Introduction
Prostate cancer presents with malignant tumors that
threaten the health of middle-aged and elderly men. Concomitant
with a global increase in the age of the population, the incidence
of prostate cancer has risen (1). No
effective method for the early diagnosis of this disease currently
exists. The decision to undergo surgical resection of prostate
tumors depends on the incidence of prostate cancer metastases; the
clinical diagnosis of metastases typically relies on bone scans,
but these cannot detect metastases in the sentinel lymph nodes or
soft tissue (2). However,
human-derived antibody preparation, together with
radioimmunoimaging technology have emerged as potential molecular
tools for the early detection and diagnosis of prostate cancer
metastases (3,4).
Prostate-specific membrane antigen (PSMA) is a type
II transmembrane glycoprotein located in the prostate epithelial
cells, and is comprised of 750 amino acid residues across 3
domains; these are an intracellular domain of 19 amino acids, a
transmembrane domain of 24 amino acids and an ectodomain of 707
amino acids. PSMA is a more sensitive and specific marker of
prostate tumors compared with prostate-specific antigen (4,5), as it
is highly expressed in prostate cancer, particularly
androgen-independent and metastatic prostate cancer, but is rarely
expressed in normal non-prostate tissue. PSMA has high tissue
specificity, making it an ideal target protein for the diagnosis
and treatment of prostate cancer (6); however, it also has an N-terminal
transmembrane domain with marked hydrophobicity, making it
unsuitable for use as an immunogen. To the best of our knowledge,
no study of PSMA ectodomain immunogenic sites has previously been
reported. As prokaryotically-expressed PSMA protein fragments
contain linear protein molecules that are not folded in
physiologically-appropriate three-dimensional conformations, the
immunogenicity of these linear protein molecules is unclear. In
addition, to the best of our knowledge, there have been no previous
reports on the binding specificity of prepared anti-PSMA. These
unknowns have hindered the study of the molecular diagnostic
imaging and radioimmunotherapy in the diagnosis and treatment of
prostate cancer. The present study used bioinformatics to predict
the immunogenicity of fragments of PSMA ectodomain polypeptide, and
the immune binding activity of these prokaryotically-expressed
fragments was characterized.
Materials and methods
Plasmids and strains
pET-32a (cat. no. 69015; Novagen, Inc., Madison, WI,
USA) plasmids acted as vectors. DH5α and BL21 (DE3) pLysS strains
(cat. no. CB106; Tiangen Biotech Co., Ltd., Beijing, China) of
competent Escherichia coli (E. coli) were used for
inducible protein expression.
Cells
LNCaP (androgen-sensitive human prostate
adenocarcinoma) and PC-3 (human prostate adenocarcinoma) cell lines
(Shanghai Institute of Biochemistry and Cell Biology, Chinese
Academy of Science, Shanghai, China) were used for verification of
polyclonal antibody binding.
Experimental animals
A total of three 6–8-week-old BALB/c male mice
(Shanghai Institute of Materia Medica, Chinese Academy of Science,
Shanghai, China), weighing 18–20 g, were maintained under specific
pathogen-free conditions in the present study. The protocol of the
present study was approved by the Institutional Animal Care and Use
Committee of Shanghai University of Traditional Chinese Medicine
(Shanghai, China).
Reagents
Reagents used were sourced as follows: Restriction
endonucleases KpnI and XhoI, RPMI-1650, fetal bovine
serum (Thermo Fisher Scientific, Inc., Waltham, MA, USA); T4
ligase, isopropyl-β-D-thiogalactopyranoside (IPTG), a 10 kb DNA
ladder (cat. no. B600024l; Sangon Biotech Co., Ltd., Shanghai,
China) a protein marker (cat. no. SM0431; Thermo Fisher Scientific,
Inc.), an EZNA plasmid extraction kit (cat. no. D6943; Omega
Bio-Tek Inc., Norcross, GA, USA); a plasmid purification kit (Merck
Sharpe & Dohme, Shanghai, China); acrylamide and methylene
bis-acrylamide (Genview Scientific Inc., El Monte, CA, USA); a
3,000-unit dialysis bag, sodium dodecyl sulfate (SDS) and Tris base
(Sino-American Biotechnology Co., Ltd., Shanghai, China); anti-PSMA
(YPSMA-1) mouse monoclonal antibody (cat. no. ab19071, Abcam,
Shanghai, China); and Freund's complete adjuvant and fluorescein
isothiocyanate (FITC)-labeled goat anti-mouse polyclonal
immunoglobulin G (cat. no. F9006; Sigma-Aldrich, St. Louis, MO,
USA). All reagents were of analytical grade.
Prediction of the PSMA ectodomain
polypeptide antigen
Using the primary structure of PSMA reported in
National Center for Biotechnology Information GenBank (AAA60209.1),
the hydrophilicity, antigen indices and homology of PSMA ectodomain
amino acid sequences were analyzed using ExPASy, Protean
(http://www.expasy.org/) and BLASTN software
(https://blast.ncbi.nlm.nih.gov/)
(7). Amino acid sequences with high
hydrophilicity and antigen indices, and low homology were selected
as polypeptide antigen sequences; specific immunogenic peptides
were then selected by determining the most appropriate polypeptide
antigen sequence and, thus, expression region of PSMA to be used.
The selected polypeptide immunogenic fragment is referred to as the
recombinant PSMA antigen ectodomain fragment
(r-ectodomain-PSMA).
Synthesis of the PSMA antigen
ectodomain fragment-encoding gene
Using the PSMA gene sequence (GenBank M9948.1) and
the prokaryotic expression host, the nucleotide sequence of the
PSMA ectodomain was optimized to account for preferred codon usage
in the expression host (7). The
nucleotide sequence was generated by Sangon Biotech Co., Ltd. The
synthesized gene fragment was then inserted into the pET-32a vector
at restriction enzyme sites KpnI and XhoI.
Construction of prokaryotic expression
vector
Double restriction endonuclease digestion using
KpnI and XhoI was performed in the target gene and
the pET-32a expression vector; this was followed by digested
product purification using TIANgel Extraction kit (cat. no. DP209;
Tiangen Biotech Co., Ltd.). The digested target fragment and
plasmid were ligated at 16°C for 4 h using T4 DNA ligase (cat. no.
RT406; Tiangen Biotech Co., Ltd.) and transformed into a DH5α
bacterial strain by a heat shock pulse at 42°C for 90 s. The
successfully transformed plasmid colonies growing on the agar plate
were selected and individually inoculated into fresh culture tubes,
which were incubated at 37°C in a 220 rpm shaker overnight. This
was followed by plasmid extraction using a EZNA plasmid extraction
kit. KpnI and XhoI double restriction endonuclease
digestions and recombinant plasmid sequencing were used to confirm
target gene insertion.
Inducible expression of recombinant
proteins
pET-32a-r-ectodomain-PSMA plasmid was transformed
into E. coli BL21 (DE3) pLysS. A single colony of positive
pET-32a-r-ectodomain-PSMA/E. coli BL21 (DE3) pLysS was
inoculated into 5 ml LB medium containing ampicillin (100 µg/ml)
and incubated overnight at 37°C in a 220-rpm shaker. The overnight
culture was inoculated into fresh LB medium (1% volume/volume)
containing a final concentration of 100 µg/ml ampicillin and
incubated at 37°C at 220 rpm until the culture reached an optical
density at 600 nm of 0.6–0.8; this was followed by addition of IPTG
solution, at a final concentration of 0.5 mM, to induce protein
expression. A number of incubation temperatures and durations (15
or 25°C overnight or 37°C for 5 h) of recombinant protein
expression induction were evaluated. Subsequent to induction,
recombinant protein expression was terminated by centrifuging the
solution at 15,294 × g, at 4°C for 5 min to collect the cells. The
harvested cells were then resuspended in phosphate-buffered saline
(PBS) and lysed by ultrasonication, and the lysate was centrifuged
at 20,817 × g, at 4°C for 30 min. The supernatant and pellet of the
lysate were analyzed using 10% SDS-polyacrylamide gel
electrophoresis (PAGE).
Purification using affinity
chromatography
Nickel ion affinity chromatography was performed in
accordance with a previous study by Liu et al (8). PBS at pH 7.0 was used to wash the
bacterial culture pellet. PBS (10 ml) and an additional 10 ml PBS
containing 8 mol/l urea were then added to resuspend the bacteria,
which was followed by ultrasonication and centrifugation at 20,817
× g, 4°C for 30 min. The pellet was dissolved in binding buffer in
a chromatography cabinet overnight. Dissolved pellet was then
centrifuged at 20,817 × g, 4°C for 30 min to collect the
supernatant. The supernatant was subsequently used for affinity
chromatography, following filtration through a 0.45 µm microporous
membrane. Eluted proteins were collected and dialysed successfully
using a 3,000-unit dialysis bag. The purity of the target protein
was evaluated using SDS-PAGE.
Western blot analysis
10% SDS-PAGE was used to separate the proteins,
which were then transferred to a 0.45-µm polyvinylidene fluoride
(PVDF) membrane using a semi-dry transfer device at 12 V for 40
min. The PVDF membrane was then blocked in Tris-buffered saline
with 0.1% Tween 20 (TBS-T), containing 5% bovine serum albumin
(BSA; cat. no. ST023; Beyotime Institute of Biotechnology, Haimen,
China), at 4°C overnight. Subsequent to being washed with TBS-T 4
times (10 min each), the membrane was incubated with a 1:500
dilution of YPSMA-1 (in 5% BSA-containing TBS-T) at room
temperature for 2 h. Subsequent to additional TBS-T washes, a
1:10,000 dilution of alkaline phosphatase-labeled goat anti-mouse
IgG (diluted in TBS-T containing 5% BSA) was incubated with the
membrane at room temperature for 2 h. A 5-bromo-4-chloro-3-indolyl
phosphate/nitro blue tetrazolium chloride kit (cat. no. C3206;
Beyotime Institute of Biotechnology) was used for the colorimetric
development of the PVDF membrane following additional washing
steps.
Preparation of the polyclonal
antibody
A mixture of 0.5 ml purified recombinant protein
(250 µg) and an equal volume of Freund's complete adjuvant were
used to immunize 3 male BALB/c mice at 6–8 weeks of age. Each mouse
was immunized subcutaneously at three sites on the back with 15 µg
protein in each injection site, 4 times at 20-day intervals,
followed by an intraperitoneally-administered booster dose (5th
dose) containing 23 µg protein. Serum from each animal was
collected 2 weeks after this immunization for measurement of the
titer using an indirect enzyme-linked immunosorbent assay (9).
Flow cytometry
The prostate cancer LNCaP cell line, expressing
PSMA, and the PC-3 cell line, not expressing PSMA, were cultured as
described in a previous study (10).
LNCaP and PC-3 cells were harvested during the logarithmic growth
phase in order to prepare live cell suspensions. Subsequent to
being washed in PBS, the cell suspension was diluted to
1×109 cells/l in PBS; 100 µl of cell suspension from
each cell line was then collected and incubated with a 1:2,500
dilution of recombinant PSMA mouse polyclonal antibodies at room
temperature for 30 min. Cells were washed 3 times with PBS and
centrifuged at 500 × g for 5 min at room temperature prior to
incubation with FITC-labeled goat anti-mouse IgG at room
temperature for a further 30 min, in the dark. The cells were
washed 3 subsequent times in PBS and centrifuged. Following this,
0.5 ml PBS buffer was added to resuspend the cells prior to
measurements and flow cytometry analyses.
Statistical analysis
Statistical analyses were performed using SPSS 15.0
software (SPSS, Inc., Chicago, IL, USA). Data were expressed as the
mean ± standard deviation. Differences were analyzed for
significance using a two-tailed Student's t-test for independent
means. P<0.05 was considered to indicate a statistically
significant difference.
Results
Prediction of the PSMA ectodomain
polypeptide immunogen
BLASTN, Protean and ExPASy software was used to
determine the hydrophilicity, immunogenic indices and amino acid
homology of 707 amino acids of the PSMA ectodomain. Table I demonstrates the 3 immunogenic
polypeptide sequences selected for high hydrophilicity and
immunogen indices, and low amino acid homology. The polypeptide
immunogens 1 and 2, which had higher priority scores and were in
the C-terminal region of PSMA were therefore selected. The
expression region containing polypeptide immunogen 1 and 2 is
referred to as the recombinant PSMA ectodomain polypeptide
immunogenic fragment, and this contained a total of 310 amino acids
(amino acids 440–750).
 | Table I.Predicted polypeptide immunogens. |
Table I.
Predicted polypeptide immunogens.
| Polypeptide
immunogen | Amino acid
sequence | Sequence
beginning | Sequence end | Length, amino
acids |
|---|
| Polypeptide immunogen
1 | ESWTKKSPSPEFSGM | 495 | 509 | 15 |
| Polypeptide immunogen
2 | APSSHNKYAGESFPG | 693 | 707 | 15 |
| Polypeptide immunogen
3 |
KMGGSAPPDSSWRGS | 308 | 322 | 15 |
Construction of the expression
vector
Using the whole gene sequence of PSMA, the
polypeptide immunogenic fragment sequence was optimized to account
for preferred codon usage, and this was followed by oligonucleotide
synthesis. The synthesized target DNA fragment was ligated into the
pET-32a vector in order to construct the expression vector
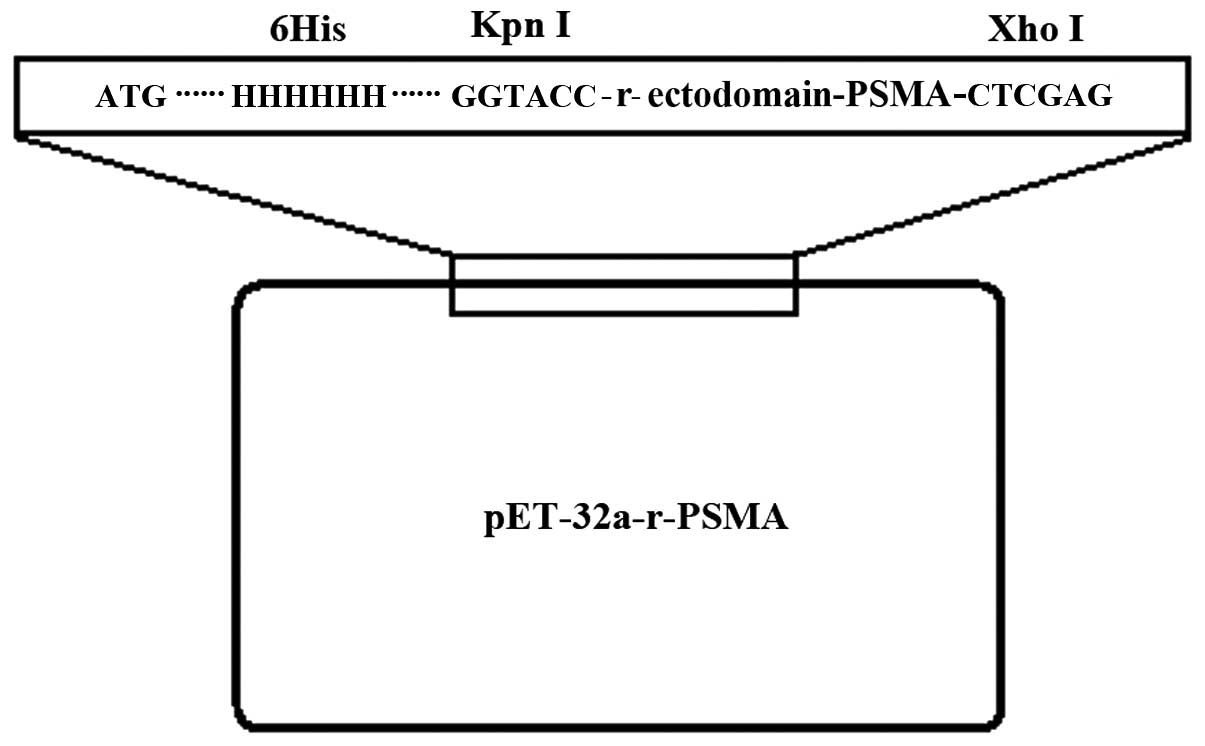
pET-32a-r-ectodomain-PSMA (Fig. 1).
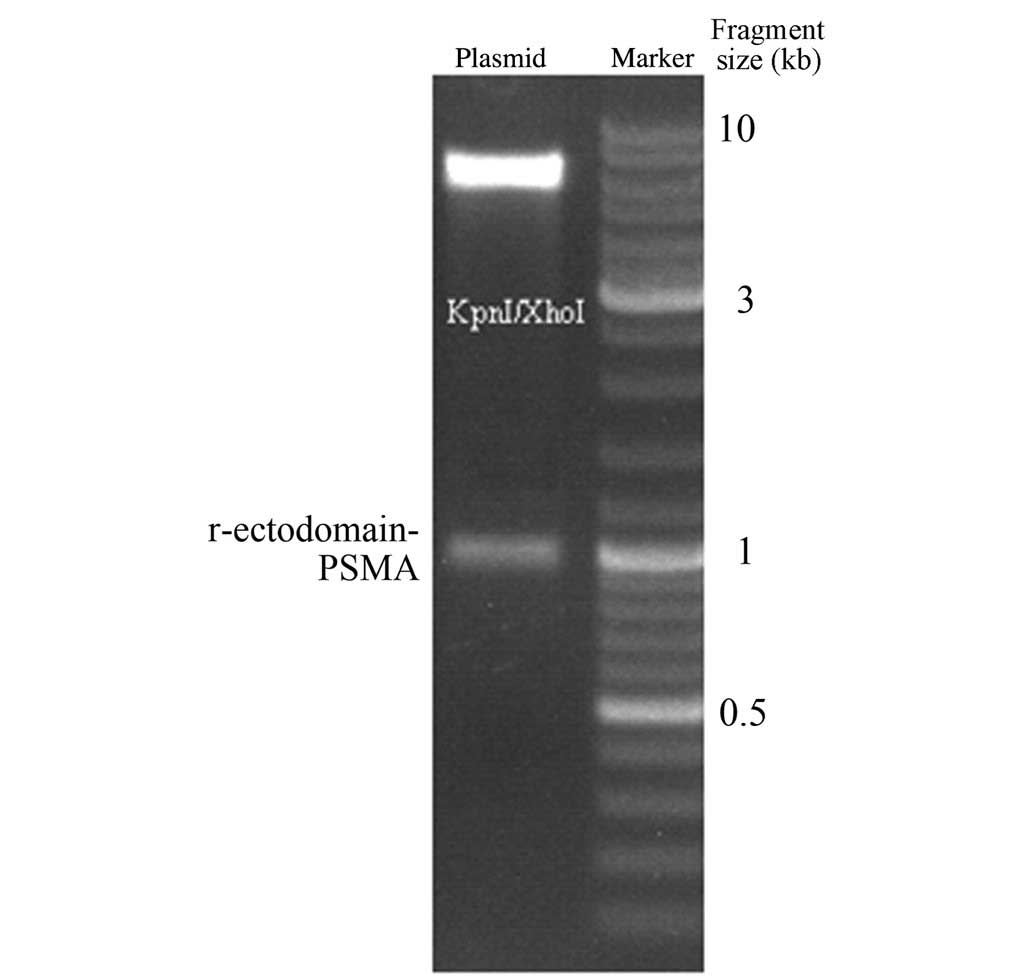
The pET-32a-r-ectodomain-PSMA was then digested by restriction
endonuclease KpnI and XhoI. Gel electrophoresis
revealed that the cleavage of KpnI and XhoI generated
the target fragment, which was consistent with the expected
fragment size of 930 bp (Fig. 2).
The endonuclease cleavage fragment was verified by sequencing, and
the results of this were consistent with the PSMA sequence,
indicating successful construction of the expression vector.
Expression of the r-ectodomain-PSMA
recombinant protein
Subsequent to transforming the
pET-32a-r-ectodomain-PSMA construct into E. coli BL21 (DE3)
pLysS, protein expression was induced using 0.5 mM IPTG under
varying conditions. Upon terminating the expression, 10% SDS-PAGE
was used to verify differing protein expression. The target protein
was expressed in the pellet at 15°C, 25°C and 37°C (Fig. 3). The most abundant protein
expression was induced by 0.5 mM IPTG overnight at 25°C.
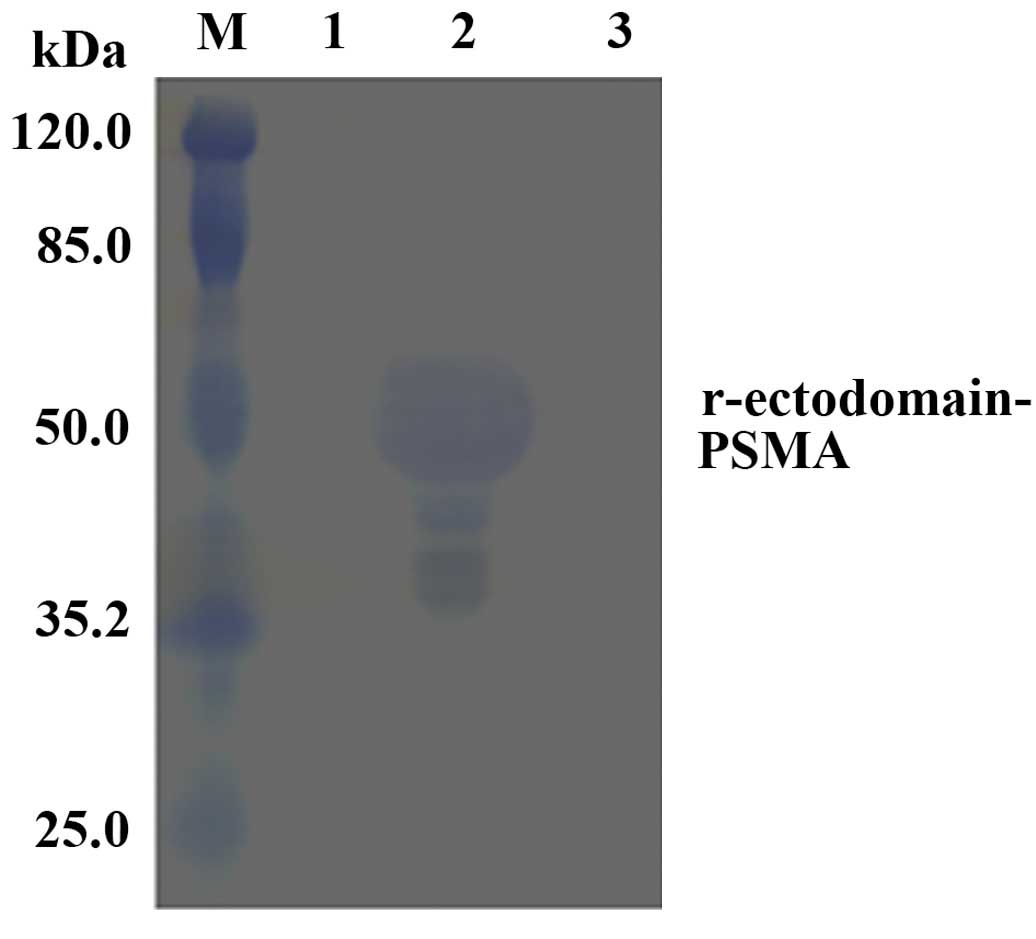
Purification of the r-ectodomain-PSMA
recombinant protein
Nickel ion affinity chromatography was conducted
using marker protein 6His to purify the target protein and collect
imidazole-eluted protein. SDS-PAGE revealed that the size of the
purified target protein matched its predicted size of 50 kDa
(Fig. 4), with ~95% purity.
Western blot analysis of the
r-ectodomain-PSMA recombinant protein
Anti-YPSMA-1 ectodomain monoclonal antibody was used
in western blot analysis to assess the expressed r-ectodomain-PSMA
recombinant protein. The fusion protein was demonstrated to
specifically bind the anti-PSMA ectodomain monoclonal antibody
(Fig. 5).
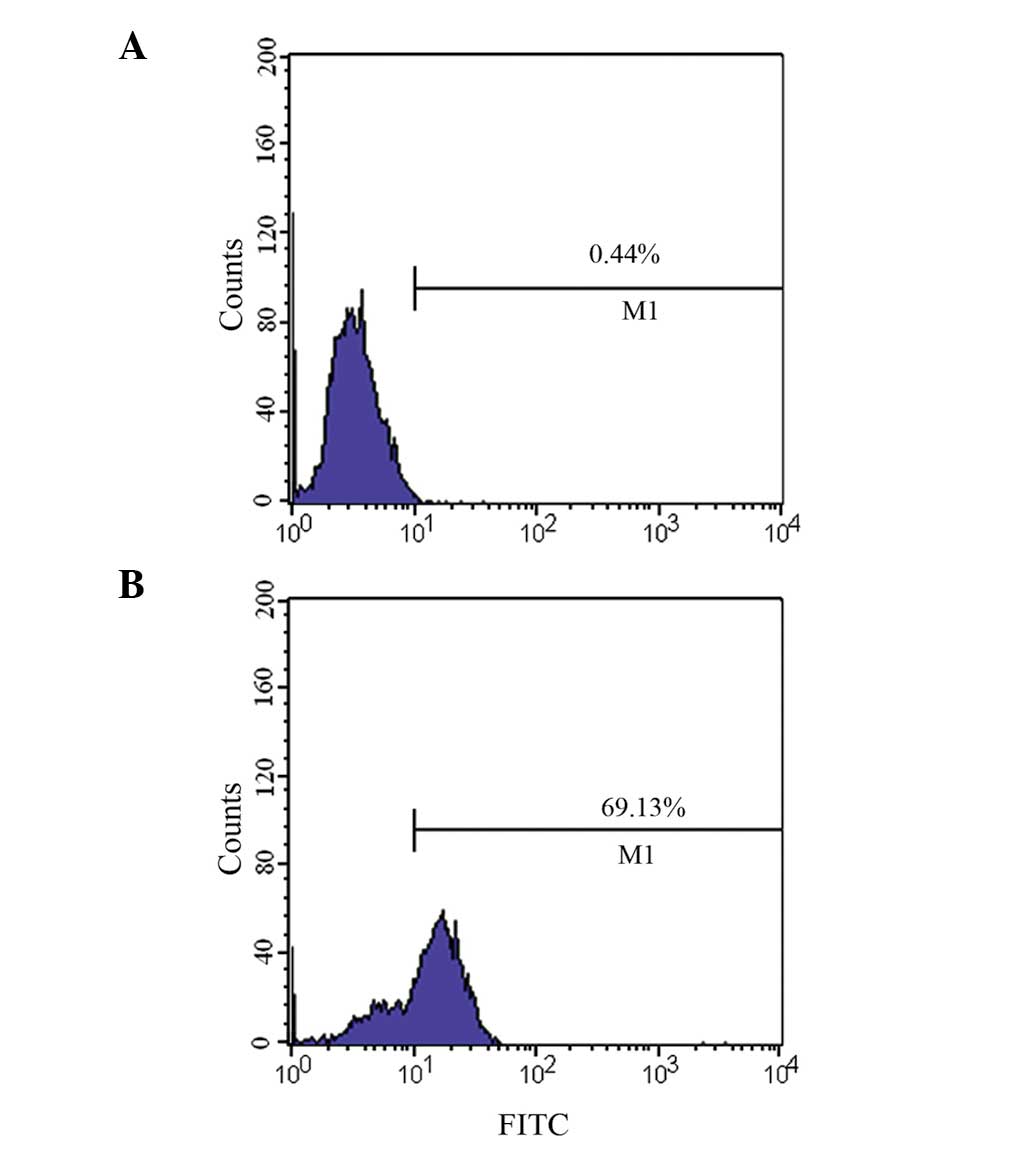
Verification of binding activity of
polyclonal antibodies using flow cytometry
Flow cytometry (Fig.
6) revealed that the binding rate of PSMA-positive LNCaP cells
to the recombinant protein polyclonal antibody was 69.1%, whilst
the binding rate of PSMA-negative PC-3 to the recombinant protein
polyclonal antibody was 0.44% (basically no binding).
Discussion
Antibody recognition of an antigen does not involve
recognition of the whole molecule but rather the immunogenic
epitopes (11). For immunogenic
recognition, an immunogen does not necessarily have to express the
full-length protein; 300–400 amino acids in the N-terminal and
C-terminal regions of proteins are frequently recognized as
immunogenic fragments. In accordance with this, the present study
predicted 3 PSMA immunogenic fragments with high immunogenicity
(Table I). Peptides in the
N-terminal and C-terminal regions are preferable to peptides in the
middle of a protein for immunogen selection, as peptides in the
middle of a protein are more likely to reside inside the protein
upon folding; in the present study, polypeptide immunogen 3 was
eliminated for this reason. The entire protein was also too large
to be suitable for prokaryotic expression. However, the length of
the C-terminal region containing the predicted polypeptide
immunogens 1 and 2 is between 300–400 amino acids, and for this
reason, a fragment of 310 amino acids at the C-terminal of PSMA
ectodomain (amino acids 440–750) was selected for prokaryotic
expression.
Whole sequence synthesis of the 310 amino acids of
the C-terminal region was performed by ligating the peptide into a
pET-32a vector, thereby constructing the expression vector of
pET-32a-r-ectodomain-PSMA. Subsequent to the restriction
endonuclease KpnI and XhoI cleavage of
pET-32a-r-ectodomain-PSMA, the size of the target fragment was
verified using gel electrophoresis; this was confirmed to be
consistent with the expected fragment size of 930 bp, indicating a
successful construction of the recombinant plasmid. As demonstrated
by Fig. 3, the optimal conditions
for recombinant protein induction were overnight and at 25°C.
Following nickel ion affinity chromatography, SDS-PAGE analysis
revealed that the purified protein size (~50 kDa) was consistent
with the predicted value.
The present study used YPSMA-1 ectodomain monoclonal
antibody to verify the target protein expression. The binding site
of this antibody was within the 716–723 amino acids of the PSMA
ectodomain (12). In the current
study, the expressed PSMA ectodomain protein fragment also
contained the amino acids of the antibody-binding site. Application
of YPSMA-1 in western blot analysis revealed the expression of
recombinant protein as a clear 50-kDa protein band (Fig. 5), indicating immunogenic activity of
the prepared recombinant protein. Furthermore, recombinant protein
addition combined with adjuvant immunization in BALB/c mice
produced a high-titer polyclonal antibody. Flow cytometry confirmed
that this antibody could bind to PSMA-positive LNCaP cells, but not
to PSMA-negative PC-3 cells (Fig.
6), indicating that the polyclonal antibodies prepared using
the immunogenic fragments produced a specific immune response.
The present study demonstrated that the anti-PSMA
ectodomain monoclonal antibody has considerable value in the
diagnosis of prostate cancer in radioimmunoimaging. However, the
penetrability and clearance of large monoclonal antibodies is
generally poor in vivo. Repeated use of murine monoclonal
antibodies can lead to human anti-mouse antibody reactions,
limiting clinical applications (13–15). For
this reason, small humanized antibodies have emerged as possible
diagnostic tools, and the PSMA ectodomain recombinant protein
generated in the current study provides a basis for screening and
production of small humanized antibodies.
References
|
1
|
Yu SQ and Xia SJ: Development of molecular
targeted treatment for prostate cancer. Zhong Hua Yi Xue Za Zhi
Bian Ji Bu. 87:718–719. 2007.(In Chinese).
|
|
2
|
Ross JS, Gray KE, Webb IJ, Gray GS, Rolfe
M, Schenkein DP, Nanus DM, Millowsky MI and Bander NH:
Antibody-based therapeutics: Focus on prostate cancer. Cancer
Metastasis Rev. 24:521–537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou LL and Wang XS: Overview of
prostate-specific membrane antigen. An Hui Hua Gong Bian Ji Bu.
35:28–31, 34. 2010.(In Chinese).
|
|
4
|
You J, Cozzi P, Walsh B, Willcox M,
Kearsley J, Russell P and Li Y: Innovative biomarkers for prostate
cancer early diagnosis and progression. Crit Rev Oncol Hematol.
73:10–22. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perner S, Hofer MD, Kim R, Shah RB, Li H,
Möller P, Hautmann RE, Gschwend JE, Kuefer R and Rubin MA:
Prostate-specific membrane antigen expression as a predictor of
prostate cancer progression. Hum Pathol. 38:696–701. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mhawech-Fauceglia P, Zhang S, Terracciano
L, Sauter G, Chadhuri A, Herrmann FR and Penetrante R:
Prostate-specific membrane antigen (PSMA) protein expression in
normal and neoplastic tissues and its sensitivity and specificity
in prostate adenocarcinoma: An immunohistochemical study using
mutiple tumour tissue microarray technique. Histopathology.
50:472–483. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao Z, Jiang X, Beckett ML and Wright GL
Jr: Generation of a baculovirus recombinant prostate-specific
membrane antigen and its use in the development of a novel protein
biochip quantitative immunoassay. Protein Expr Purif. 19:12–21.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu T, Toriyabe Y and Berkman CE:
Purification of prostate-specific membrane antigen using
conformational epitope-specific antibody-affinity chromatography.
Protein Expr Purif. 49:251–255. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zuo X, Liu C, Tang G and Jin Y: Mouse PD-1
gene fragment expression of extracellular domain, purification, and
its polyclonal antibody preparation. Zhong Guo Mian Yi Xue Za Zhi
Bian Ji Bu. 10:919–922. 2010.(In Chinese).
|
|
10
|
Tu SH, Shen JF, Tao R, Ji XW and Wang YC:
The preparation of 99Tcm-J591 and its imaging of nude mice bearing
human prostate cancer. Ji Chu Yi Xue Yu Lin Chuang. 33:284–288.
2013.(In Chinese).
|
|
11
|
Liu H, Liu B, He X and Zhao XT:
Expression, purification, and identification the major epitope
region of herpes simplex virus type 1 glycoprotein D. Ji Chu Yi Xue
Yu Lin Chuang Bian Ji Bu. 33:350–355. 2013.(In Chinese).
|
|
12
|
Tykvart J, Navrátil V, Sedlák F, Corey E,
Colombatti M, Fracasso G, Koukolík F, Bařinka C, Šácha P and
Konvalinka J: Comparative analysis of monoclonal antibodies against
prostate-specific membrane antigen (PSMA). Prostate. 74:1674–1690.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Osborne JR, Akhtar NH, Vallabhajosula S,
Anand A, Deh K and Tagawa ST: Prostate-specific membrane
antigen-based imaging. Urol Oncol. 31:144–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holland JP, Divilov V, Bander NH,
Smith-Jones PM, Larson SM and Lewis JS: 89Zr-DFO-J591 for immunoPET
of prostate-specific membrane antigen in vivo. J Nucl Med.
51:1293–1300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kampmeier F, Williams JD, Maher J, Mullen
GE and Blower PJ: Design and preclinical evaluation of a
99mTc-labelled diabody of mAb J591 for SPECT imaging of
prostate-specific membrane antigen (PSMA). EJNMMI Res. 4:132014.
View Article : Google Scholar : PubMed/NCBI
|