Introduction
Tumor hypoxia was first described in the 1950s, and
there is currently increasing evidence to indicate that it is a
common feature in numerous types of cancer, including
hepatocellular carcinoma (HCC) (1,2).
Although HCC is among the most hypervascular tumor types, it
contains hypoxic regions due to rapid cell proliferation and
aberrant formation of blood vessels (3). The effects of hypoxia on cells are
predominantly mediated by hypoxia-inducible factors (HIF), which
consist of an oxygen-regulated subunit (HIF-1α, HIF-2α or HIF-3α)
and a constitutively expressed HIF-1β subunit (4).
Under normal oxygen pressure and in the presence of
Fe2+ and acetone dicarboxylic acid, prolyl hydroxylase
domain (PHD) catalyzes the hydroxylation of key amino acid residues
in the HIF-α oxygen-dependent degradation domain (5). Hydroxylated HIF-α binds to the Von
Hippel-Lindau tumor suppressor and is rapidly degraded via a
ubiquitin-proteasome pathway (6).
Under hypoxic conditions, HIFs are not modified by PHDs, but
dimerize with the aryl-hydrocarbon receptor nuclear translator
(ARNT)/HIF-1β via interactions with helix-loop-helix and
Per/Arnt/Sim domains (5). The HIF
heterodimers are translocated to the nucleus, and co-activators
such as CBP/p300 are recruited (5).
The HIF heterodimers recognize and bind hypoxia response elements
(HREs) that contain a consensus sequence (G/A) CGTG within the
promoter regions of target genes to drive adaptive gene
transcription (7,8). It has been reported that HIF-1 and
HIF-2 regulate the expression of hundreds of genes that are
involved in numerous processes associated with cancer biology,
including cell survival, tumor angiogenesis, metastasis and
resistance to radiation and chemotherapy (8,9).
HIF-3α is a member of the HIF family and was
initially discovered by Gu et al (10) in 1998. HIF-3α has relatively low
sequence identities with HIF-1α and HIF-2α (10). HIF-1α and HIF-2α have two
transactivation domains (TADs) (11), while HIF-3a has only one TAD
(12). HIF-3α has a unique leucine
zipper domain and an LXXLL (L is Leucine and X is any amino acid)
motif (10). These unique structural
features are evolutionarily conserved. Compared with HIF-1α and
HIF-2α, which have been studied extensively (5), little is known about the regulation and
function of HIF-3α. Recent studies have indicated that hypoxia
induces HIF-3α expression, and that HIF-3α may be a target gene of
HIF-1 and HIF-2 (13–15). HIF-3 may suppress the expression of
genes that are typically inducible by HIF-1α and HIF-2α in tumor
cells, and therefore, it may be a negative regulator of gene
expression in response to hypoxia (12,14).
The expression pattern of HIF-3α in HCC tissues is
currently unknown, and only a few studies have investigated the
association between HIF-3α expression and the expression of HIF-1α
and HIF-2α (13–15); however, the results are inconsistent
or even conflicting. To determine the role of HIF-3α in
hepatocarcinogenesis, immunostaining was used herein to compare
HIF-3α expression in HCC and paired peritumoral tissues obtained
from 126 clinical samples. Furthermore, the association between the
expression of HIF-3α and HIF-1α/HIF-2α was assessed in HCC clinical
tissues and the human cell lines PLC/PRF/5 and Hep3B.
Materials and methods
Patients and specimens
Tissue samples from a total of 126 patients with HCC
that underwent a surgical liver resection were obtained between
October 2005 and June 2009. Tissue samples for 76 patients were
obtained from the Department of Hepatobiliary Surgery of the
Affiliated Hospital of Guiyang Medical College (Guiyang, China) and
the remaining samples were sourced from 50 patients at the
Department of General Surgery of Center Hospital of Huanggang
(Huanggang, China). Informed consent was obtained from all
patients. The research protocol was approved by the Human Ethics
Committees of the Guiyang Medical College and the Center Hospital
of Huanggang. All tissue specimens were obtained from the patients
prior to any medical treatments. Peritumoral tissues were obtained
from at least 2 cm away from the primary tumor site. All patients
tested positively for the hepatitis B antigen HBsAg and negatively
for hepatitis C virus and human immunodeficiency virus. The cohort
had 110 males and 16 females, with an average age of 48.8 years and
an age range of 19–66 years. The maximum diameter of HCC tissue was
<5.0 cm in 60 patients and was ≥5.0 cm in 66 patients. Data from
follow-up examinations following liver resection were collected for
all patients. The clinical pathological features of the 126 HCC
patients are listed in Table I.
 | Table I.Correlations between HIF-3α protein
expression in surgical specimens of HCC and clinicopathological
characteristics. |
Table I.
Correlations between HIF-3α protein
expression in surgical specimens of HCC and clinicopathological
characteristics.
|
| HIF-3α |
|
|---|
|
|
|
|
|---|
| Parameter | Low | High | P-value |
|---|
| Age (years) |
|
| 0.200 |
|
<50 | 13 | 21 |
|
|
≥50 | 47 | 45 |
|
| Gender |
|
| 0.202 |
|
Male | 50 | 60 |
|
|
Female | 10 | 6 |
|
| Cirrhosis |
|
| 0.595 |
|
Absent | 40 | 41 |
|
|
Present | 20 | 25 |
|
| Tumor size
(cm) |
|
| 0.114 |
|
<5 | 33 | 27 |
|
| ≥5 | 27 | 39 |
|
| AFP (µg/l) |
|
| 0.377 |
|
<400 | 32 | 30 |
|
|
≥400 | 28 | 36 |
|
| Histological
grade |
|
| 0.676 |
|
Well | 9 | 8 |
|
|
Moderate | 43 | 46 |
|
|
Poor | 8 | 12 |
|
| Capsular
infiltration |
|
| 0.526 |
|
Absent | 44 | 45 |
|
|
Present | 16 | 21 |
|
| Vascular
invasion |
|
| 0.098 |
|
Absent | 42 | 59 |
|
|
Present | 15 | 10 |
|
Cell culture and transfection
The human HCC cell lines PLC/PRF/5 and Hep3B were
purchased from the Institute of Biochemistry & Cell Biology of
the Shanghai Institutes for Biological Sciences (Chinese Academy of
Sciences, Shanghai, China). HIF-1α and HIF-2α expression plasmids
and the control plasmid pcDNA3.1 were purchased from Shanghai Gene
Chem Co., Ltd. (Shanghai, China). Cells were grown in six-well
plates with Dulbecco's modified Eagle's medium (DMEM; GE Healthcare
Life Sciences, Logan, UT, USA) containing 10% fetal bovine serum
(FBS), 2 mmol/l L-glutamine, 50 units/ml penicillin and 50 g/ml
streptomycin (all GE Healthcare Life Sciences) at 37°C and in 5%
CO2. PLC/PRF/5 and Hep3B cells at 70–80% confluence were
transfected with 1.2 µg plasmids using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc., Waltham, MA, USA), following
the manufacturer's protocol. Transfected cells were incubated at
37°C for 6 h, and were then cultured for a further 16 h with fresh
DMEM medium containing 10% FBS. The mRNA and protein expression
levels of the target genes in the transfected cells were analyzed
using quantitative reverse transcription polymerase chain reaction
(RT-qPCR) and western blot analysis, respectively.
RT-qPCR
Total RNA was isolated from the PLC/PRF/5 and Hep3B
cells using TRIzol reagent (Thermo Fisher Scientific, Inc.), as
previously described (16,17). To avoid genomic DNA contamination,
RNA samples were treated with RNase-free DNase I ((Takara Bio,
Inc., Shiga, Japan) for 20 min at 37°C. The quantity and quality of
total RNA were determined using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc.). cDNA synthesis was performed at
42°C for 60 min in a total volume of 25 µl containing 2 µg RNA, 1.6
µM oligo(dT)18, 0.6 µM dNTPs, 200 U M-MLV reverse transcriptase,
and 10X Reaction buffer (all Promega Corporation, Madison, WI,
USA). All researchers received biosafety training and were screened
and vaccinated against hepatitis B virus. qPCR analysis was
performed as described previously (16,18),
using a reaction mixture consisting of 10 µl 2X SYBR Green mix
(Invitrogen; Thermo Fisher Scientific, Inc.), 2 µl cDNA template,
0.6 µl each of forward and reverse primers (10 µM) and double
distilled H2O, to a final volume of 20 µl. qPCR was
performed on a ABI 7500 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with the following cycling program:
Denaturation at 94°C for 1 min, followed by 40 cycles at 94°C for 1
min, 55°C for 1 min and 72°C for 1 min. The primer sequences were
as follows: HIF-1α forward, 5′-ACTTCTGGATGCTGGTGATTTG-3′ and
reverse, 5′-GCTTCGCTGTGTGTTTTGTTCT-3′; HIF-2α forward,
5′-TCATGCGACTGGCAATCAGC-3′ and reverse, 5′-GTCACCACGGCAATGAAACC-3′;
HIF-3α forward, 5′-CCTGGACATGAAGTTCACCTACTG-3′ and reverse,
5′-GGAAGCGATACTGCCCTGTTA-3′; and β-actin forward,
5′-AGTTGCGTTACACCCTTTCTTGAC-3′ and reverse,
5′-GCTCGCTCCAACCGACTGC-3′. The number of replications was three for
each sample. Reactions without template cDNA were used as negative
control. The cycle quantification (Cq) values were determined and
the data were analyzed using the 2−ΔΔCq method (19), following normalization to of
β-actin.
Immunohistochemical (IHC) analysis and
scoring of protein expression
IHC analysis of the tissue samples was performed as
described previously (18). Briefly,
the tissue samples were fixed using 10% formaldehyde, embedded in
paraffin (both Boster Biological Technology, Ltd., Wuhan, China)
and rehydrated using ethanol, after which endogenous peroxidase
activity was blocked using 3% H2O2 in
methanol solution (Boster Biological Technology, Ltd.).
Subsequently, the tissue samples were cut into 5 µm sections using
a microtome (Leica RM2155; Leica Microsystems GmbH, Wetzlar,
Germany). The sections were then immersed in citrate buffer (pH
6.0; Boster Biological Technology, Ltd.) and heated in a microwave
oven for 15 min in order to unmask the antigens, after which the
sections were incubated with mouse anti-HIF-3α (1:250; cat. no.
NBP2-45735; Novus Biologicals LLC, Littleton, CO, USA) at 37°C for
1 h and then overnight at 48°C. After washing three times with
phosphate-buffered saline, the sections were incubated with
biotin-conjugated goat anti-mouse immunoglobulin G (IgG) secondary
antibody (1:200; cat. no. sc-2075; Santa Cruz Biotechnology, Inc.,
Dallas, Texas, USA), followed by incubation with horseradish
peroxidase (HRP)-conjugated streptavidin (Boster Biological
Technology, Ltd.) for 20 min. Detection of immunoreactivity was
performed using 3,3′-diaminobenzidine (Boster Biological
Technology, Ltd.) under a FV300 fluorescent microscope (Olympus
Corporation, Tokyo, Japan).
The protein expression level of HIF-3α was assessed
using the following classification system based on the number of
cells with cytoplasmic and nuclear staining: I) No staining; II)
nuclear staining in <10% of cells and/or with weak cytoplasmic
staining; III) nuclear staining in 10–50% of cells and/or with
distinct cytoplasmic staining; and IV) nuclear staining in >50%
of cells and/or with strong cytoplasmic staining (20–23). The
staining scores I and II were considered to be low expression,
while the scores III and IV were considered as high expression
(20–23). The protein level of HIF-1α and HIF-2α
in HCC and paired peritumoral tissues has been described in our
previous study (18).
Protein preparation and western blot
analysis
Cells were lysed in lysis buffer (50 mM Tris, pH
7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 500 mM
NaCl, 10 mM MgCl2, 10 µg/ml leupeptin, 10 µg/ml
aprotinin, 10 ml/l NP-40, 0.2 g/l NaN3 and 1 mM PMSF;
Bio-Rad Laboratories, Inc., Hercules, CA, USA) and the protein
concentration was quantified using a Bradford Protein Assay
(Bio-Rad Laboratories, Inc.). A total of 30–50 µg protein from each
sample was separated using 10% SDS-PAGE, transferred to
polyvinylidene fluoride membranes, and blocked with 5% nonfat milk
in Tris-buffered saline and Tween 20 (Boster Biological Technology,
Ltd.) for 2 h (24). The membranes
were then incubated with mouse anti-HIF-3α (1:250; cat. no.
NBP2-45735; Novus Biologicals LLC), HIF-1α (1:1,000; cat. no.
sc-53546; Santa Cruz Biotechnology, Inc.), HIF-2α (1:250; cat. no.
sc-13596; Santa Cruz Biotechnology, Inc.) and β-actin (1:3,000;
cat. no. sc-47778; Santa Cruz Biotechnology, Inc.) monoclonal
antibodies for 1 h at 37°C and at 4°C overnight. The membranes were
washed and then incubated with HRP-conjugated goat anti-mouse IgG
monoclonal antibody (1:2,000, cat. no. sc-2005; Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. The bands were
visualized using an enhanced chemiluminescence detection system
(SuperSignal West Pico Chemiluminescent Substrate; cat. no. 34080;
Thermo Fisher Scientific, Inc.) and band intensities were
calculated using ImageJ software, version 1.41 (https://imagej.nih.gov/ij/).
Statistical analysis
Statistical analysis was performed using SPSS
software, version 16.0 (SPSS, Inc., Chicago, IL, USA). Continuous
variables were expressed as the mean ± standard deviation and
analyzed using a two-tailed Student's t-test. The correlation
analyses between HCC clinicopathological parameters and HIF-3α
expression were conducted using a two-tailed Mann-Whitney U-test.
Survival curves were computed by Kaplan-Meier analysis. Linear
associations were evaluated using Spearman's Rank or Pearson's
correlation coefficients. Prognostic significance was analyzed
using a log-rank test. P<0.05 was considered to indicate a
statistically significant difference.
Results
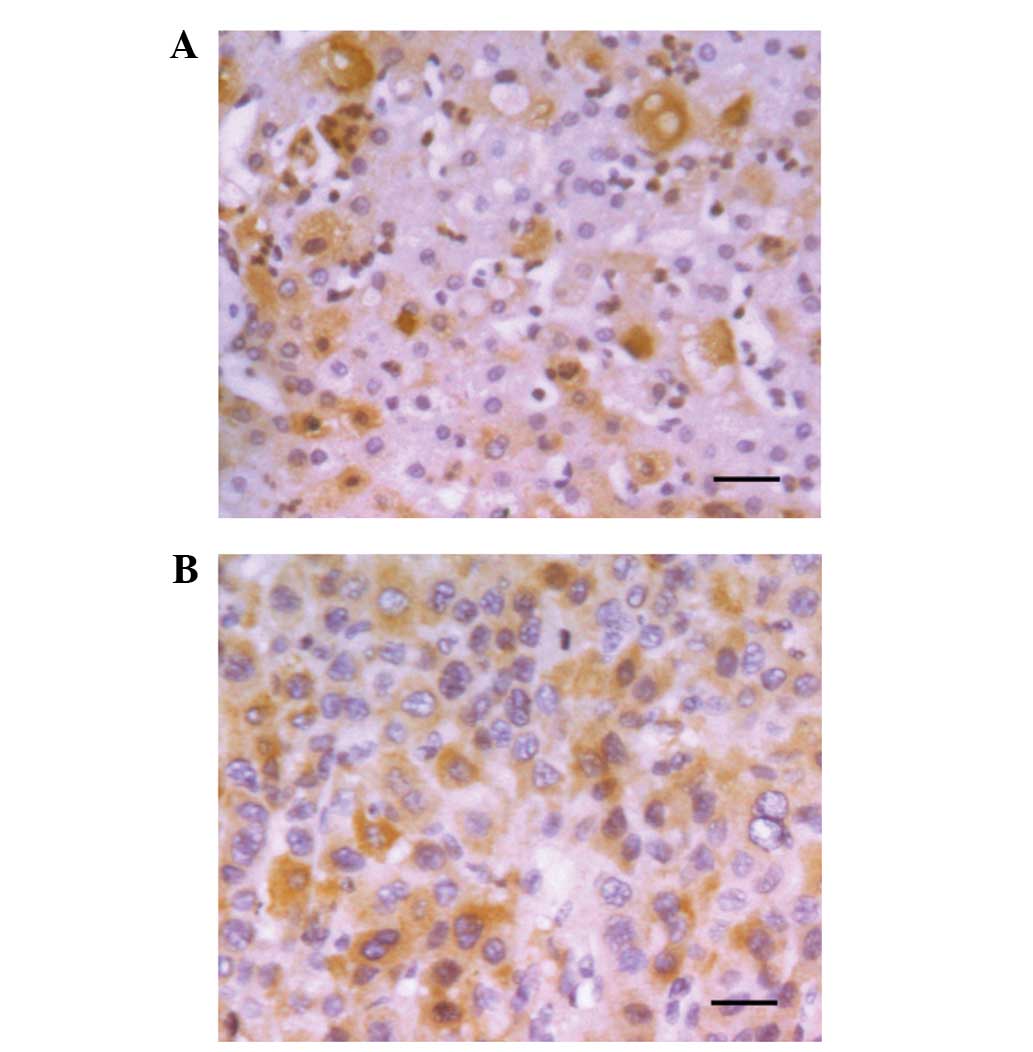
Expression of HIF-3α in HCC
tissues
The protein level of HIF-3α was measured using IHC
analysis on paraffin-embedded sections of 126 human HCC samples and
paired peritumor tissues. Positive staining of HIF-3α was located
in cytoplasm and/or nuclei of HCC tissues, with representative
staining shown in Fig. 1A. High
expression of HIF-3α was found in 66/126 tumor tissues (52.3%) and
63/126 peritumoral tissues (50.0%). The expression of HIF-3α was
upregulated in 46.0% (58/126) and downregulated in 42.9% (54/126)
of tumor tissues, when compared with the peritumoral tissues. No
obvious difference in HIF-3α expression was identified between the
remaining 11.1% (14/126) of tumor and peritumoral tissues. Our
previous study showed that HIF-1α was higher in HCC tissues
compared with peritumoral tissues (18). Therefore, as opposed to HIF-1α, the
hypoxic microenvironment in liver cancer did not increase HIF-3α
protein expression.
Next, we analyzed the association between HIF-3α
expression and pathological features of HCC. As shown in Table I, no significant correlation was
found between HIF-3α expression and the clinicopathological
features of HCC, including age, gender, presence of liver
cirrhosis, tumor size, serum α-fetoprotein level, tumor
differentiation grade, capsular infiltration and portal vein
invasion.
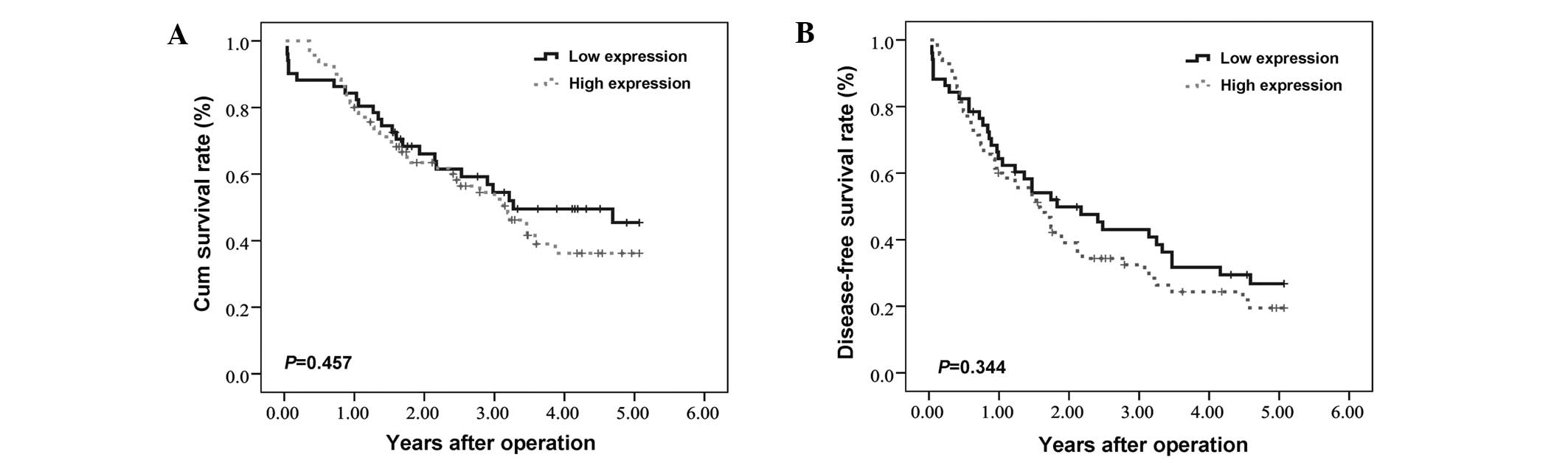
Disease-free survival (DFS) and
overall survival (OS) of HCC patients
The correlation between HIF-3α expression and
long-term patient survival following hepatectomy was analyzed. No
significant correlation was found between HIF-3α expression levels
in HCC tissues and the OS or DFS times of HCC patients. The mean OS
periods for patients with high and low HIF-3α expression levels in
their tumor tissues were 36.5±2.7 and 39.0±3.3 months (P=0.457),
respectively. The mean DFS period of the patients with high and low
HIF-3α expression levels in their tumor tissues were 25.9±2.6 and
29.9±3.3 months (P=0.344, Fig. 2),
respectively.
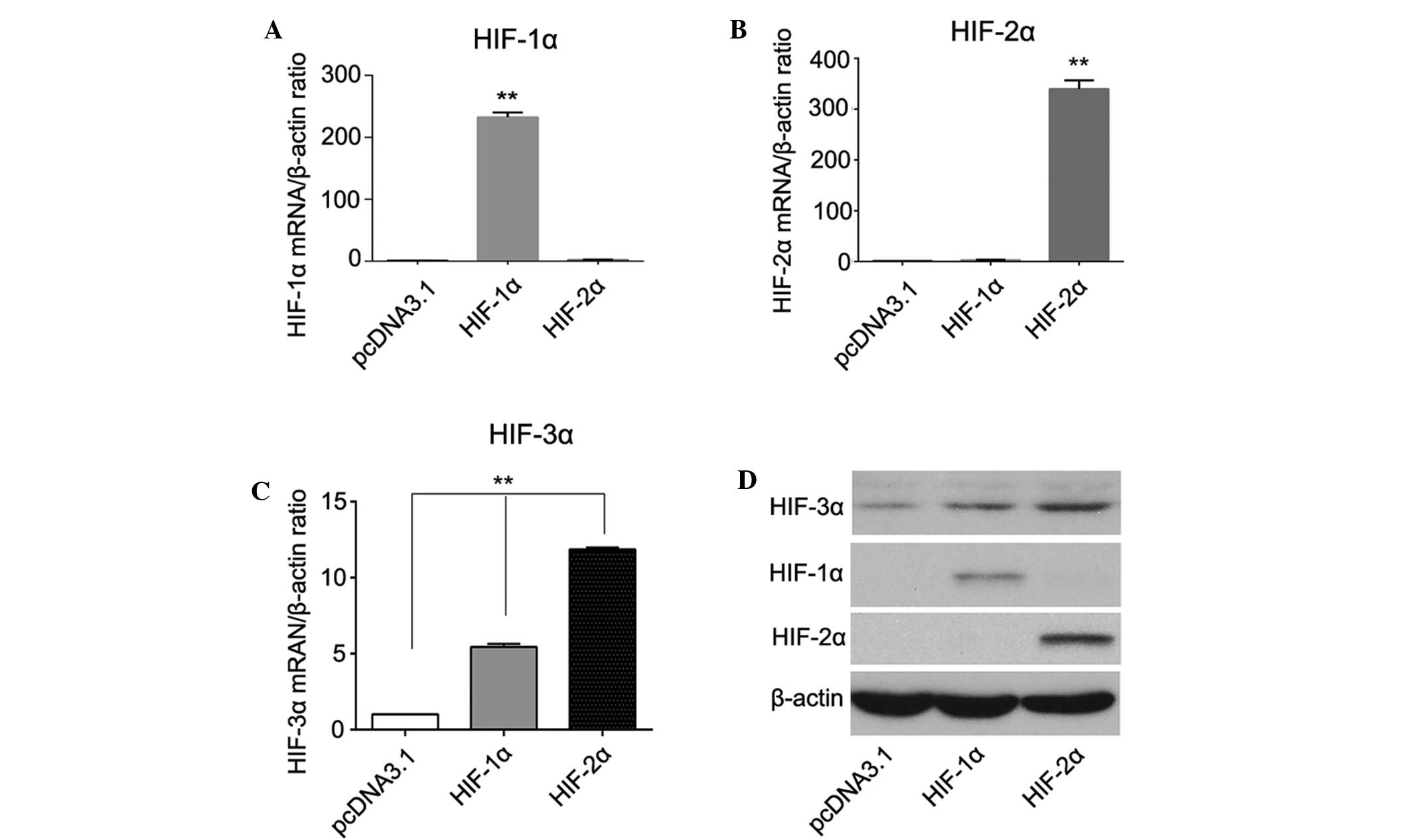
Association between HIF-3α expression
and the expression of HIF-1α and HIF-2α
A few studies have reported an association between
HIF-3α expression and the expression of HIF-1α and HIF-2α (13–15);
however, the results are inconsistent or even conflicting.
Therefore, we investigated their relationship both in vivo
and in vitro. In HCC tissues, Spearman correlation analysis
revealed that the expression of HIF-3α was significantly correlated
with the expression of HIF-2α (rs=0.198, P=0.030), but
not with HIF-1α expression (rs=0.045, P=0.855) (data not
shown). In addition, PLC/PRF/5 and Hep3B cells transfected with
either HIF-1α or HIF-2α plasmids expressed a higher level of HIF-3α
compared with the pcDNA2.1 control cells. The elevation of HIF-3α
was higher in the HIF-2α overexpressing cells compared with the
HIF-1α overexpressing cells, indicating that HIF-3α may be a target
gene of HIF-1α and HIF-2α in PLC/PRF/5 and Hep3B cells, but HIF-3α
is regulated more effectively by HIF-2α (Fig. 3).
Discussion
In general, hypoxia is the most important factor
involved in the regulation of the expression of HIF-α members. It
has been reported that the mRNA and protein levels of HIF-α members
are increased in hypoxic environments, and are often overexpressed
in hypoxic solid tumors (25).
Unlike the observation of consistent increase of HIF-2α in the
majority of tumors, our previous data have shown that the
expression patterns of HIF-1α and HIF-2α are opposite in HCC and
paired peritumoral tissues (18).
The level of HIF-1α in HCC tissues is significantly higher compared
with that in peritumoral tissues, whereas the level of HIF-2α is
markedly lower in tumor tissues compared with peritumoral tissues
(18). Notably, we did not identify
any obvious differences in HIF-3α expression between HCC and
peritumoral tissues in this study. The expression of HIF-3α protein
was increased in ~50% of the HCC specimens compared with
peritumoral tissues, but was decreased or unaltered in the other
~50%. This discrepancy may be attributed to different sensitivities
of HIF-1α, HIF-2α and HIF-3α in response to hypoxia. Furthermore,
although all HIF-α members are predominantly regulated by oxygen
pressure, they are additionally regulated by other factors in the
tumor microenvironment. These factors include glucose metabolism
and mutations in proto-oncogenes and tumor suppressor genes
(26–29). These factors are likely to exert
different effects on the expression of HIF-α factors, leading to
the upregulation of HIF-1α, the downregulation of HIF-2α and the
inconsistent expression of HIF-3α in HCC tissues. Therefore, the
regulation of HIF-α expression is complicated, and further
investigations are required to determine the underling mechanisms
that control HIF-3a expression in HCC tissues.
The varying expression patterns of HIF-α factors in
HCC tissues indicate that they may serve different functions in
response to hypoxia. The transactivation activity of HIF-3α is
different from those of HIF-1α and HIF-2α (30). Previous studies have suggested that
HIF-3α may suppress HIF-1α and HIF-2α mediated gene expression when
the expression of ARNT is limited (13,31,32).
Hara et al (12) transfected
expression vectors containing HIF-1α, HIF-2α or HIF-3α genes into
COS-7 cells and found that HIF-1α and HIF-2α upregulated the
transcription of HRE-driven genes, whereas HIF-3α inhibited their
expression. Recent research has indicated that HIF-3α is an
oxygen-dependent transcription activator, and serves a crucial
function in the transcriptional response to hypoxia by binding to
target gene promoters, including LC3C, REDD1 and SQRDL, thus
stimulating their expression (33).
Therefore, further studies are required to investigate the role of
HIF-3α in response to hypoxia.
To reveal the role of HIF-3α in the development of
HCC, we divided the human samples into two groups based on the
score of HIF-3α expression. However, no significant correlation was
identified between HIF-3α expression and the clinicopathological
characteristics of HCC samples. Furthermore, no significant
correlation was detected between the expression of HIF-3α in HCC
tissues and the OS or DFS of HCC patients. No statistically
significant correlation was detected between HIF-3α expression and
the prognosis of HCC patients. However, larger population-based
studies are required to confirm the inconsistent expression
patterns of HIF-3α in HCC and to identify the underlying
causes.
Previous studies have reported associations among
the expression levels of HIF-1α, HIF-2α, and HIF-3α; however, their
results are inconsistent or conflicting (13–15,34).
Tanaka et al (13) have found
that the siRNA-mediated knockdown of HIF-1α in human renal cell
carcinoma notably reduced the 2,2-dipyridyl-induced expression of
HIF-3α protein. In addition, an IHC study revealed an overlapping
region with positive HIF-1α and HIF-3α expression within the cells
(13). These results indicate that
HIF-3α is a target gene of HIF-1α. However, the expression of a
stabilized form of HIF-1α did not alter HIF-3α mRNA levels in
either zebrafish embryos (34) or
3T3-L1 cells (15). Hatanaka et
al (15) have observed that
HIF-2α specifically binds to the sequence between −251 and −228 bp
upstream of the transcription start site of mouse HIF-3α, which is
essential in the response to HIF-2α stimulation. In human umbilical
venous endothelial cells, HIF-3α expression is promoted by HIF-1
and HIF-2 (14). All these
inconsistent results regarding the HIF-1α- and HIF-2α-mediated
regulation of HIF-3α induction may be due to the different cell
types used in the experiments. In HCC cell lines, HIF-1α and HIF-2α
increased the expression of HIF-3α in PLC/PRF/5 and Hep3B cells.
The correlation between HIF-2α and HIF-3α expression is more
marked, indicating that HIF-3α may be a target gene of HIF-2α in
both PLC/PRF/5 and Hep3B cells.
In conclusion, despite the structural similarities
between HIF-1α, HIF-2α and HIF-3α, their expression patterns
notably differ, indicating that they may play different roles in
the development of HCC. The expression of HIF-3α protein was not
associated with histopathological features, OS or DFS in HCC
patients. However, HIF-3α is a potential target gene of HIF-2α in
PLC/PRF/5 cells. Further studies are required to confirm the direct
regulation of HIF-3α by HIF-2α.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81160311).
References
|
1
|
Mucaj V, Shay JE and Simon MC: Effects of
hypoxia and HIFs on cancer metabolism. Int J Hematol. 95:464–470.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao S, Yang S, Wu C, Wang Y, Jiang J and
Lu Z: Protein expression of hypoxia-inducible factor-1 alpha and
hepatocellular carcinoma: A systematic review with meta-analysis.
Clin Res Hepatol Gastroenterol. 38:598–603. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravalli RN, Cressman EN and Steer CJ:
Cellular and molecular mechanisms of hepatocellular carcinoma: An
update. Arch Toxicol. 87:227–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Semenza GL: Oxygen sensing,
hypoxia-inducible factors and disease pathophysiology. Ann Rev
Pathol. 9:47–71. 2014. View Article : Google Scholar
|
|
5
|
Yang SL, Wu C, Xiong ZF and Fang X:
Progress on hypoxia-inducible factor-3: Its structure, gene
regulation and biological function (Review). Mol Med Rep.
12:2411–2416. 2015.PubMed/NCBI
|
|
6
|
Haase VH: The VHL tumor suppressor: Master
regulator of HIF. Curr Pharm Des. 15:3895–3903. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Majmundar AJ, Wong WJ and Simon MC:
Hypoxia-inducible factors and the response to hypoxic stress. Mol
Cell. 40:294–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Semenza GL: Hypoxia-inducible factors:
Mediators of cancer progression and targets for cancer therapy.
Trends in pharmacological sciences. 33:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Sun M, Wang L and Jiao B: HIFs,
angiogenesis and cancer. J Cell Biochem. 114:967–974. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gu YZ, Moran SM, Hogenesch JB, Wartman L
and Bradfield CA: Molecular characterization and chromosomal
localization of a third alpha-class hypoxia inducible factor
subunit, HIF3alpha. Gene Expr. 7:205–213. 1998.PubMed/NCBI
|
|
11
|
Tian H, McKnight SL and Russell DW:
Endothelial PAS domain protein 1 (EPAS1), a transcription factor
selectively expressed in endothelial cells. Genes Dev. 11:72–82.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hara S, Hamada J, Kobayashi C, Kondo Y and
Imura N: Expression and characterization of hypoxia-inducible
factor (HIF)-3alpha in human kidney: Suppression of HIF-mediated
gene expression by HIF-3alpha. Biochem Biophys Res Commun.
287:808–813. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tanaka T, Wiesener M, Bernhardt W, Eckardt
KU and Warnecke C: The human HIF (hypoxia-inducible factor)-3alpha
gene is a HIF-1 target gene and may modulate hypoxic gene
induction. Biochem J. 424:143–151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Augstein A, Poitz DM, Braun-Dullaeus RC,
Strasser RH and Schmeisser A: Cell-specific and hypoxia-dependent
regulation of human HIF-3α: Inhibition of the expression of HIF
target genes in vascular cells. Cell Mol Life Sci. 68:2627–2642.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hatanaka M, Shimba S, Sakaue M, Kondo Y,
Kagechika H, Kokame K, Miyata T and Hara S: Hypoxia-inducible
factor-3alpha functions as an accelerator of 3T3-L1 adipose
differentiation. Biol Pharm Bull. 32:1166–1172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang X, Dong W, Thornton C, Scheffler B
and Willett KL: Benzo(a)pyrene induced glycine N-methyltransferase
messenger RNA expression in fundulus heteroclitus embryos. Mar
Environ Res. 69(Suppl): S74–S76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang X, Thornton C, Scheffler BE and
Willett KL: Benzo[a]pyrene decreases global and gene specific DNA
methylation during zebrafish development. Environ Toxicol
Pharmacol. 36:40–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang SL, Liu LP, Jiang JX, Xiong ZF, He QJ
and Wu C: The correlation of expression levels of HIF-1α and HIF-2α
in hepatocellular carcinoma with capsular invasion, portal vein
tumor thrombi and patients' clinical outcome. Jpn J Clin Oncol.
44:159–167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dai CX, Gao Q, Qiu SJ, Ju MJ, Cai MY, Xu
YF, Zhou J, Zhang BH and Fan J: Hypoxia-inducible factor-1 alpha,
in association with inflammation, angiogenesis and MYC, is a
critical prognostic factor in patients with HCC after surgery. BMC
Cancer. 9:4182009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiang ZL, Zeng ZC, Fan J, Tang ZY, He J,
Zeng HY and Chang JY: The expression of HIF-1α in primary
hepatocellular carcinoma and its correlation with radiotherapy
response and clinical outcome. Mol Biol Rep. 39:2021–2029. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zeng HY
and Gao DM: Gene expression profiling of fixed tissues identified
hypoxia-inducible factor-1α, VEGF and matrix metalloproteinase-2 as
biomarkers of lymph node metastasis in hepatocellular carcinoma.
Clin Cancer Res. 17:5463–5472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie H, Song J, Liu K, Ji H, Shen H, Hu S,
Yang G, Du Y, Zou X, Jin H, et al: The expression of
hypoxia-inducible factor-1alpha in hepatitis B virus-related
hepatocellular carcinoma: Correlation with patients' prognosis and
hepatitis B virus X protein. Dig Dis Sci. 53:3225–3233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang SL, Yu C, Jiang JX, Liu LP, Fang X
and Wu C: Hepatitis B virus X protein disrupts the balance of the
expression of circadian rhythm genes in hepatocellular carcinoma.
Oncol Lett. 8:2715–2720. 2014.PubMed/NCBI
|
|
25
|
Semenza GL: HIF-1 mediates metabolic
responses to intratumoral hypoxia and oncogenic mutations. J Clin
Invest. 123:3664–3671. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tsapournioti S, Mylonis I, Hatziefthimiou
A, Ioannou MG, Stamatiou R, Koukoulis GK, Simos G, Molyvdas PA and
Paraskeva E: TNFα induces expression of HIF-1α mRNA and protein but
inhibits hypoxic stimulation of HIF-1 transcriptional activity in
airway smooth muscle cells. J Cell Physiol. 228:1745–1753. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kilic-Eren M, Boylu T and Tabor V:
Targeting PI3K/Akt represses hypoxia inducible factor-1α activation
and sensitizes rhabdomyosarcoma and Ewing's sarcoma cells for
apoptosis. Cancer Cell Int. 13:362013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen C, Cai S, Wang G, Cao X, Yang X, Luo
X, Feng Y and Hu J: c-Myc enhances colon cancer cell-mediated
angiogenesis through the regulation of HIF-1α. Biochem Biophys Res
Commun. 430:505–511. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agani F and Jiang BH: Oxygen-independent
Regulation of HIF-1: Novel Involvement of PI3K/AKT/mTOR pathway in
cancer. Curr Cancer Drug Targets. 13:245–251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li QF, Wang XR, Yang YW and Lin H: Hypoxia
upregulates hypoxia inducible factor (HIF)-3alpha expression in
lung epithelial cells: Characterization and comparison with
HIF-1alpha. Cell Res. 16:548–558. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maynard MA, Evans AJ, Hosomi T, Hara S,
Jewett MA and Ohh M: Human HIF-3alpha4 is a dominant-negative
regulator of HIF-1 and is down-regulated in renal cell carcinoma.
FASEB J. 19:1396–1406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maynard MA, Evans AJ, Shi W, Kim WY, Liu
FF and Ohh M: Dominant-negative HIF-3 alpha 4 suppresses VHL-null
renal cell carcinoma progression. Cell Cycle. 6:2810–2816. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang P, Yao Q, Lu L, Li Y, Chen PJ and
Duan C: Hypoxia-inducible factor 3 is an oxygen-dependent
transcription activator and regulates a distinct transcriptional
response to hypoxia. Cell Rep. 6:1110–1121. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang P, Lu L, Yao Q, Li Y, Zhou J, Liu Y
and Duan C: Molecular, functional and gene expression analysis of
zebrafish hypoxia-inducible factor-3α. Am J Physiol Regul, Integr
Comp Physiol. 303:R1165–R1174. 2012. View Article : Google Scholar
|