Introduction
Cervical cancer is the second most prevalent type of
cancer among women worldwide (1),
and is a complex disease involving numerous oncogenes or the
abnormal expression of tumor suppressors (2,3).
Currently, the phosphoinositide 3-kinase(PIK3)/protein kinase B
signaling pathway is considered crucial to the pathogenesis of
cervical cancer. The PIK3 catalytic subunit α gene is upregulated
in cervical cancer due to the amplification of the chromosome
3q26.3 locus (4). In addition, tumor
suppressor genes, such as phosphatase and tensin homolog, may be
downregulated due to genetic mutations or deletions, which
contribute to the development of cervical cancer (5). However, the precise molecular
mechanisms underlying the pathogenesis of cervical carcinogenesis
remain unclear. Therefore, it is crucial to identify specific
molecular markers and mechanisms for use in cervical cancer
detection.
C-C chemokine receptor type 5 (CCR5) belongs to the
chemokines, a family of structurally-related proteins that were
initially recognized as mediators of chemotaxis and cellular homing
(6). This family is loosely divided
into three groups: Homeostatic/constitutive chemokines,
inflammatory/inducible chemokines (7,8) and dual
function chemokines. To date, CCR5 has been demonstrated to be
involved in a variety of biological processes, including tumor
development. For example, the expression of chemokine (C-C motif)
ligand 5 (CCL5), a ligand that binds with CCR5, correlates with
breast cancer stage (9) and is
associated with enhanced melanoma formation in nude mice (10). Furthermore, treatment with a CCL5
antagonist was observed to decrease tumor growth in a breast cancer
model (11). Ng-Cashin et al
demonstrated that CCR5 knockout was able to inhibit local tumor
growth and improved responses to cancer vaccines in mice (12). In addition, van Deventer et al
showed that the expression of CCR5 in stromal cells promoted
pulmonary metastasis (13). However,
few studies have investigated the association between CCR5 and
cervical cancer development.
The aim of the present study was to investigate the
expression of CCR5 in human cervical cancer cells, and to evaluate
the effect of CCR5 knockdown on the viability, colony formation and
invasiveness of the cells. Furthermore, the potential of micro RNA
(miR)-107 as a regulator of CCR5 expression in the cervical cancer
cells was evaluated.
Materials and methods
Human tissue samples
A total of 28 pairs of human cervical cancer and
adjacent normal tissues were obtained from the Department of
Gynecology, Yi-Du Central Hospital of Weifang, (Weifang, China).
Informed consent was obtained from all patients. The majority of
the cancer was stage IIa or lower according to the International
Federation of Gynecology and Obstetrics (FIGO) staging system
(14). Histologically, all included
biopsies were squamous cell carcinoma. All use of human specimens
was approved and supervised by the Ethics Committee of Jinan
Maternity and Child Care Hospital. The specimens were frozen in
liquid nitrogen and stored at −80°C until required.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using Invitrogen TRIzol
reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. Oligo (dT) primers and
M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA)
were applied to reverse transcribe the 1 µg total RNA into cDNA.
qPCR was performed to detect the CCR5 mRNA expression level using a
SYBR Premix Ex Taq™ kit (Takara Bio, Dalian, China) according to
the manufacturer's protocol. β-actin was used as the reference
gene. qPCR cycling was performed using the iQ5 real-time PCR
detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA),
under the following conditions: Denaturing at 94°C for 4 min,
followed by 40 cycles of amplification including 94°C for 60 sec,
58°C for 60 sec, and 72°C for 60 sec. Primers used for the qPCR
were as follows: CCR5 forward, 5′-GAGACTCTTGGGATGACGC-3′ and
reverse, 5′-GTTTGGCAATGTGCTTTTG-3′; and β-actin forward,
5′-TGCGTGACATTAAGGAGAAGC-3′ and reverse, 5′-TCCATGCCCAGGAAGGAA-3′
(Genewiz, Inc., Beijing, China). All qPCR experiments were
conducted in triplicate.
Western blot analysis
Cells were homogenized in radioimmunoprecipitation
assay (RIPA) lysis buffer (BioVision, Inc., Milpitas, CA, USA) in
the presence of 1% (v/w) protease inhibitor cocktail (Pierce
Biotechnology, Inc., Rockford, IL, USA). Total protein was isolated
and, following the measurement of its concentration using a
Bicinchoninic acid assay (Qcbio Science & Technologies, Co.,
Ltd., Shanghai, China), 20 µg protein was separated by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (ZhiYou
Biotechnology Co., Ltd., Guangzhou, China) and blotted onto
nitrocellulose membranes (EMD Millipore, Billerica, MA, USA). The
membranes were subsequently incubated with polyclonal rabbit
anti-human CCR5 antibody (1:1,000; ab65850; Abcam, Cambridge, MA,
USA) and monoclonal rabbit anti-human β-actin antibody (1:1,000;
ab181602; Abcam) in blocking solution overnight at 4°C. After
washing five times with phosphate-buffered saline solution, the
membranes were probed with a secondary horseradish
peroxidase-conjugated goat anti-rabbit IgG antibody (1:1,000;
ab150077; Abcam) for 2 h at room temperature. The relative amount
of protein was normalized to β-actin and analyzed with a Gel-Pro
Analyzer, version 4.0 (Media Cybernetics, Inc., Rockville, MD,
USA).
Small interfering RNA (siRNA) for CCR5
knockdown
siRNA used for the knockdown of CCR5 (siRNA-CCR5)
was purchased from Chang Jing Bio-Tech, Ltd. (Changsha, China). The
CCR5 knockdown siRNA sequence was as follows: siRNA-CCR5 (top),
5′-GATCCGTCCAATCTATGACATCAATTCAAGAGATTGATGTCATAGATTGGACTTTTTTGGAAGAATTCA-3′
and siRNA-CCR5 (bottom),
5′-AGCTTGAATTCTTCCAAAAAAGTCCAATCTATGACATCAATCTCTTGAATTGATGTCATAGATTGGACG-3′.
A scrambled siRNA (siRNA-NC) was used as a control.
Cell culture and transfection
Human cervical cancer cell lines HeLa and C33A
(ZhiYou Biotechnology) were maintained in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) supplemented with 10% (v/v)
heat-inactivated fetal bovine serum (FBS), 100 IU/ml penicillin and
100 µg/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C
in a humidified atmosphere with 5% CO2. Transfection was
performed using Lipofectamine 2000 reagent (Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol.
MTT and colony formation assays
Transfected cells were plated in 96-well plates at a
density of 5,000 cells/well. At 48 h after transfection, the cells
were incubated with MTT for 4 h at 37°C. The cells were then
agitated with MTT solvent on an orbital shaker at 377 × g for 10
min in the dark at room temperature (Thermo Fisher Scientific,
Inc.). The absorbance was measured at 570 nm using a
spectrophotometer (M5/M5a; Xinhua, Guangdong, China). In the colony
formation assay, cells were seeded into a 12-well plate at a
density of 200 cells/well, with a change of medium every 3 days.
After ~10 days, the majority of the cell clones contained >50
cells. The clones were washed with 1X phosphate-buffered saline and
stained with crystal violet (Yuanye Bio-Technology, Co., Ltd.,
Shanghai, China) for ~5 min.
Cell invasion assay
Invasion assays were performed in 24-well Transwell
chambers (Corning Incorporated, Corning, NY, USA). The upper
compartments of the chambers were filled with 100 µl pre-chilled
serum-free RPMI-1640 mixed with Matrigel (1:7; BD Biosciences,
Franklin Lakes, NJ, USA). The Matrigel remained at room temperature
for 4 h for solidification. Subsequently, 5×104 HeLa
cells or 8×104 C33A cells were trypsinized, washed and
resuspended in serum-free RPMI-1640, then seeded in the upper
chamber. An additional 500 µl RPMI-1640 containing 10% FBS was
added to the lower chamber as a chemoattractant. The chambers were
incubated at 37°C in 5% CO2 for 24 h (HeLa cells) or 48
h (C33A cells), then fixed in 100% methanol. Fixed cells were
stained with crystal violet and the number of invasive cells was
counted. Five random fields in each chamber were analyzed. Assays
were performed in triplicate.
Identification of a CCR5-targeting
miRNA using multiple miRNA target prediction algorithms
In order to identify a sequence potentially capable
of inhibiting the expression of CCR5, a number of miRNA target
prediction software packages were used, namely Targetscan
(http://www.targetscan.org),
microRNA.org, DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index)
and miRwalk (www.umm.uni-heidelberg.de/apps/zmf/mirwalk/).
Luciferase assay of the effect of
miR107 on CCR5 expression
A pcDNA3/enhanced green fluorescent protein
(EGFP)-CCR5-3′ untranslated region (UTR) vector and the mutant
pcDNA3/EGFP-CCR5-3′UTR, in which a number of nucleotides within the
binding sites were mutated, were purchase from ZhiYou Biotechnology
Co., Ltd. For the luciferase reporter assay, the HeLa cells were
co-transfected with a miR-107 mimic (Genewiz, Inc.) and
pcDNA3/EGFP/CCR5-3′UTR or mutant 3′UTR, whereas the control cells
were transfected with pcDNA3/EGFP-CCR5-3′UTR or mutant 3′UTR only.
The plasmid expressing red fluorescent protein (RFP) was
transfected as the spike-in control. At 48 h after transfection,
the cells were lysed using RIPA buffer, and the EGFP and RFP
intensities were measured using an F-4500 fluorescence
spectrophotometer (Hitachi, Ltd., Tokyo, Japan).
Statistical analysis
All data are presented as the mean ± standard
deviation, and the difference between groups was determined using
the two-tailed Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
CCR5 is upregulated in human cervical
cancer tissues
In order to determined the role of CCR5 in cervical
cancer, the mRNA expression levels of CCR5 were evaluated in 28
pairs of human cervical cancer samples and adjacent normal tissues
using RT-qPCR. The results revealed that CCR5 mRNA expression was
increased in the majority of cancer tissues compared with the
matched normal control tissues (Fig.
1A). To further confirm the upregulation of CCR5, western blot
analysis was performed to detect the CCR5 protein expression levels
in samples 015 and 043 and confirmed that it was higher in the
tumor tissue than in the adjacent normal tissue (Fig. 1B). These results indicate that CCR5
is upregulated in human cervical cancer, suggesting that CCR5 may
exert an oncogenic effect in cervical cancer development.
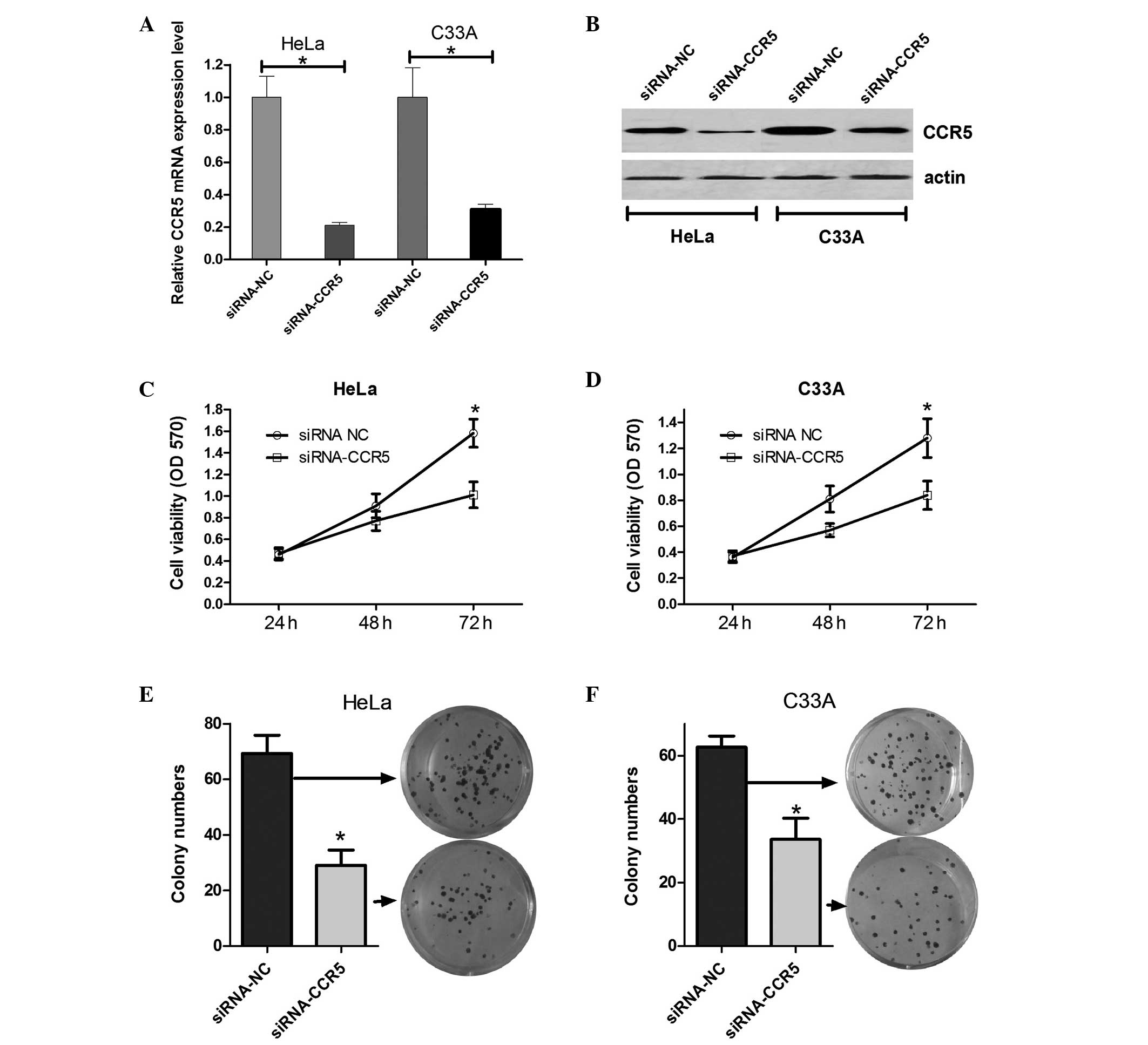
Downregulation of CCR5 affects
cervical cancer cell growth
On the basis of the results demonstrating that CCR5
is upregulated in cervical cancer cells, it was speculated that
CCR5 may affect cervical cancer cell growth. Firstly, siRNA-CCR5
(for knockdown of CCR5) or an siRNA control vector were transfected
into HeLa and C33A cells, and the mRNA and protein expression
levels of CCR5 were evaluated using RT-qPCR and western blot
analysis. As shown in Fig. 2A and B,
CCR5 mRNA and protein expression levels in HeLa and C33A cells
transfected with siRNA-CCR5 were evidently decreased compared with
those in the cells transfected with the scrambled siRNA. Next, an
MTT assay was performed to determine the effect of CCR5 on HeLa and
C33A cell viability. The results indicate that cells transfected
with siRNA-CCR5 had decreased cell viability at 48 and 72 h
compared with the cells transfected with scramble siRNA (Fig. 2C and D). In addition, colony
formation assays were performed to assess the effect of CCR5 on the
long-term proliferative capacity of HeLa and C33A cells. As shown
in Fig. 2E and F, the colony number
of HeLa and C33A cells treated with siRNA-CCR5 decreased by
approximately half compared with the colony number in the control
group. These results suggest that the downregulation of CCR5 is
able to inhibit cervical cancer cell growth and proliferation.
Downregulation of CCR5 inhibits the
invasion of cervical cancer cells
Previous studies have shown that CCR5 is associated
with tumor cell invasiveness (15).
Therefore, a cell invasion assay was performed to determine whether
CCR5 was associated with cervical cancer cell invasion. Compared
with the control group, the invasive cell number decreased ~2-fold
in the HeLa and C33A cells transfected with siRNA-CCR5 (Fig. 3), which suggests that the
downregulation of CCR5 is able to inhibit cervical carcinoma cell
invasion.
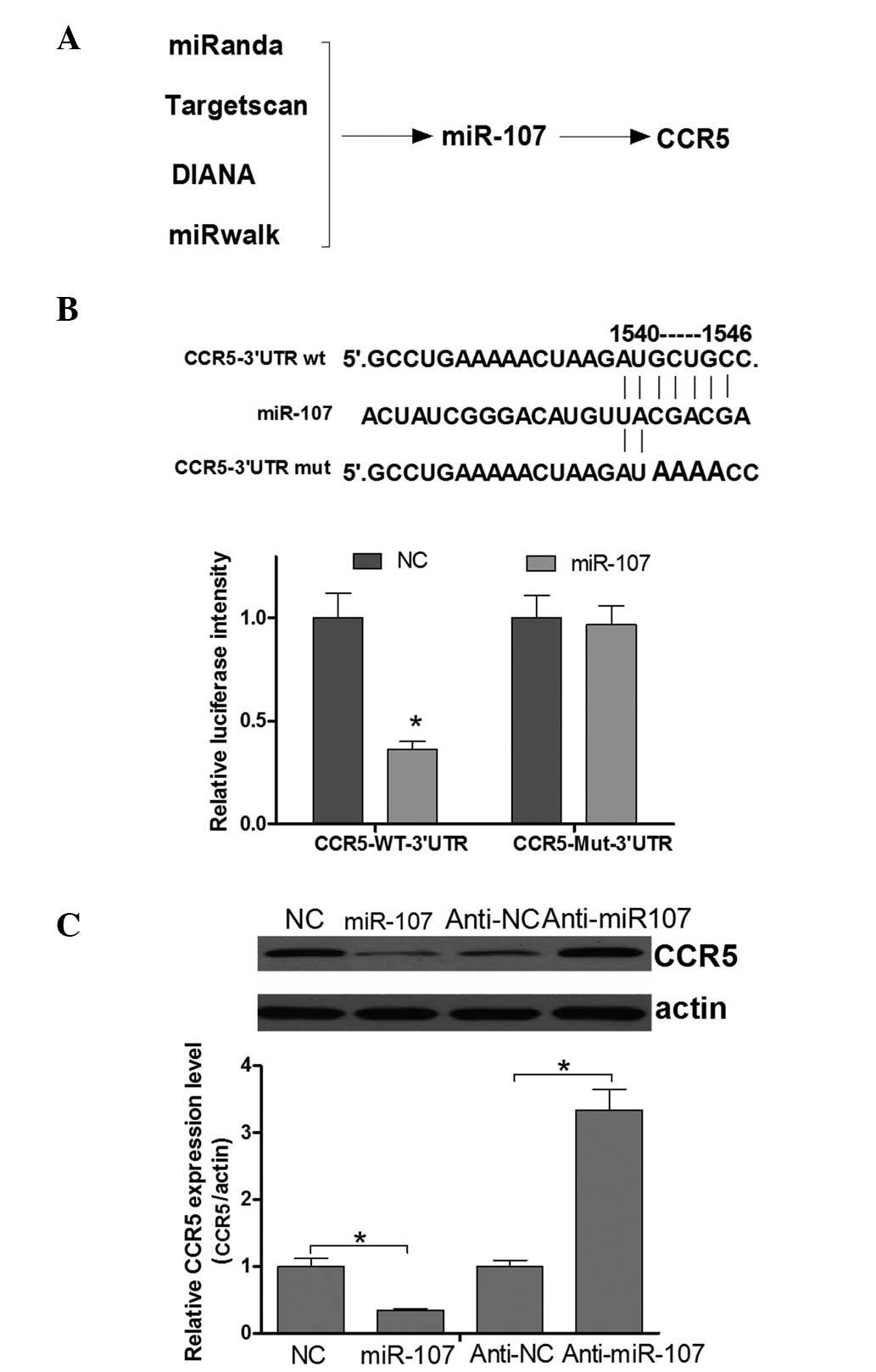
miR-107 is a candidate regulator of
CCR5 in cervical cancer cells
miRNAs function as tumor suppressors or oncogenes
via the direct regulation of associated oncogenes or tumor
suppressor genes. Tumor suppressor miRNAs are usually downregulated
in tumors and may result in the reduced expression of tumor
oncogenes and contribute to the tumorigenesis of cancers. The
upregulation of CCR5 in cervical cancer tissues suggests that
miRNAs may be crucially involved in the regulation of CCR5 in
cervical cancer development. Improved specificity of miRNA
prediction may be attained by the consensus of multiple algorithms.
Therefore, four programs (Targetscan, miRanda, DIANA and miRwalk)
were used in the present study to predict an miRNA sequence able to
target the CCR5 3′UTR and regulate its expression. Finally,
miR-107, the mRNA 3′UTR of which contains a putative CCR5 binding
site (Fig. 4A), was identified as a
candidate for directly targeting CCR5. This analysis is consistent
with a model in which tumor suppressor miRNAs negatively regulate
tumor oncogenes during tumor development. To confirm that CCR5
expression was directly regulated by miR-107, a luciferase reporter
system was applied in HeLa cells. Fig.
4B shows that miR-107 was able to directly target the CCR5 mRNA
3′UTR. Next, whether the endogenous CCR5 was regulated by miR-107
in cervical cancer cells was investigated. The results of a western
blot assay demonstrated that the CCR5 protein expression level was
negatively regulated by miR-107 (Fig.
4C). Thus, it was concluded that CCR5 is negatively regulated
by miR-107 in cervical cancer cells.
Discussion
CCR5 has been demonstrated to promote tumor growth
in cancer cell in vitro and metastasis in a mouse model
(16,17). Furthermore, a prior study indicated
that CCR5 heterozygous genotype (+/Δ32) may have a significant
effect on the early stage of cervical cancer development (18). However, there is no direct evidence
for the involvement of CCR5 in cervical cancer tumorigenesis.
Therefore, the aim of the present study was to determine whether
CCR5 participates in cervical cancer tumorigenesis. Firstly, CCR5
mRNA expression levels were evaluated using RT-qPCR and western
blot analysis in cervical cancer tissues and matched adjacent
normal control tissues. The results showed that the mRNA expression
of CCR5 was upregulated in 21 cancer tissues compared with the
matched normal tissues (28 pairs of specimens in total). Next, the
effect of CCR5 on cervical cancer cell lines was investigated using
an siRNA to knockdown CCR5. MTT and colony formation assays
indicate that knockdown of CCR5 had an inhibitory effect on the
growth of these cells. Thus, it may be speculated that CCR5 is able
to promote cervical cancer proliferation. In addition, the results
of cell invasion assays indicate that knockdown of CCR5 is able to
inhibit HeLa and C33A cell invasion. Collectively, the present
results suggest that CCR5 may function as an oncogene during
cervical cancer development.
Accumulating evidence indicates that the
downregulation of tumor suppressor miRNAs may be a common mechanism
in the tumorigenesis of cervical cancer though target oncogenes.
For example, miR-99a and −99b have been found to inhibit cervical
cancer cell proliferation and invasion by targeting the mechanistic
target of rapamycin signaling pathway (19). Another miRNA, miR-506 functions as a
tumor suppressor by targeting the hedgehog signaling pathway
transcription factor GLI3 in human cervical cancer cells (20). In the present study, CCR5 was
upregulated in all of the cervical cancer tissues tested,
indicating that it may serve an oncogenic function in tumorigenesis
(Figs. 1–3). Therefore, in order to determine the
association between the miRNA-mediated suppression of CCR5 and the
expression of CCR5 in cervical cancer development, a bioinformatics
approach was used in the present study to predict miRNAs that could
bind to the CCR5-3′UTR. Four independent miRNA target prediction
algorithms indicated that miR-107 was a potential candidate.
Furthermore, a luciferase reporter assay showed that miR-107
significantly decreased the luciferase activity of CCR5-3′UTR,
while RT-qPCR and western blot analyses indicated that the miR-107
was able to directly repress endogenous CCR5 mRNA and protein
expression. These results indicate that the upregulation of CCR5
may at least be partly attributed to downregulation of miR-107.
miR-107 has been a widely researched miRNA in the
development of various types of cancer (21). To date, a number of studies have
indicated the involvement of miR-107 in cell cycle arrest and
growth suppression in lung and pancreatic cancer (22,23).
However, miR-107 has additionally been shown to promote
invasiveness and metastatic dissemination in breast cancer cells
(24). In addition, a previous study
suggested that the upregulation of miRNA-107 exerts an inductive
effect on the proliferation of human gastric cancer cells by
targeting the transcription factor forkhead box protein O1
(25). Thus, it is apparent that
miR-107 is able to function as a tumor suppressor or as an oncomiR,
depending on the type of cell. The results of the present study
indicate that miR-107 directly targets CCR5 and represses its
expression in cervical cancer cells, which implies that the
miR-107/CCR5 axis may contribute to the development of cervical
cancer. However, the exact mechanism by which miR-107 affects the
cervical cancer cell phenotype is unclear and requires further
study.
In conclusion, CCR5 is overexpressed in cervical
cancer cells and may function as a tumor oncogene. Furthermore, the
knockdown of CCR5 was able to repress cervical cancer cell
proliferation and invasion. In addition, it was identified and
experimentally validated that miR-107 directly targets CCR5,
potentially providing a molecular mechanism for the upregulation of
CCR5 in cervical cancer. Therefore, miR-107 and CCR5 may be of use
as novel therapeutic targets for the treatment of cervical
cancer.
Acknowledgements
The authors of the present study would like to thank
ZhiYou Biotechnology Co., Ltd. (Guangzhou, China) for their
technical support.
References
|
1
|
Bedkowska GE, Ławicki S and Szmitkowski M:
Molecular markers of carcinogenesis in the diagnostics of cervical
cancer. Postepy Hig Med Dosw (Online). 63:99–105. 2009.(In Polish).
PubMed/NCBI
|
|
2
|
Rajkumar T, Sabitha K, Vijayalakshmi N,
Shirley S, Bose MV, Gopal G and Selvaluxmy G: Identification and
validation of genes involved in cervical tumourigenesis. BMC
Cancer. 11:802011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hu X, Schwarz JK, Lewis JS Jr, Huettner
PC, Rader JS, Deasy JO, Grigsby PW and Wang X: A microRNA
expression signature for cervical cancer prognosis. Cancer Res.
70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ma YY, Wei SJ, Lin YC, Lung JC, Chang TC,
Whang-Peng J, Liu JM, Yang DM, Yang WK and Shen CY: PIK3CA as an
oncogene in cervical cancer. Oncogene. 19:2739–2744. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Su TH, Chang JG, Perng LI, Chang CP, Wei
HJ, Wang NM and Tsai CH: Mutation analysis of the putative tumor
suppressor gene PTEN/MMAC1 in cervical cancer. Gynecol Oncol.
76:193–199. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ben-Baruch A, Michiel DF and Oppenheim JJ:
Signals and receptors involved in recruitment of inflammatory
cells. J Biol Chem. 270:11703–11706. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cyster JG: Chemokines and the homing of
dendritic cells to the T cell areas of lymphoid organs. J Exp Med.
189:447–450. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cyster JG: Chemokines and cell migration
in secondary lymphoid organs. Science. 286:2098–2102. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luboshits G, Shina S, Kaplan O, Engelberg
S, Nass D, Lifshitz-Mercer B, Chaitchik S, Keydar I and Ben-Baruch
A: Elevated expression of the CC chemokine regulated on activation,
normal T cell expressed and secreted (RANTES) in advanced breast
carcinoma. Cancer Res. 59:4681–4687. 1999.PubMed/NCBI
|
|
10
|
Mrowietz U, Schwenk U, Maune S, Bartels J,
Küpper M, Fichtner I, Schröder JM and Schadendorf D: The chemokine
RANTES is secreted by human melanoma cells and is associated with
enhanced tumour formation in nude mice. Br J Cancer. 79:1025–1031.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Robinson SC, Scott KA, Wilson JL, Thompson
RG, Proudfoot AE and Balkwill FR: A chemokine receptor antagonist
inhibits experimental breast tumor growth. Cancer Res.
63:8360–8365. 2003.PubMed/NCBI
|
|
12
|
Ng-Cashin J, Kuhns JJ, Burkett SE,
Powderly JD, Craven RR, van Deventer HW, Kirby SL and Serody JS:
Host absence of CCR5 potentiates dendritic cell vaccination. J
Immunol. 170:4201–4208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van Deventer HW, O'Connor W Jr, Brickey
WJ, Aris RM, Ting JP and Serody JS: C-C chemokine receptor 5 on
stromal cells promotes pulmonary metastasis. Cancer Res.
65:3374–3379. 2005.PubMed/NCBI
|
|
14
|
Son JH, Kong TW, Kim SH, Paek J, Chang SJ,
Lee EJ and Ryu HS: Prediction of lymph node metastasis in patients
with apparent early endometrial cancer. Obstet Gynecol Sci.
58:385–390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, He Q, Shao YG and Ji M: Chemokines
fluctuate in the progression of primary breast cancer. Eur Rev Med
Pharmacol Sci. 17:596–608. 2013.PubMed/NCBI
|
|
16
|
Lin S, Wan S, Sun L, Hu J, Fang D, Zhao R,
Yuan S and Zhang L: Chemokine C-C motif receptor 5 and C-C motif
ligand 5 promote cancer cell migration under hypoxia. Cancer Sci.
103:904–912. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mango RL, Wu QP, West M, McCook EC, Serody
JS and van Deventer HW: C-C chemokine receptor 5 on pulmonary
mesenchymal cells promotes experimental metastasis via the
induction of erythroid differentiation regulator 1. Mol Cancer Res.
12:274–282. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Singh H, Sachan R, Jain M and Mittal B:
CCR5-Delta32 polymorphism and susceptibility to cervical cancer,
Association with early stage of cervical cancer. Oncol Res.
17:87–91. 2008.PubMed/NCBI
|
|
19
|
Wang L, Chang L, Li Z, Gao Q, Cai D, Tian
Y, Zeng L and Li M: miR-99a and −99b inhibit cervical cancer cell
proliferation and invasion by targeting mTOR signaling pathway. Med
Oncol. 31:9342014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou C, Li G, Zhou J, Han N, Liu Z and Yin
J: miR-107 activates ATR/Chk1 pathway and suppress cervical cancer
invasion by targeting MCL1. PLoS One. 9:e1118602014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roldo C, Missiaglia E, Hagan JP, Falconi
M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, et
al: MicroRNA expression abnormalities in pancreatic endocrine and
acinar tumors are associated with distinctive pathologic features
and clinical behavior. J Clin Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: MiR-107 and MiR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martello G, Rosato A, Ferrari F, Manfrin
A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T,
et al: A microRNA targeting dicer for metastasis control. Cell.
141:1195–1207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li F, Liu B, Gao Y, Liu Y, Xu Y, Tong W
and Zhang A: Upregulation of microRNA-107 induces proliferation in
human gastric cancer cells by targeting the transcription factor
FOXO1. FEBS Lett. 588:538–544. 2014. View Article : Google Scholar : PubMed/NCBI
|