chromatin condensation,
nuclear fragmentation and scattered apoptotic bodies (Fig. 1).
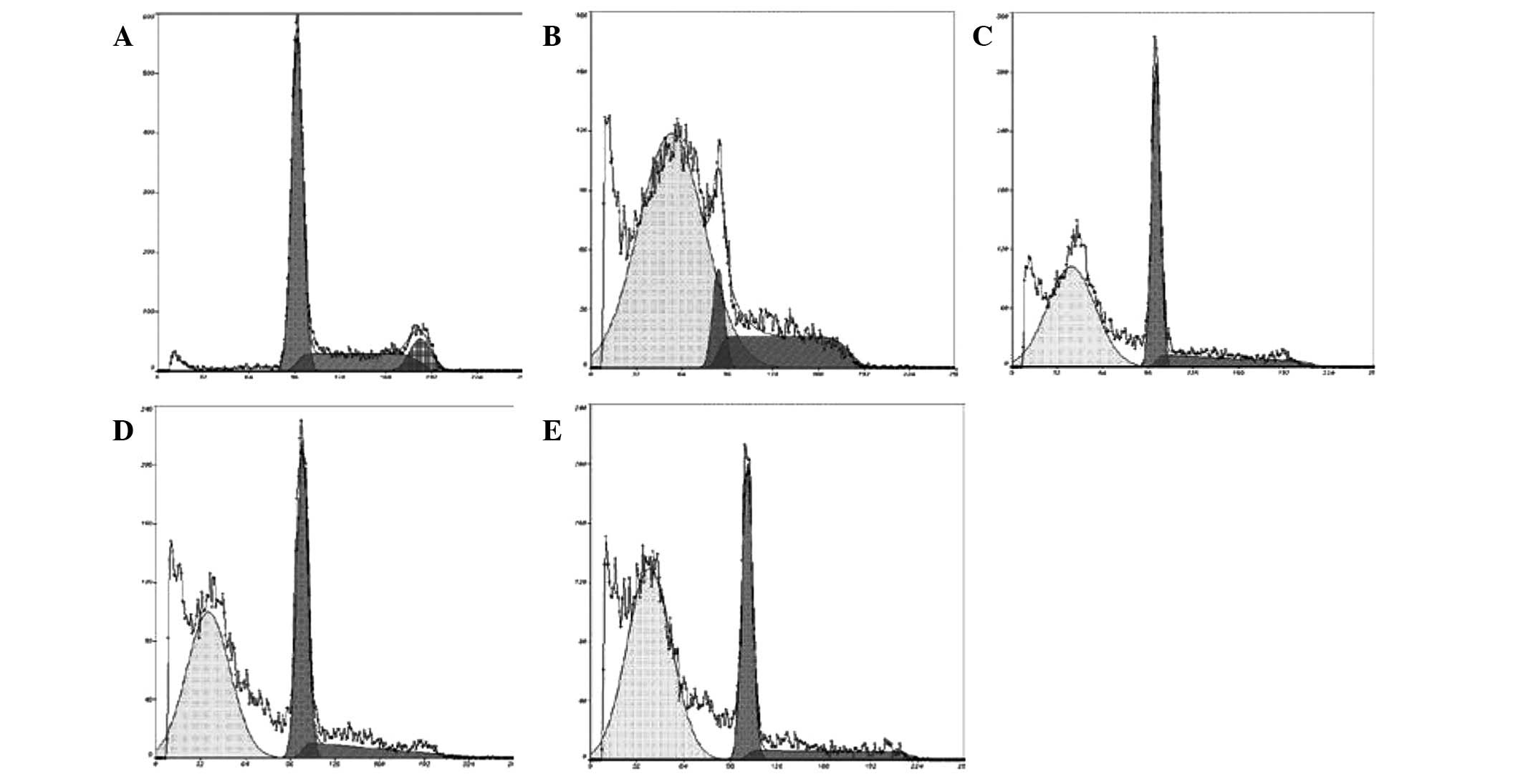
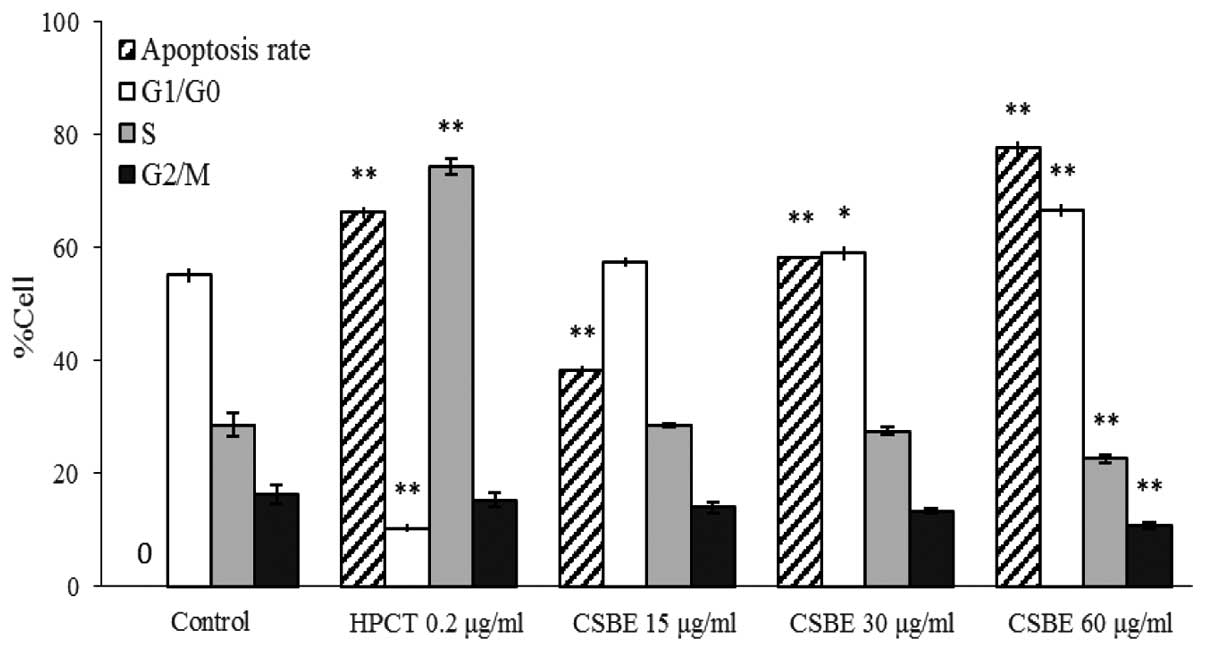
Effect of CSBE on apoptosis and cell
cycle distribution of SGC-7901 cells
PI-mediated fluorescence-activated cell sorting was
used to observe CSBE-induced SGC-7901 cell apoptosis. Following
administration of CSBE, the total number of apoptotic SGC-7901
cells significantly increased in a dose-dependent manner (Fig. 2). The proportion of cells undergoing
late apoptosis was significantly higher, as compared with the
controls. In order to determine whether cell growth inhibition was
associated with cell cycle alterations, cell cycle distribution was
also determined. CSBE markedly increased the proportion of
G1 cells, and reduced the proportion of SGC-7901 cells
in the G2/M phase, which indicated the occurrence of
G0/G1 phase arrest (Fig. 3).
Effect of CSBE on the mitochondrial
membrane potential of SGC-7901 cells
The mitochondrial membrane potential decreased in a
dose-dependent manner following CSBE exposure, which was indicated
by a decrease in the fluorescence intensity (Table I). This was associated with the
opening of the mitochondrial permeability transition pore (PTP),
which may have resulted in the release of apoptotic factors.
 | Table I.Effect of CSBE on the MMP of SGC-7901
cells. |
Table I.
Effect of CSBE on the MMP of SGC-7901
cells.
| Group | Concentration
(µg/ml) | Intracellular MMP
levels (%) |
|---|
| Control | 0 | 49.8±0.18 |
| CSBE | 15 |
40.7±0.31a |
|
| 30 |
38.8±0.21a |
|
| 60 |
8.1±0.12a |
| HCPT | 0.2 |
4.0±0.12a |
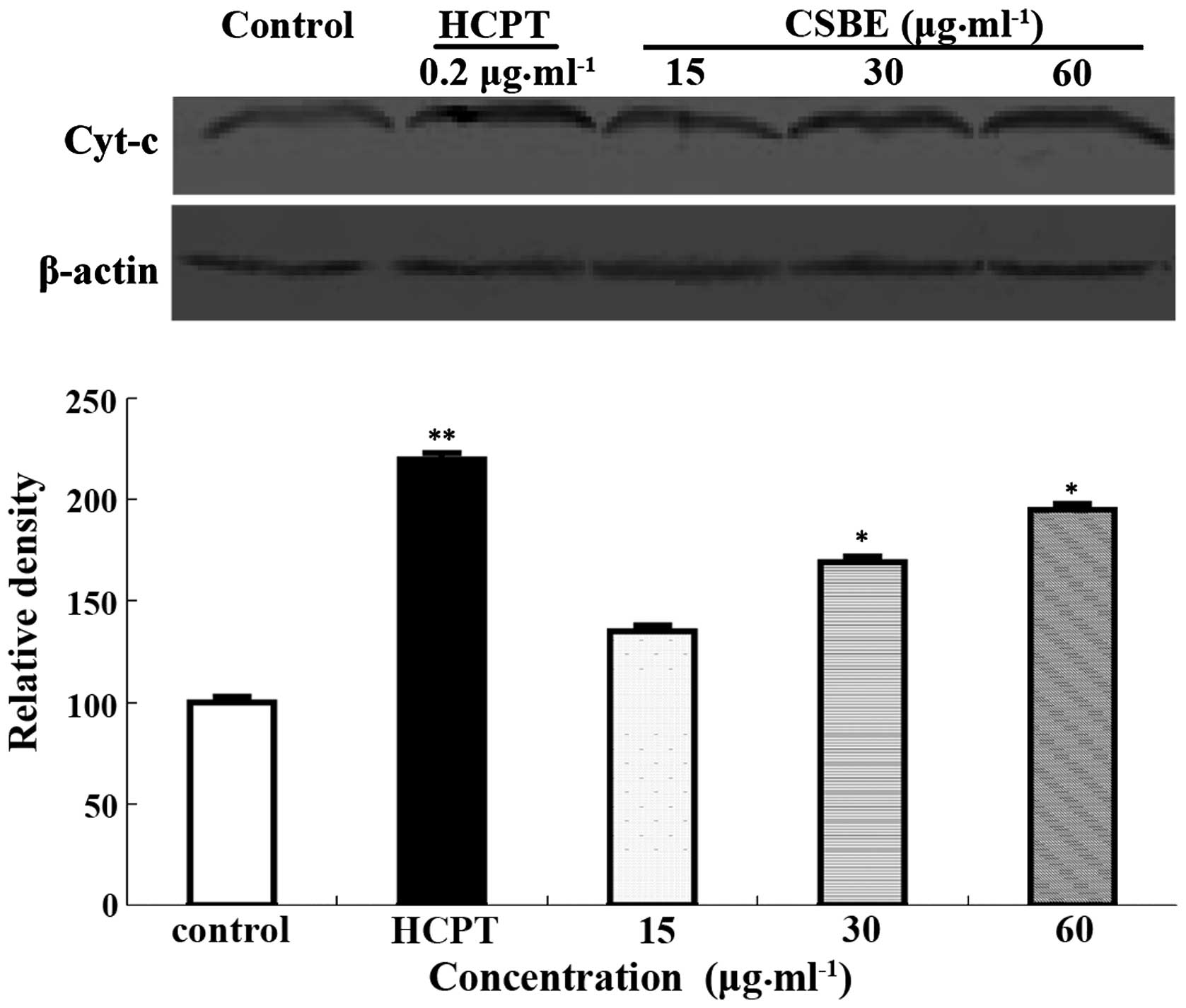
Effect of CSBE on cytochrome c
release
Western blotting was used to detect cytochrome
c release into the cytoplasm, following CSBE exposure. The
levels of cytochrome c in the cytoplasm increased in a
dose-dependent manner (Fig. 4),
which may indicate that cytochrome c is involved in
CSBE-induced SGC-7901 apoptosis.
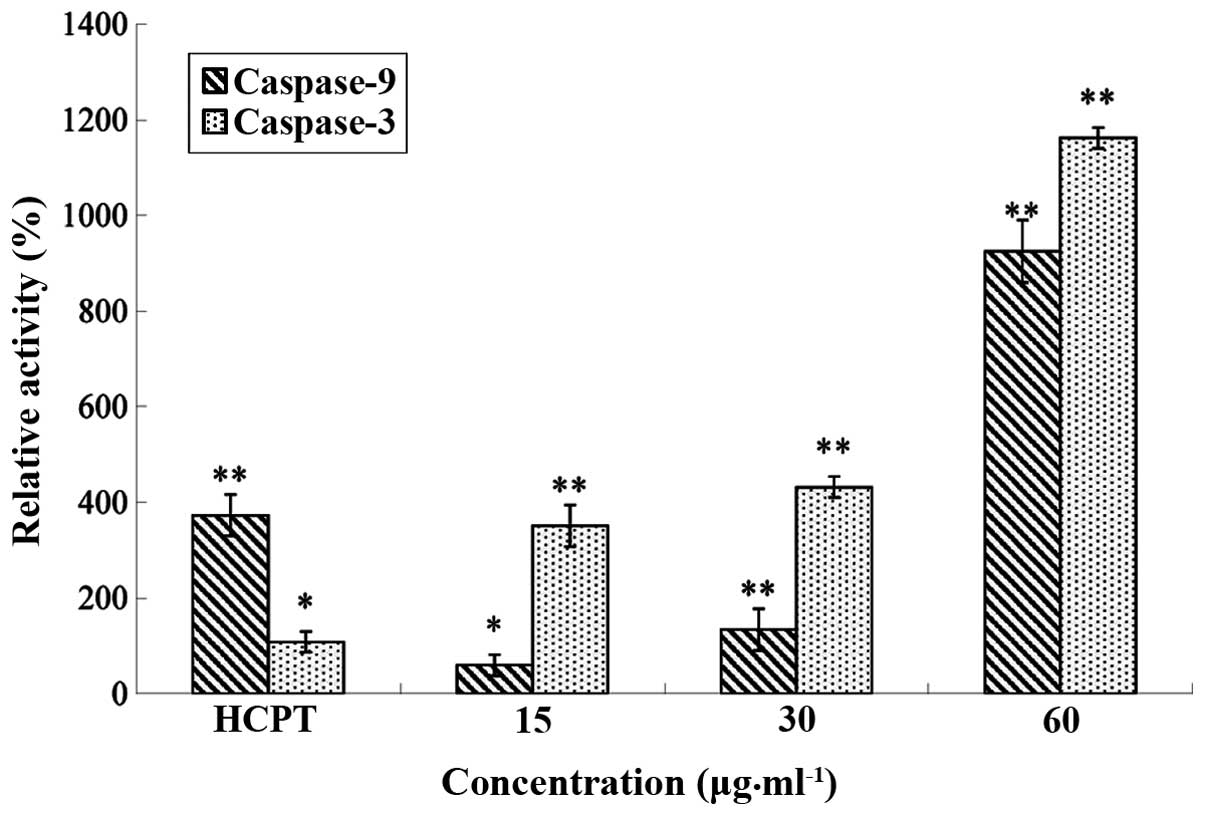
Effect of CSBE on caspase-9 and
caspase-3 activity
The relative activity of caspase-9/caspase-3 was
detected using a caspase-9 and caspase-3 test kit. Following
treatment with CSBE, the relative activity of caspase-9/caspase-3
in SGC-7901 cells increased in a dose-dependent manner (Fig. 5).
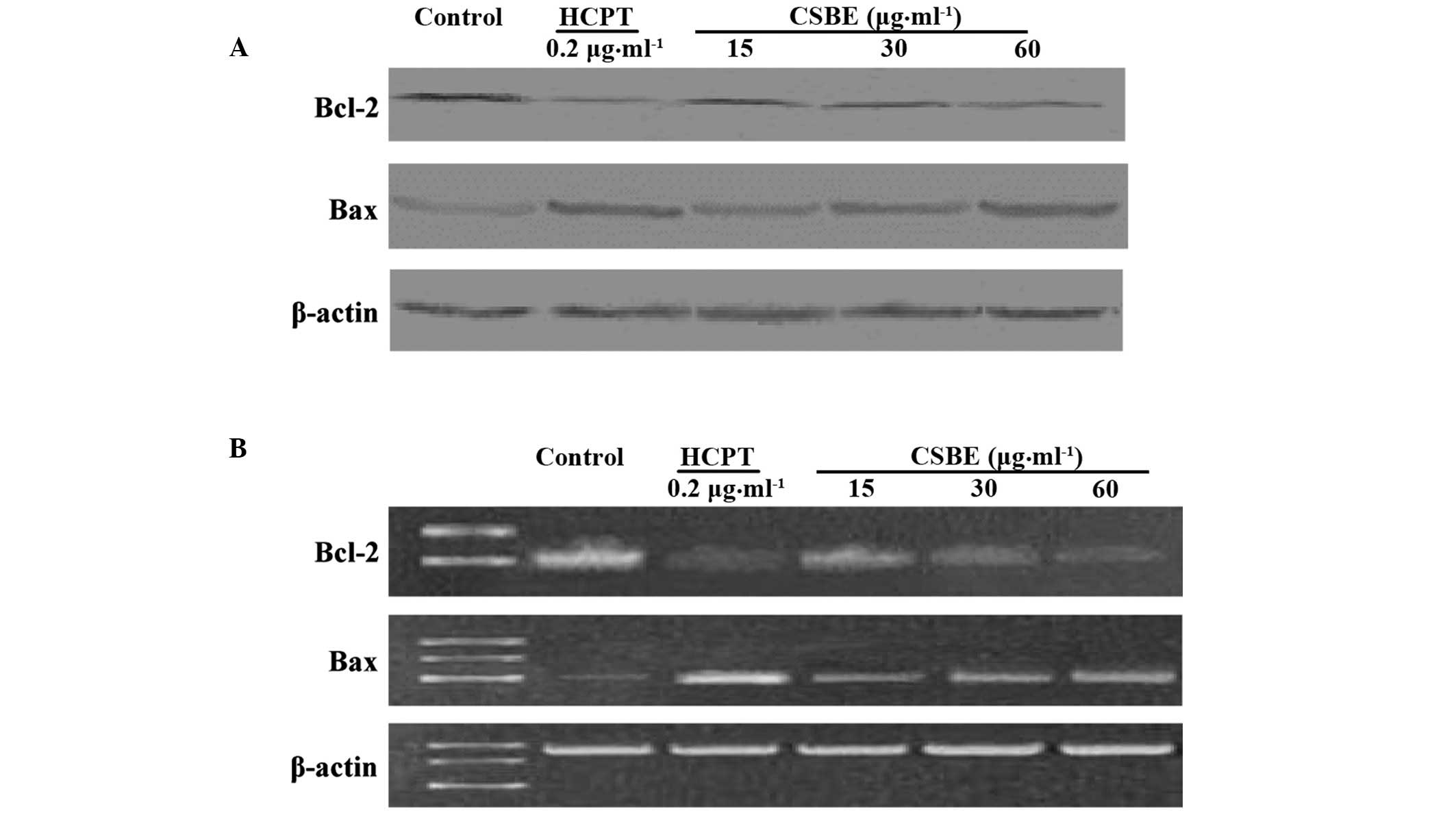
Effect of CSBE on BCL-2 and BAX
protein and mRNA expression levels
Western blotting was used to detect the BCL-2 and
BAX protein expression levels in SGC-7901 cells (Fig. 6A). CSBE was able to inhibit BCL-2
protein expression in a dose-dependent manner, with the inhibition
rate reaching 56.4%, following exposure to 60 µg/ml CSBE.
Conversely, CSBE was demonstrated to increase the levels of BAX
protein. RT-PCR was used to detect the effects of CSBE on the
transcription of BCL-2 and BAX (Fig.
6B). The results suggest that CSBE may significantly reduce
BCL-2 mRNA expression levels and increase BAX mRNA expression
levels 24 h following administration.
Discussion
CSBE has previously been demonstrated to have
dose-dependent, anti-proliferative effects on SGC-7901 cells
(21); however, the underlying
mechanism remains poorly understood. Previous studies (22,23)
evaluated the morphological alterations in SGC-7901 cells
undergoing apoptosis using a fluorescence microscope. Furthermore,
an analysis of cell cycle distribution in SGC-7901 cells undergoing
apoptosis reported that the apoptotic peak, consisting of cells
that had lost their DNA content, occurred to the left of the normal
genomic peak (G1) (24).
In the present study, exposure of SGC-7901 cells to CSBE was
associated with: A dose-dependent accumulation of SGC-7901 cells in
the G0/G1 phase of the cell cycle;
morphological changes to the cells; and the presence of an
apoptotic peak. The findings of the present study suggested that
CSBE may have been able to induce SGC-7901 cell apoptosis.
The mitochondrial membrane potential assay is a
sensitive indicator of mitochondrial function in early apoptosis.
The mitochondrial membrane potential of SGC-7901 cells markedly
decreased following CSBE exposure, and this may have promoted
cytochrome c release into the cytoplasm; this in turn may
have activated apoptosis via the caspase cascade reaction (25,26). In
the present study, CSBE administration in SGC-7901 cells was
associated with mitochondrion PTP opening, a decrease in the
mitochondrion membrane potential and cytochrome c release
into the cytoplasm.
The disruption of the mitochondrial membrane
potential is an early event in apoptosis, which may result in
activation of apoptotic cascades (27,28).
This cascade activation triggers the release of cytochrome
c, and other apoptotic factors, including second
mitochondria-derived activator of caspase/direct inhibitor of
apoptosis-binding protein with low pI, into the cytosol.
Subsequently, cytochrome c is able to bind apoptotic
protease activating factor-1 and activated procaspase-9 (29). Activated caspase-9 cleaves caspase-3,
which is an executioner of apoptosis (30). In the present study, CSBE may have
induced the release of cytochrome c into the cytoplasm,
which may have subsequently activated caspase-9 and the downstream
caspase-3, leading to SGC-7901 cell apoptosis. Therefore, the
results of the present study suggested that CSBE may have
indirectly induced SGC-7901 cell apoptosis.
The BCL-2 family consists of pro-apoptotic and
anti-apoptotic members, which are associated with the mitochondrial
apoptosis pathway. BCL-2 has been demonstrated to preserve
mitochondrial integrity, in order to prevent apoptosis, whereas BAX
has been demonstrated to promote the release of cytochrome c
from the mitochondria, which in turn activates downstream caspases
(31,32). In the present study, CSBE was
demonstrated to decrease the ratio of BCL-2:BAX, which may have
induced SGC-7901 cell apoptosis. The inhibition of BCL-2 protein
expression may have prevented BCL-2 and BAX from forming a
heterologous dimer, which in turn may have enhanced SGC-7901 cell
apoptosis. The results of RT-PCR suggested that CSBE was able to
alter the expression levels of BCL-2 and BAX by regulation at the
transcriptional level.
In conclusion, CSBE was able to inhibit the
proliferation of SGC-7901 cells, induce apoptosis, and initiate
G0/G1 phase arrest. Apoptosis may have been
induced via CSBE-mediated upregulation of the pro-apoptotic protein
BAX, and downregulation of the anti-apoptotic protein BCL-2, which
in turn may have induced a reduction in the mitochondrial membrane
potential, leading to mitochondrial cytochrome c release and
subsequent activation of caspase-9 and caspase-3. Therefore, CSBE
may have induced SGC-7901 cell apoptosis via the mitochondrial
apoptosis pathway.
Acknowledgements
The present study was supported in part by The Open
Research Program for Key Laboratory (College of Heilongjiang,
China; grant no. CPAT-2012003); The Natural Science Item of
Department of Education (Heilongjiang, China; grant no. 12541205);
and The Innovation Talents Item of Science and Technology (Harbin,
China; grant no. 2014RFQXJ154).
References
|
1
|
Germanò MP, De Pasquale R, D'Angelo V,
Catania S, Silvari V and Costa C: Evaluation of extracts and
isolated fraction from Capparis spinosa L. buds as an antioxidant
source. J Agric Food Chem. 50:1168–1171. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bonina F, Puglia C, Ventura D, Aquino R,
Tortora S, Sacchi A, Saija A, Tomaino A, Pellegrino ML and de
Caprariis P: In vitro antioxidant and in vivo phytoprotective
effects of a lyophilized extract of Capparis spinosa L buds. Cosmet
Sci. 53:321–335. 2002.
|
|
3
|
Siracusa L, Kulisic-Bilusic T, Politeo O,
Krause I, Dejanovic B and Ruberto G: Phenolic composition and
antioxidant activity of aqueous infusions from Capparis spinosa L.
and Crithmum maritimum L. before and after submission to a two-step
in vitro digestion model. J Agric Food Chem. 59:12453–12459. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tlili N, Khaldi A, Triki S and Munné-Bosch
S: Phenolic compounds and vitamin antioxidants of caper (Capparis
spinosa). Plant Foods Hum Nutr. 65:260–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abraham Issac SV, Palani A, Ramaswamy BR,
Shunmugiah KP and Arumugam VR: Antiquorum sensing and antibiofilm
potential of Capparis spinosa. Arch Med Res. 42:658–668. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boga C, Forlani L, Calienni R, Hindley T,
Hochkoeppler A, Tozzi S and Zanna N: On the antibacterial activity
of roots of Capparis spinosa L. Nat Prod Res. 25:417–421. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gadgoli C and Mishra SH: Antihepatotoxic
activity of p-methoxy benzoic acid from Capparis spinosa. J
Ethnopharmacol. 66:187–192. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
al-Said MS, Abdelsattar EA, Khalifa SI and
el-Feraly FS: Isolation and identification of an anti-inflammatory
principle from Capparis spinosa. Pharmazie. 43:640–641.
1988.PubMed/NCBI
|
|
9
|
Zhou HF, Xie C, Jian R, Kang J, Li Y,
Zhuang CL, Yang F, Zhang LL, Lai L, Wu T and Wu X: Biflavonoids
from Caper (Capparis spinosa L.) fruits and their effects in
inhibiting NF-kappa B activation. J Agric Food Chem. 59:3060–3065.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ziyyat A, Legssyer A, Mekhfi H, Dassouli
A, Serhrouchni M and Benjelloun W: Phytotherapy of hypertension and
diabetes in oriental Morocco. J Ethnopharmacol. 58:45–54. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eddouks M, Lemhadri A and Michel JB:
Hypolipidemic activity of aqueous extract of Capparis spinosa L. in
normal and diabetic rats. J Ethnopharmacol. 98:345–350. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huseini HF, Hasani-Rnjbar S, Nayebi N,
Heshmat R, Sigaroodi FK, Ahvazi M, Alaei BA and Kianbakht S:
Capparis spinosa L. (Caper) fruit extract in treatment of type 2
diabetic patients: A randomized double-blind placebo-controlled
clinical trial. Complement Ther Med. 21:447–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lam SK and Ng TB: A protein with
antiproliferative, antifungal and HIV-1 reverse transcriptase
inhibitory activities from caper (Capparis spinosa) seeds.
Phytomedicine. 16:444–450. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bai HJ, Zhou ZB and Zhao XL: Study on
activity of anti-H22 of xinjiang province plant Capparis spinosa.
Hei Long Jiang Xu Mu Shou Yi. 12:100–101. 2007.(In Chinese).
|
|
15
|
Caldas C, Carneiro F, Lynch HT, Yokota J,
Wiesner GL, Powell SM, Lewis FR, Huntsman DG, Pharoah PD, Jankowski
JA, et al: Familial gastric cancer: Overview and guidelines for
management. J Med Genet. 12:873–880. 1999.
|
|
16
|
Dicken JL, van de Velde CJH, Coit DG, Shah
MA, Verheij M and Cats A: Treatment of resectable gastric cancer.
Therap Adv Gastroenterol. 5:49–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li WL, Yu L and Ji YB: Chemical
constituents of n-butanol extract of Capparis spinosa L. Asian J
Chem. 26:3435–3437. 2014.
|
|
18
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55–63. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gong J, Traganos F and Darzynkiewicz Z: A
selective procedure for DNA extraction from apoptotic cells
applicable for gel electrophoresis and flow cytometry. Anal
Biochem. 218:314–319. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nicoletti I, Migliorati G, Pagliacci MC,
Grignani F and Riccardi C: A rapid and simple method for measuring
thymocyte apoptosis by propidium iodide staining and flow
cytometry. J Immunol Methods. 139:271–279. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji YB and Yu L: N-butanol extract of
Capparis spinosa L. induces apoptosis primarily through a
mitochondrial pathway involving mPTP open, cytochrome c release and
caspase activation. Asian Pac J Cancer Prev. 15:9153–9157. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramadevi Mani S and Lakshmi BS: G1 arrest
and caspase-mediated apoptosis in HL-60 cells by dichloromethane
extract of Centrosema pubescens. Am J Chin Med. 38:1143–1159. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raposa T and Natarajan AT: Fluorescence
banding pattern of human and mouse chromosomes with a benzimidazol
derivative (Hoechst 33258). Humangenetik. 21:221–226.
1974.PubMed/NCBI
|
|
24
|
Lecellier G and Brenner C: Genomic and
proteomic screening of apoptosis mitochondrial regulators for drug
target discovery. Curr Med Chem. 14:875–881. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Im AR, Kim YH, Uddin MR, Chae S, Lee HW,
Kim YS and Lee MY: Neuroprotective effects of Lycium chinense
Miller against rotenone-induced neurotoxicity in PC12 cells. Am J
Chin Med. 41:1343–1359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen KH, Chen ZT and Duh PD: Cytotoxic
effect of Eucalyptus citriodora resin on human hepatoma HepG2
cells. Am J Chin Med. 40:399–413. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Desagher S, Osen-Sand A, Nichols A, Eskes
R, Montessuit S, Lauper S, Maundrell K, Antonsson B and Martinou
JC: Bid-induced conformational change of Bax is responsible for
mitochondrial cytochrome c release during apoptosis. J Cell Biol.
144:891–901. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng EH, Kirsch DG, Clem RJ, Ravi R,
Kastan MB, Bedi A, Ueno K and Hardwick JM: Conversion of Bcl-2 to a
Bax-like death effector by caspases. Science. 278:1966–1968. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Green DR and Reed JC: Mitochondria and
apoptosis. Science. 281:1309–1312. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thornberry NA and Lazebnik Y: Caspases:
Enemies within. Science. 281:1312–1316. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin XM, Oltvai ZN and Korsmeyer SJ: BH1
and BH2 domains of Bcl-2 are required for inhibition of apoptosis
and heterodimerization with Bax. Nature. 369:321–323. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kluck RM, Bossy-Wetzel E, Green DR and
Newmeyer DD: The release of cytochrome c from mitochondria: A
primary site for Bcl-2 regulation of apoptosis. Science.
275:1132–1136. 1997. View Article : Google Scholar : PubMed/NCBI
|