Introduction
Long non-coding RNAs (lncRNAs) are transcripts
composed of >200 nucleotides (1),
which do not contain functional open reading frames, cannot encode
proteins (2), and are pervasively
transcribed throughout the entire genome (3). Several lncRNAs have been found to serve
a critical role in embryonic development (4), while the dysfunction of lncRNAs is
associated with widespread conditions, such as birth defects
(5). Cleft palate (CP) is the most
common congenital malformation in the oral and craniofacial region,
and may occur at any stage of the palate development, including the
palatal shelf growth, elevation or fusion (6). In addition, the loss of medial edge
epithelial cells or failure of mesenchymal consolidation can cause
CP. However, the etiology of CP is complex and remains poorly
understood.
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is
a persistent environmental contaminant (7) that is a teratogen (8) and has been found to cause CP at high
rates in mice and humans (8–12). Low levels of TCDD are able to inhibit
medial edge epithelial cell apoptosis and suppress the mesenchymal
growth of the palatal shelves, which leads to the failure of
palatal fusion (13–15). The identification of lncRNAs has
provided novel opportunities for the investigation of the precise
etiology and pathogenesis of CP, which remain unclear. However,
although a great number of studies have investigated the underlying
mechanisms of TCDD-induced CP (8,14,16–18),
the involvement of lncRNAs in TCDD-induced CP has been rarely
reported (6).
lncRNA H19 is a 2,300 bp non-coding RNA (ncRNA),
which is transcribed from the maternal allele, with a high
expression observed prenatally and a low expression observed
postnatally. lncRNA H19 is evolutionarily conserved at the
nucleotide level of humans and rodents, and is not translated into
a protein (19). The lncRNA H19 gene
is pervasively expressed in various tissues in embryos, including
the palate (20). However, whether
the expression of lncRNA H19 in TCDD-induced mouse CP varies with
different developmental stages has yet to be examined. lncRNA H19
is expressed from the imprinted gene locus that also contains the
reciprocally imprinted insulin-like growth factor 2 (IGF2) gene
(21). Genetic evidence indicates
that the parent of origin-dependent expression patterns of the
lncRNA H19 and IGF2 genes is coordinated in mice (22). The IGF2 gene codes for a growth
factor that serves an important role during prenatal development
(23).
In the present study, we speculated that lncRNA H19
may have an important role in the process of TCDD-induced mouse CP
and thus investigated this effect in a mouse model of CP.
Materials and methods
Animals
Pregnant Kunming mice (n=48; age, 8–12 weeks;
weight, ~25 g) were obtained from the Henan Laboratory Animal
Center of Zhengzhou University (Zhengzhou, China). All experiments
were performed in accordance with the Experimental Animal Center
Guide for the Care and Use of Laboratory Animals, and the
Institutional Ethical Guidelines for Experiments with Animals. The
day of vaginal plug confirmation was determined as the embryonic
day 0 (E0). The mice were housed at a constant temperature of
28±2°C and relative humidity of 60±10% with a 12 h light/dark
cycle.
Construction of CP model and sample
collection
The pregnant mice were divided into control
(untreated) and TCDD-treated groups (n=24 in each group). TCDD
(dissolved in 100 µg/ml dimethyl sulfoxide suspension;
Sigma-Aldrich, St. Louis, MO, USA) was dissolved in corn oil and
was prepared into a 6.0-mg/ml suspension. This suspension was
administered orally to each pregnant mouse (64 µg/kg body weight)
in the TCDD-treated group on embryonic day 10 (E10). The same
volume of corn oil (cat no. 383756; Jinlongyu Oil And Grain Food
Convenience Store, Qinghuangdao, China) was administered to the
control mice. On time points E12, E13, E13.5, E14, E14.5, E15,
E15.5, E16 and E17, pregnant mice from the two experimental groups
were sacrificed (4 mice were sacrificed at each time point from
E12-E17). The fetuses were first removed from the uterus of the
sacrificed mice. Subsequently, the palates of the fetuses were
harvested from the two experimental groups under a stereomicroscope
(model no. SZ61TRC-SET; Olympus Corporation, Tokyo, Japan) and
immediately preserved in RNAlater solution (AM7020 product no.
AM7020; Ambion; Thermo Fisher Scientific, Inc., Austin, TX, USA) at
4°C and stored at −80°C for further RNA isolation. Certain palate
samples (n=5) from fetuses at E13.5 up to E16.5 from the control
and TCDD groups were fixed with 10% formalin for histological
examination. The lengths of fetuses was measured by vernier caliper
(cat no. 500-752-10, Mitutoyo, Tokyo, Japan), and the weights of
the fetuses were measured by electronic balance (cat no. AX224ZH,
Ohaus Corporation, Parsippany, NJ, USA) on E12, E13, E14, E15, E16
and E17, respectively.
Histological analysis
After palate samples were fixed with 10% formalin
for 2 days, then flushed with water overnight. The samples were
then dehydrated as follows: Firstly, samples were treated in 70%
ethanol for ~5 min and then transferred to 80% ethanol for 5 min.
Next, samples were transferred to 95% ethanol for 5 min.
Subsequently, samples were transferred to 100% ethanol for 5 min
and then transferred again into fresh 100% ethanol for 5 min,
followed by two changes of xylene for 5 min per change. Finally,
the samples were fixed in paraffin (cat no. 8002-74-2, Kunlun,
Daqin, China) at 60°C for 3 h. Then, samples were cut into 5 µm
thick sections, and placed on glass slides (cat no. HDMED7101,
Yancheng, Jiangsu, China). Samples were deparaffinized in three
changes of xylene for a duration of 5 min per change. The samples
were dehydrated as follows: Firstly, slides were transferred
through two changes of 100% ethanol for 5 min per change and then
transferred to 95% ethanol for 5 min. Subsequently, samples were
transferred to 80 and 70% ethanol, respectively, each for 5 min.
Slides were rinsed in running distilled tap water at room
temperature for at least 5 min. Samples were then stained in
hematoxylin (cat no. H9627) solution for 3 min, then stained the
samples in working eosin solution (cat no. H004642), both
Sigma-Aldrich, for 2 min. Slides were rinsed with water at room
temperature for at least 5 min. Finally, a drop of neutral balsam
(cat no. 33645; Ningbo, Shanghai, China) was administered to the
tissue on each slide, and a coverslip was placed on top. Slides
were viewed on an electron microscope (model no. CX41; Olympus
Corporation; magnification, ×40).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Palates (n=10) were harvested from the TCDD-treated
and control mice from E13.5-E15.5 due to the formation of bud
palatal shelves, growth and perfusion at the aforementioned time
points, which are important to the development of palates. Palates
were lysed in TRIzol lysis solution (cat no. 15596-018; Invitrogen;
Thermo Fisher Scientific, Inc., Carlsbad, CA, USA). Total RNA of
the palates was isolated according to the manufacturer's
instructions, and then dissolved in nuclease-free water. In order
to detect the expression of lncRNA H19, first-strand cDNA was
synthesized using a PrimeScript II 1st strand cDNA Synthesis kit
(cat no. 6210A; Takara Bio Inc., Otsu, Japan) and then amplified by
qPCR with the SYBR Premix Ex Taq kit (cat no. DRR420A, Takara)
using an ABI 7900 Prism Real-Time PCR system (model no. 7900HT;
Applied Biosystems; Thermo Fisher Scientific, Inc.). 18s rRNA was
used as an internal control. The conditions of qPCR for lncRNA H19
and IGF2 amplification were as follows: Polymerase activation for
15 min at 95°C, followed by 40 cycles at 95°C for 15 sec, 56°C for
20 sec and 72°C for 30 sec. The threshold cycle (Cq) value of the
PCR amplification curve of the target gene was analyzed using the
2−ΔΔCq method (24). All
primers were synthesized by Invitrogen (Thermo Fisher Scientific,
Inc.) and were as follows: Forward, 5′-CGGACATCTAAGGGCATCA-3′ and
reverse, 5′-AAGACGGACCAGAGCGAAA-3′ for 18s rRNA; forward,
5′-TACCCCGGGATGACTTCATC-3′ and reverse, 5′-TATCTCCGGGACTCCAAACC-3′
for lncRNA H19; and forward, 5′-TATCTCCGGGACTCCAAACC-3′ and
reverse, 5′-CAAATGTGGGGACACAGAGG-3′ for IGF2. The final PCR product
were confirmed to be lncRNA H19 and IGF2 by agarose gel
electrophoresis.
Statistical analysis
All data were compared using double-sided Student's
t test or one-way analysis of variance. Tukey's post hoc test was
used to determine the differences between the two groups. The
choice of tests was performed automatically using the SPSS
software, version 13.0 (SPSS, Inc., Chicago, IL, USA). All data are
presented as the mean ± standard deviation of two or three
independent experiments. P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of TCDD on fetal mice
The appearance, feeding, drinking and activity of 12
pregnant mice in the control and TCDD-treated groups did not
present any evident differences. At time-points E12, E13, E14, E15,
E16 and E17, the fetuses were removed from the uterus of control
and TCDD-treated mice. There were no differences in tail length and
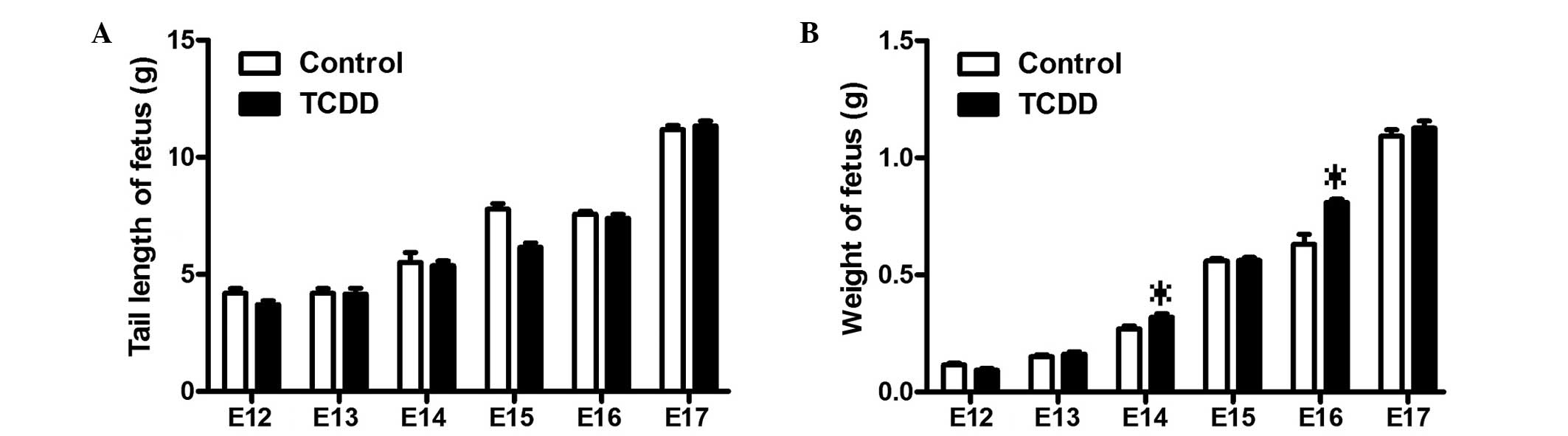
birth rates between control and TCDD-treated mice. As shown in
Fig. 1, there were also no
statistically significant differences in the tail length and body
weight of the fetal mice at the various time points.
Morphological and histological
observations of palatal development
For the fetuses from TCDD-treated mice, the detected
rate of CP was 80% (272/340). Their morphological and histological
alterations were examined and the results are shown in Fig. 2. Formation of the bud of palatal
shelves began on E13.5 in the fetuses from the control group
(Fig. 2A). By contrast, the
formation of palatal shelves in the fetuses from TCDD-treated mice
was interrupted on E13.5 (Fig. 2B).
The construction of palate shelves in normal fetuses began on E14.5
(Fig. 2C), whereas the palate
shelves remained separated on E14.5 and E15.5 in the fetuses from
TCDD-treated mice (Fig. 2D). On
E15.5, the palatal shelves of the fetuses from the untreated mice
were completely fused, which was not observed in the fetuses from
TCDD-treated mice, indicating CP formation (Fig. 2E and F).
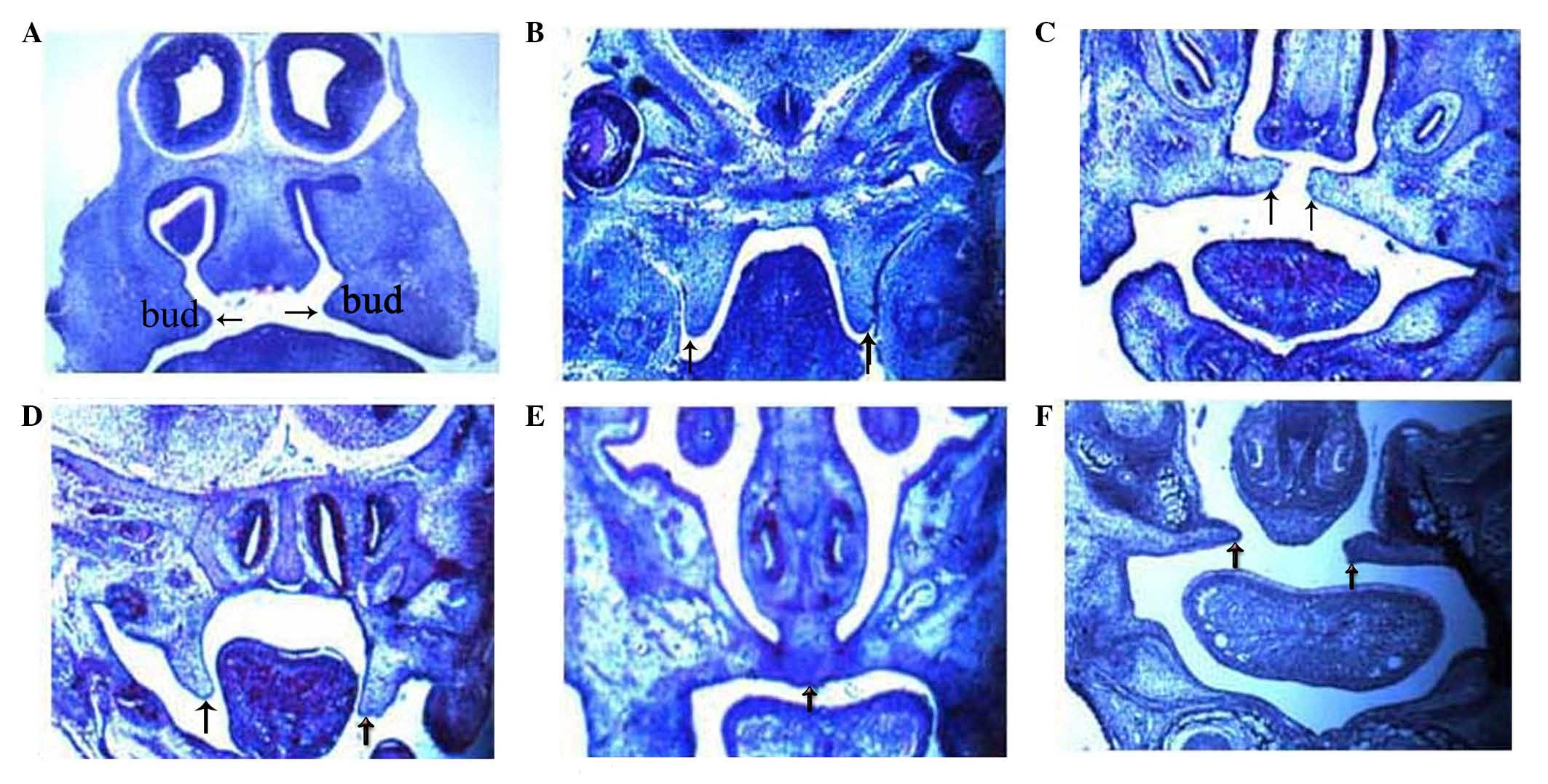 | Figure 2.Histological analysis of palatal
shelves in the control and TCDD-treated group between the time
points E13.5 and E15.5. On E13.5, (A) the palate shelves of the
fetuses elevated in the control group, whereas (B) failing to
elevate in the TCDD-treated group. On E14.5, (C) the palate shelves
of the fetuses were elevated and reached a horizontal position
above the dorsum of the tongue in the control group, whereas (D)
delayed elevation was observed in the TCDD-treated group. On E15.5,
(E) the palate shelves of the fetuses were fused together in the
control group, whereas (F) failure to fuse was observed in the
TCDD-treated group, resulting in cleft palate. In the two groups,
hematoxylin and eosin staining was used (magnification, ×40). E,
embryonic day; TCDD,
2,3,7,8-tetrachlorodibenzo-p-dioxin. |
Expression levels of lncRNA H19 at
different developmental stages of CP
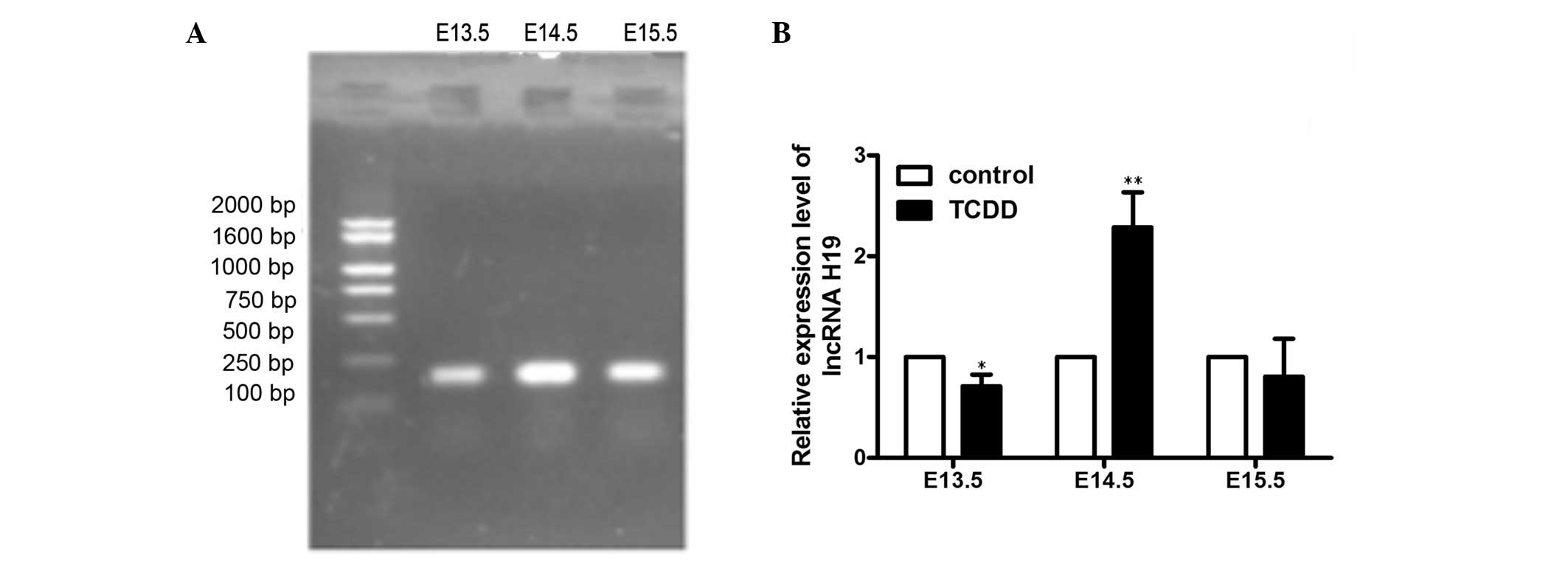
In order to investigate the expression patterns of
lncRNA H19 parallel to the CP formation induced by TCDD treatment,
RT-qPCR was performed on palate tissues from E13.5, E14.5 and
E15.5. The primers for lncRNA H19 are on an exon and cDNA
sequencing of the final PCR products confirmed lncRNA H19 (Fig. 3A) The results showed that the
expression levels of lncRNA H19 varied with the development of
palate (Fig. 3B). The relative
expression of lncRNA H19 in the TCDD-treated group was found to be
reduced on E13.5 (0.29±0.16-fold vs. control; P<0.05) and on
E15.5 (0.19±0.53-fold vs. control). However, the relative
expression levels of IGF2 were significantly increased on E14.5
(2.29±0.49; P<0.01) compared with the corresponding control
palate tissues.
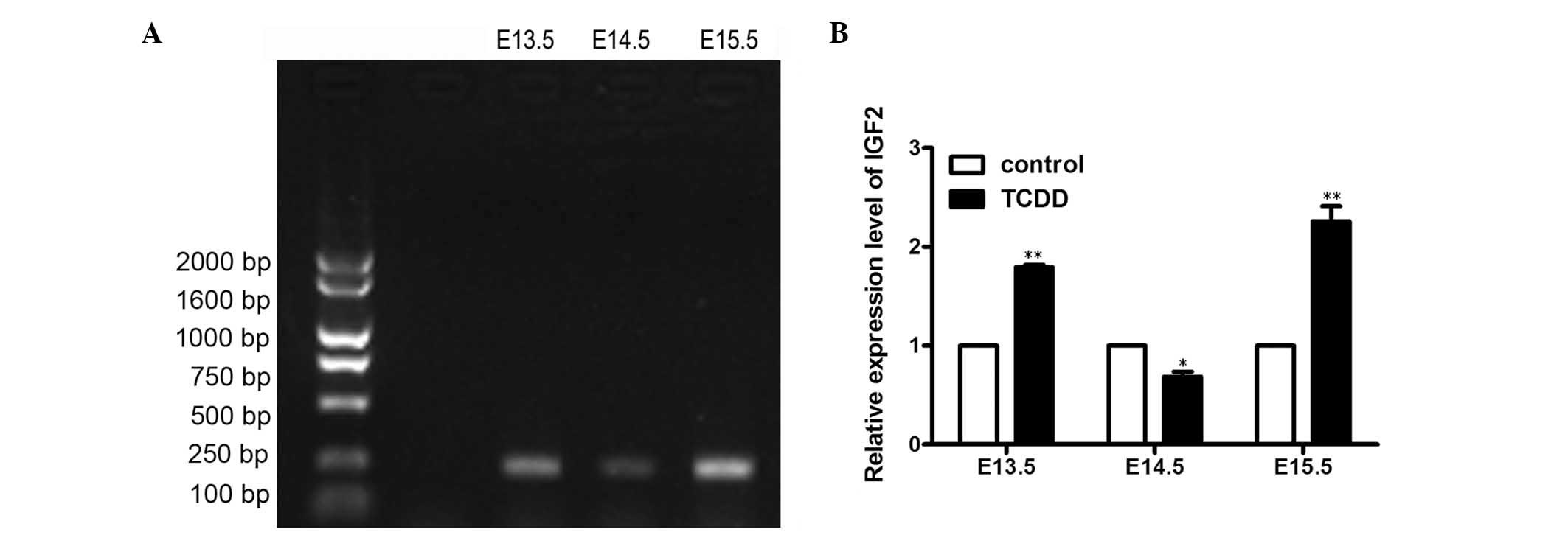
Expression of insulin-like growth
factor 2 (IGF2) in the development of CP
In order to investigate the biological function of
lncRNA H19, one of the lncRNA H19 target genes, IGF2, was also
investigated. IGF2 is a mitogen for a variety of cell types and is
required for normal embryonic growth. IGF2 was amplified by
RT-qPCR, and the final PCR product was confirmed to be IGF2. The
results indicated that the expression of IGF2 was embryo
age-specific during the development of the palate (Fig. 4A and B). More specifically, the
expression levels of IGF2 gene in the TCDD-treated group were
significantly increased on E13.5 (1.79±0.04-fold vs. control;
P<0.01) and E15.5 (2.26±0.22-fold vs. control; P<0.01).
However, the expression levels of IGF2 were significantly reduced
on E14.5 (0.69±0.07-fold; P<0.05) compared with the
corresponding control palate tissues, respectively.
Discussion
ncRNAs account for ~98% of the entire genome
(25), and lncRNAs are evidently the
most numerous and functionally diverse amongst the multiple ncRNA
classes (26). Previous studies have
shown that lncRNAs serve a vital role in the development in
embryonic development (27–29), such as heart and body-wall
development (30) and other features
of organogenetic development (31).
Cleft palate (CP) is one of the most common birth defects, with a
complex genetic and environmental etiology (32). However, few studies have investigated
the pattern of lncRNA expression during the development of the
palate. The lncRNA H19 gene was initially identified as a CP gene
in transforming growth factor-β3-knockout mice by RNA sequencing
analysis (6). However, whether
lncRNA H19 is involved in H19 gene expression in TCDD-induced CP
has yet to be investigated.
In the present study, a mouse CP model was initially
established, which was induced by oral administration of TCDD, and
subsequently the role of lncRNA H19 was determined. The results
demonstrated that TCDD resulted in CP development and the rate was
80%, which is in accordance with previous findings stating that
TCDD induced CP at a high rate in mice (33). In addition, the present study found
that there were no differences in the tail length and body weight
of the fetal mice between the control and TCDD-treated groups,
which coincided with an absence in differences in litter size and
body weight between the TCDD fetal mice and control mice (34). Furthermore, the current study showed
that the expression levels of lncRNA H19 varied with the stages of
TCDD-induced palatogenesis between the time points E13.5 and E15.5.
It has been established that the period from E13.5 to E15.5 is
important for the development of palate. For example, both sides of
the palates lift above the tongue on E13.5 and grow rapidly on
E14.5, and begin to touch each other on E15.5. Notably, the
expression levels of lncRNA H19 in TCDD-treated mice were lower on
E13.5 and E15.5 compared with those of the control, while a high
expression was observed on E14.5. These results are similar to
those of a previous study, which stated that the expression level
of lncRNA H19 increased between E14 and E15 (6). Therefore, lncRNA H19 is suggested to be
the primary contributor to the development of CP induced by TCDD,
including in the stages of palatal convergence, adhesion and
fusion.
As mentioned earlier, lncRNA H19 may play an
important role in the pathogenesis of CP induced by TCDD; however,
the mechanisms underlying lncRNA H19-regulated CP in TCDD-treated
mice remain elusive. In the present study, the expression levels of
IGF2 gene were found to be high on E13.5 and E15.5, whereas a low
expression was detected on E14.5. Thus, the IGF2 gene showed the
opposite expression pattern to that of lncRNA H19, which suggested
lncRNA H19 may function through interaction with IGF2. However, the
precise mechanisms through which lncRNA H19 regulates TCDD-induced
CP require further investigation.
In conclusion, the present study revealed that
lncRNA H19 mediates TCDD-induced CP, which provides a new insight
into the role of lncRNA in CP.
Acknowledgements
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
81502843).
References
|
1
|
Cao J: The functional role of long
non-coding RNAs and epigenetics. Biol Proced Online. 16:112014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fachel AA, Tahira AC, Vilella-Arias SA,
Maracaja-Coutinho V, Gimba ER, Vignal GM, Campos FS, Reis EM and
Verjovski-Almeida S: Expression analysis and in silico
characterization of intronic long noncoding RNAs in renal cell
carcinoma: Emerging functional associations. Mol Cancer.
12:1402013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gao L, Mai A, Li X, Lai Y, Zheng J, Yang
Q, Wu J, Nan A, Ye S and Jiang Y: LncRNA-DQ786227-mediated cell
malignant transformation induced by benzo(a)pyrene. Toxicol Lett.
223:205–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim ED and Sung S: Long noncoding RNA:
Unveiling hidden layer of gene regulatory networks. Trends in Plant
Sci. 17:16–21. 2012. View Article : Google Scholar
|
|
5
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res. 41(D1):
D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ozturk F, Li Y, Zhu X, Guda C and Nawshad
A: Systematic analysis of palatal transcriptome to identify cleft
palate genes within TGFβ3-knockout mice alleles: RNA-Seq analysis
of TGFβ3 Mice. BMC genomics. 14:1132013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boutros PC, Yan R, Moffat ID, Pohjanvirta
R and Okey AB: Transcriptomic responses to
2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in liver: Comparison of
rat and mouse. BMC Genomics. 9:4192008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pratt RM, Dencker L and Diewert VM:
2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced cleft palate in the
mouse: Evidence for alterations in palatal shelf fusion. Teratog
Carcinog Mutagen. 4:427–436. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weber H, Harris MW, Haseman JK and
Birnbaum LS: Teratogenic potency of TCDD, TCDF and TCDD-TCDF
combinations in C57BL/6N mice. Toxicol Lett. 26:159–167. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Abbott BD and Birnbaum LS: TCDD alters
medial epithelial cell differentiation during palatogenesis.
Toxicol Appl Pharmacol. 99:276–286. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Emerson JL, Thompson DJ, Strebing RJ,
Gerbig CG and Robinson VB: Teratogenic studies on
2,4,5-trichlorophenoxyacetic acid in the rat and rabbit. Food
Cosmet Toxicol. 9:395–404. 1971. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wolfe WH, Michalek JE, Miner JC, Rahe AJ,
Moore CA, Needham LL and Patterson DG Jr: Paternal serum dioxin and
reproductive outcomes among veterans of Operation Ranch Hand.
Epidemiology. 6:17–22. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abbott BD, Schmid JE, Brown JG, Wood CR,
White RD, Buckalew AR and Held GA: RT-PCR quantification of AHR,
ARNT, GR, and CYP1A1 mRNA in craniofacial tissues of embryonic mice
exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin and hydrocortisone.
Toxicol Sci. 47:76–85. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takagi TN, Matsui KA, Yamashita K, Ohmori
H and Yasuda M: Pathogenesis of cleft palate in mouse embryos
exposed to 2,3,7, 8-tetrachlorodibenzo-p-dioxin (TCDD). Teratog
Carcinog Mutagen. 20:73–86. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamada T, Mishima K, Fujiwara K, Imura H
and Sugahara T: Cleft lip and palate in mice treated with
2,3,7,8-tetrachlorodibenzo-p-dioxin: A morphological in vivo study.
Congenit Anom (Kyoto). 46:21–25. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abbott BD: Review of the interaction
between TCDD and glucocorticoids in embryonic palate. Toxicology.
105:365–373. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jacobs H, Dennefeld C, Féret B, Viluksela
M, Håkansson H, Mark M and Ghyselinck NB: Retinoic acid drives aryl
hydrocarbon receptor expression and is instrumental to
dioxin-induced toxicity during palate development. Environ Health
Perspect. 119:1590–1595. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Imura H, Yamada T, Mishima K, Fujiwara K,
Kawaki H, Hirata A, Sogawa N, Ueno T and Sugahara T: Effect of
2,3,7,8-tetrachlorodibenzo-p-dioxin suggests abnormal palate
development after palatal fusion. Congenit Anom (Kyoto). 50:77–84.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ratajczak MZ: Igf2-H19, an imprinted
tandem gene, is an important regulator of embryonic development, a
guardian of proliferation of adult pluripotent stem cells, a
regulator of longevity, and a ‘passkey’ to cancerogenesis. Folia
Histochem Cytobiol. 50:171–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brunkow ME and Tilghman SM: Ectopic
expression of the H19 gene in mice causes prenatal lethality. Genes
Dev. 5:1092–1101. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gabory A, Jammes H and Dandolo L: The H19
locus: Role of an imprinted non-coding RNA in growth and
development. BioEssays. 32:473–480. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui H, Hedborg F, He L, Nordenskjöld A,
Sandstedt B, Pfeifer-Ohlsson S and Ohlsson R: Inactivation of H19,
an imprinted and putative tumor repressor gene, is a preneoplastic
event during Wilms' tumorigenesis. Cancer Res. 57:4469–4473.
1997.PubMed/NCBI
|
|
23
|
Stewart CE and Rotwein P: Growth,
differentiation, and survival: Multiple physiological functions for
insulin-like growth factors. Physiol Rev. 76:1005–1026.
1996.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Archer K, Broskova Z, Bayoumi AS, Teoh JP,
Davila A, Tang Y, Su H and Kim IM: Long Non-Coding RNAs as Master
Regulators in Cardiovascular Diseases. Int J Mol Sci.
16:23651–23667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Derrien T, Guigó R and Johnson R: The Long
Non-Coding RNAs: A New (P)layer in the ‘Dark Matter’. Front Genet.
2:1072011.PubMed/NCBI
|
|
27
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng L, Paulson A, Li H, Piekos S, He X,
Li L and Zhong XB: Developmental programming of long non-coding
RNAs during postnatal liver maturation in mice. PLoS One.
9:e1149172014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hutchins AP and Pei D: Transposable
elements at the center of the crossroads between embryogenesis,
embryonic stem cells, reprogramming, and long non-coding RNAs. Sci
Bull-Beijing. 60:1722–1733. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grote P, Wittler L, Hendrix D, Koch F,
Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M and
Herrmann BG: The tissue-specific lncRNA Fendrr is an essential
regulator of heart and body wall development in the mouse. Dev
cell. 24:206–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yuan Q, Blanton SH and Hecht JT: Genetic
causes of nonsyndromic cleft lip with or without cleft palate. Adv
Otorhinolaryngol. 70:107–113. 2011.PubMed/NCBI
|
|
33
|
Yamada T, Hirata A, Sasabe E, Yoshimura T,
Ohno S, Kitamura N and Yamamoto T: TCDD disrupts posterior
palatogenesis and causes cleft palate. J Craniomaxillofac Surg.
42:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Miettinen HM, Huuskonen H, Partanen AM,
Miettinen P, Tuomisto JT, Pohjanvirta R and Tuomisto J: Effects of
epidermal growth factor receptor deficiency and
2,3,7,8-tetrachlorodibenzo-p-dioxin on fetal development in mice.
Toxicol Lett. 150:285–291. 2004. View Article : Google Scholar : PubMed/NCBI
|