Introduction
Hepatitis B virus (HBV) remains a major cause of
morbidity and mortality worldwide (1). It has been estimated that ~350 million
individuals worldwide are chronically infected with HBV. A
proportion of these patients will subsequently develop hepatic
cirrhosis, so therapy to control active inflammation is important
(1). The key to effective treatment
is to inhibit the replication of HBV and prevent it from
progressing to cirrhosis, hepatic failure and hepatic cell cancer
(2–4). Lamivudine (LAM) is an antiviral agent,
and large scale control studies have suggested that LAM is able to
inhibit the replication of HBV DNA, enhance hepatitis B e-antigen
loss and seroconversion and improve the normalization of alanine
aminotransferase (ALT), thus leading to histological improvement
(5–7). The
tyrosine-methionine-aspartate-aspartate (YMDD) motif of HBV is the
binding site of LAM (8). When LAM is
combined with the YMDD motif, LAM is able to inhibit the activation
of polymerase, even in viral replication (9). However, long-term LAM therapy may
induce HBV DNA polymerase (P gene) mutations and LAM resistance
(8–13). The HBV polymerase is functionally and
structurally related to human immunodeficiency virus reverse
transcriptases (14). HBV DNA
polymerase performs reversed transcription and exhibits a highly
conserved YMDD motif. This motif, which is localized in the
polymerase structural region C area, is the combing and functioning
site of Lamivudine (a nucleoside antiviral). Lamivudine-resistant
HBV strains with YMDD mutations have been implemented in the
failure of chronic hepatitis B treatment (15). It has been reported that the
seminested PCR products of the DNA polymerase gene exhibit
intergenotypic divergence of ≥4%, which is in accordance with the
HBV DNA S gene sequence, with the exception of genotypes A and G, B
and G, and F and H (16). Only
genotypes B, C, D, and A have been detected in China. Therefore, a
fragment of HBV DNA polymerase gene can be used for genotyping
hepatitis B in China (16). This
genotyping method may also be used to predict antiviral therapeutic
responses among HBV genotypes and the development of drug
resistance due to mutations. It is a valuable tool for guiding the
treatment of lamivudine-resistant HBV in the clinical setting
(16).
Different YMDD mutations can occur in different
genotypes during LAM therapy (5,14–16), and
the effect of the genotype on LAM therapy is different (5,17,18).
Therefore, the present study aimed to investigate the correlation
between feature and genotype with regard to the YMDD mutation in
chronic hepatitis B patients following LAM therapy. Clinical
features and YMDD mutations were assessed in Chinese patients with
chronic hepatitis B following lamivudine therapy using serological
tests, genotype analysis and analysis of mutation and secondary
protein structure of the P gene via an ABI sequencing system.
Materials and methods
Patients
A total of 33 patients with chronic hepatitis B (22
males and 11 females; mean age, 31.79 years) were enrolled in this
study between July 2006 and June 2011. These patients received LAM
therapy (orally, 100 mg/day; GlaxoSmithKine Plc, Brentford, UK) for
at least 12 months. All patients were born and living in Shenyang
(China). Patients who had received interferon-α or other nucleotide
analogs, or those co-infected with hepatitis C, hepatitis D and
human immunodeficiency virus were excluded from the study. This
study was approved by the Institutional Ethics Committee for
clinical study of Shengjing Hospital of China Medical University
(Shenyang, China). The patients were enrolled after providing
written consent. Serum samples were collected prior to treatment,
and at 6 and 12 months after LAM therapy and were stored at −70°C
for further examination.
Serological tests
Serum ALT was measured using standard procedures.
Hepatitis B surface antigen (HBsAg; SBJ-H0844; Nanjing Lihong
Chemical Science & Technology Co., Ltd., Nanjing, China),
hepatitis B envelop antigen (HBeAg; GFD6511) and anti-HBe were
detected using commercial enzyme immunoassay kits (both Qingdao
Jisskang Biotechnology, Co., Ltd., Qingdao, China). HBsAg, anti-HBs
and anti-HBc were purchased from Abbott Laboratories (Abbott Park,
IL, USA). HBeAg and anti-HBe were acquired from Organon Teknika
Corporation (Durham, NC, USA). HBV DNA was detected using
semi-nested polymerase chain reaction (PCR), as previously
described (16).
Analysis of genotype, mutation and
secondary protein structure of the P gene
All positive PCR products were sequenced by Shanghai
GeneCore BioTechnologies Co., Ltd., (Shanghai, China) using an ABI
sequencing system (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). With the aid of DNASTAR software (DNASTAR,
Inc., Madison, WI, USA), three primers were designed the sequence
the HBV DNA polymerase gene (accession number, AF100309): HBV381
(nt381-402), HBV840 (nt840-861), and HBV801 (nt801-822). The
following primers were synthesized by Takara Biotechnology Co.,
Ltd., (Dalian, China): 5′-TGCGGCGTTTTATCATCTTCCT-3′,
5′-GTTTAAATGTATACCCAAAGAC-3′ and 5′-CAGCGGCATAAAGGGACTCAAG-3′,
respectively. Two pairs of semi-nested PCR primers (HBV381/HBV840
and HBV381/HBV801) were utilized in the amplification reaction,
whereas primer HBV381 was used in the sequencing reaction.
Briefly, HBV DNA was extracted from 55 serum samples
obtained from infected patients. The fragment of the polymerase
gene was amplified by semi-nested PCR with two rounds of
amplification. The reaction volume was 50 µl. The first round of
amplification was performed with an initial 5 min denaturing step
at 94°C, followed by 30 cycles of denaturing for 45 sec at 94°C,
annealing for 30 sec at 50°C, and elongation for 90 sec at 72°C,
with a final extension step (10 min at 72°C) using primers HBV381
and HBV840. The second round of amplification was performed with an
initial 5 min denaturing step at 94°C, followed by 30 cycles of
denaturing for 45 sec at 94°C, annealing for 30 sec at 55°C, and
elongation for 60 sec at 72°C, with a final extension step (10 min
at 72°C) using primers HBV381 and HBV801. The reaction products of
the semi-nested PCR were visualized on a 2% agarose gel stained
with ethidium bromide. Semi-nested PCR reaction products were
subjected to purification and sequencing using the HBV381 primer
through a commercial company (Shanghai GeneCore Bio Technologies
Co., Ltd., Shanghai, China) on an ABI sequencing system (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
Purification, sequencing reaction, precipitation and
automatic sequencing were performed on the positive PCR products,
using a Wizard® PCR Preps DNA purification system kit
and an ABI PRISM BigDye™ Terminator cycle Sequencing Ready Reaction
Kit [Promega (Beijing) Biotech Co., Ltd., Beijing, China], as
previously described (16).
The reagent boxes for use in PCR course were
purchased from Fuhua Biomedical Engineering Co., Ltd. (Shanghai,
China). Reagent boxes for purification were purchased from Promega
Corp., (Madison, WA, USA). Automatic sequencing was performed by
Shanghai GeneCore BioTechnologies Co., Ltd., using an ABI
sequencing system.
First, the eight standard full-length nucleotide
sequences of HBV DNA were obtained from GenBank (http://www.ncbi.nlm.nih.gov/genbank/),
including genotypes A (accession no. AY128092), B (AB073858), C
(AF461359), D (AY090453), E (X75664), F (X75663), G (AF405706) and
H (AY090460). Sequences of the fragment of the P gene (~262 base
pairs) were genotyped as described in previously by Ma et al
(16). Then, the mutations and
secondary protein structure of the P gene were analyzed by the
CLUSTAL V method using DNASTAR 5.0 software.
Statistical analysis
The χ2 test was used to compare variables
between groups. P<0.05 was considered statistically significant.
All analyses were performed using the SPSS software, version 14.0
(SPSS, Inc., Chicago, IL, USA).
Results
Outcome of LAM therapy
The mean ALT values prior to treatment and at 6 and
12 months after treatment were 130.26 U/l (15–386 U/l), 27.48 U/l
(8–64 U/l) and 27.06 U/l (8–110 U/l), respectively. No HBsAg loss
or HBsAg seroconversion was detected following treatment. As shown
in Table I, ALT normalization, HBV
DNA loss and YMDD mutation after 6 months of treatment were 90.91,
33.33 and 0%, respectively. ALT normalization, HBV DNA loss, YMDD
mutation, HBeAg loss and HBeAg seroconversion [HBeAg (−), HBeAb
(+)] after 12 months of treatment were 84.85, 33.33, 12.12, 72.41
and 13.79%, respectively. Four groups of patients prior to
treatment were classified according ALT level: ≥5× Upper limit of
normal (ULN), 2–5× ULN, 1–2× ULN and ≤1× ULN. No significant
differences were detected between these four groups in ALT level,
and there were no differences in HBeAg loss and HBeAg
seroconversion after 12 months of treatment between the four groups
(P>0.05). However, following 12 months of treatment, the rate of
HBeAg loss increased and the rate of HBeAg seroconversion decreased
linearly (Table I).
 | Table I.Outcome of LAM therapy and association
between ALT level at baseline and ALT level following
treatment. |
Table I.
Outcome of LAM therapy and association
between ALT level at baseline and ALT level following
treatment.
| ALT level | ≥5× ULN | 2–5× ULN | 1–2× ULN | ≤1 ULN | P-value | Total |
|---|
| Baseline |
|
| Patients,
n | 7 | 11 | 13 | 2 | – | 33 |
| HBeAg+,
n | 6/7 | 9/11 | 12/13 | 2/2 | – | 29/33 |
| 6 months, n
(%) |
|
| ALT
normalization | 7/7 (100) | 9/11 (81.82) | 12/13 (92.31) | 2/2 (100.00) | 0.566 | 30/33 (90.91) |
| HBV DNA
loss | 2/7
(28.57) | 3/11 (27.27) | 6/13 (46.15) | 0/2 (0) | 0.529 | 11/33 (33.33) |
| 12 months, n
(%) |
|
| ALT
normalization | 6/7
(85.71) | 10/11 (90.91) | 10/13 (76.92) | 2/2 (100.00) | 0.727 | 28/33 (84.85) |
| HBV DNA
loss | 2/7
(28.57) | 4/11 (36.36) | 5/13
(38.46) | 0/2 (0) | 0.736 | 11/33 (33.33) |
| YMDD
mutation | 1/7 | 2/11 | 1/12 | 0/2 (0) | 0.235 | 4/33
(12.12) |
| HBeAg
loss | 6/6
(100.00) | 7/9
(77.78) | 7/12
(58.33) | 1/2 (50.00) | 0.250 | 21/29 (72.41) |
| HBeAg
seroconversion | 0/6
(0) | 1/9
(11.11) | 2/12
(16.67) | 1/2 (50.00) | 0.347 | 4/29
(13.79) |
Genotype and the association of
genotype with the outcome after 12 months of LAM therapy
As shown in Table
II, only genotypes B, C and D were detected in the 33 patients
with chronic hepatitis B prior to treatment, with 7 cases of
genotype B (21.21%), 18 cases of genotype C (54.55%) and 8 cases of
genotype D (24.24%), respectively. The difference of intergenotypic
distribution in these three genotypes was significant (P=0.001) and
the differences of distribution between genotype C and genotypes B
or D were also statistically significant (P<0.05). Table II also shows the association between
genotype and the outcome of 12 months of LAM therapy. ALT
normalization and HBeAg seroconversion were highest in genotype B
and HBeAg loss and HBV DNA loss were highest in genotype C. All
therapeutic effects were most marked in genotype D, including ALT
normalization, HBeAg loss, HBeAg seroconversion and HBV DNA loss;
however, the difference of intergenotypic distribution in these
three genotypes was not significant (P>0.05). Both rtM204V
mutations (two patients) were detected in genotype C. A total of
four rtL180M mutations appeared with three cases in genotype C (two
mutations occur simultaneously with rtM204V), and one in genotype D
(Table II).
 | Table II.Genotype at baseline and the effect
of genotype on the outcome of 12 months LAM therapy. |
Table II.
Genotype at baseline and the effect
of genotype on the outcome of 12 months LAM therapy.
| Parameter | Genotype B | Genotype C | Genotype D | P-value |
|---|
| Genotype at
baseline, n (%) | 7
(21.21) | 18
(54.55) | 8
(24.24) | 0.001; BC 0.005; CD
0.012 |
| ALT normalization,
n (%) | 7/7 (100) | 14/18 (77.78) | 7/8 (87.50) | 0.369; BC 0.294; CD
1.0; BD 1.0 |
| HBeAg
seroconversion, n (%) | 1/6
(16.67) | 2/15
(13.33) | 1/8 (12.50) | 0.973; BC 1.0; CD
1.0; BD 1.0 |
| HBeAg loss, n
(%) | 4/6
(66.67) | 12/15 (80) | 5/8 (62.50) | 0.630; BC 0.598; CD
0.621; BD 1.0 |
| HBV DNA loss, n
(%) | 0/7 (0) | 8/18
(44.44) | 3/8 (37.50) | 0.102; BC 0.032; CD
1.0; BD 0.070 |
| rtM204V, n | 0 | 2 | 0 | 0.412; BC 0.359; CD
0.326 |
| rtL180M, n | 0 | 3 | 1 | 0.518; BC 0.250; CD
0.786; BD 1.0 |
Mutations detected during LAM therapy
and clinical symptoms
DNASTAR software analysis indicated no rtM204V/I or
rtL180M mutations prior to treatment or at 6 months after LAM
therapy. As shown in Table I, the
rate of YMDD mutations after 12 months LAM therapy was 12.12%.
Fig. 1 showed the alignment of amino
acid sequences of HBV DNA P gene encoded after 12 months LAM
therapy, which showed that there were two patients with rtM204V +
rtL180M and two patients with rtL180M alone. Other mutations
detected uncommon mutations were also listed in Fig. 1, such as rtV207 L (1 case), rtC188S
(1 case), rtP177R (1 case), rtS176R (1 case), rtV173 L (1 case),
rtG172R (1 case), rtH160Q (1 case), rtF151Y (5 cases) and rtL145 M
(4 cases).
As shown in Tables I
and II, the two patients with
rtM204V + rtL180M belonged to genotype C, with ALT levels 2–5× ULN
at baseline. One patient (HBeAg positive and abnormal ALT at
baseline) was HBeAg positive, with ALT increasing and LAM
resistance appearing after 12 months of LAM therapy. The other
patient (HBeAg negative at baseline) was HBeAg negative, HBeAb
negative, normal ALT and exhibited no drug resistance after 12
months of LAM therapy. The other two patients with rtL180M alone
belonged to genotype C and D, respectively. No drug resistance
appeared to develop following 12 months of LAM therapy (Fig. 1).
Secondary protein structure prediction
of YMDD motif prior to and following the occurrence of
mutations
Using DNASTAR software, the corresponding sequences
were analyzed when rtM204V/I or rtL180M mutations did not appear at
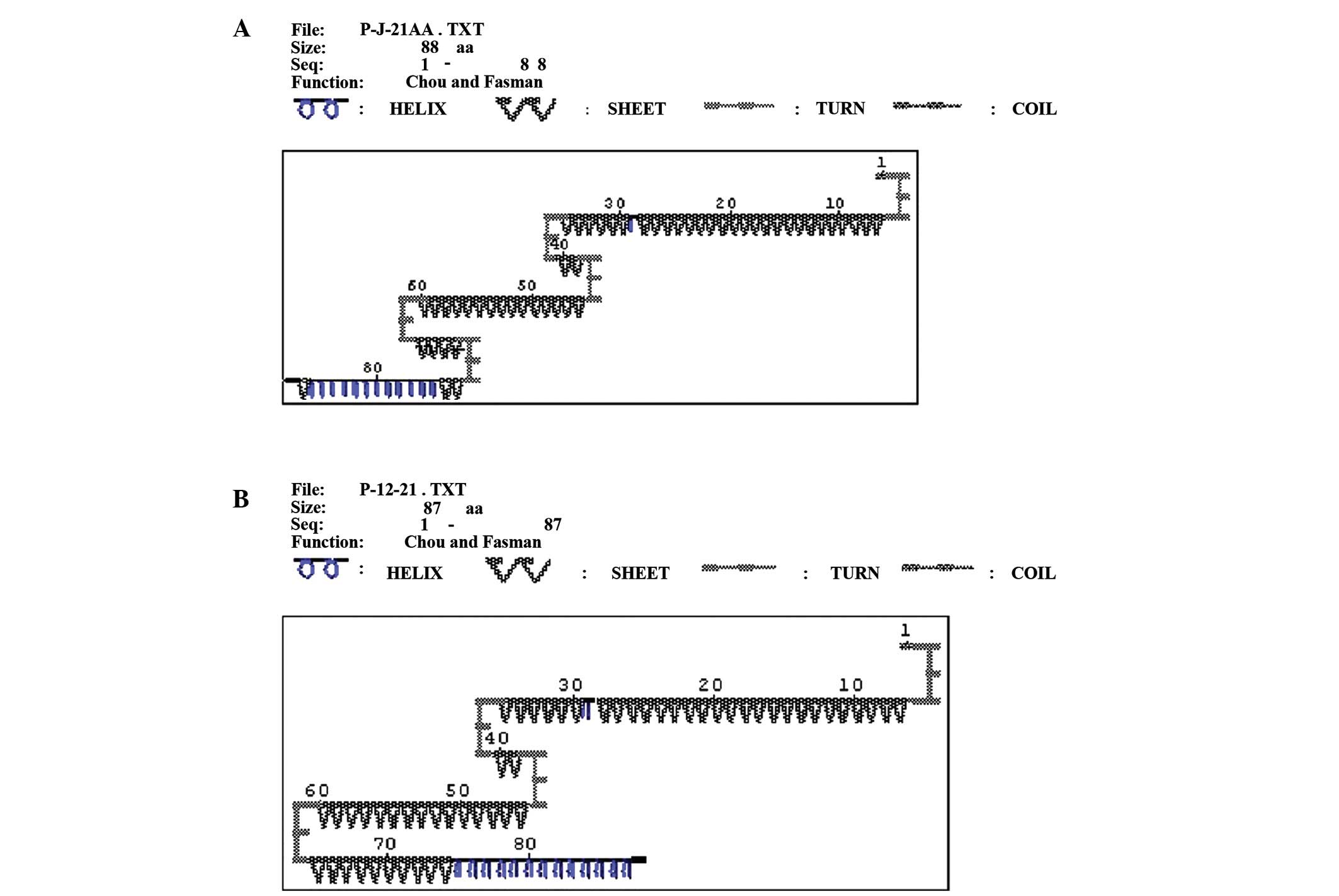
baseline and appeared after 12 months of LAM therapy. As shown in
Fig. 2, the secondary protein
structure prediction of YMDD motif prior to and following the
emergence of rtM204V + rtL180M mutations, 69–72 amino acids (AA)
incurred a rtM204V mutation, whereas 46 AA changed to rtL180M. When
there was no mutation at baseline, the secondary protein structure
of 46 AA and 69–72 AA was β turn (Fig.
2A). When mutations appeared, the β turn of 69–72 AA changed to
β sheet; however, the β turn of 46 AA and the remainder of the
secondary structure did not change (Fig.
2B).
Discussion
The results of the present study indicate that one
year of LAM therapy can improve ALT normalization. The rate of
HBeAg seroconversion was 13.79%, which is similar to the rate of
reported after one year LAM therapy in Asia by Lai et al
(10), and lower than the documented
rate of Schalm et al (20)
and Dienstag et al (21).
Chien et al (22) suggested
that ALT level at baseline could predict HBeAg seroconversion
independently, as the rates of HBeAg seroconversion in patients
with ALT >5× ULN, 2–5× ULN and <2× ULN before treatment were
64, 26 and 5%, respectively. In the present study, the rate of
HBeAg loss was consistent with previous findings (23,24);
however, the rate of HBeAg seroconversion was different, which may
be associated with the small sample size. This study suggested that
there were no significant differences among the genotypes in terms
of improvement of ALT normalization, HBeAg seroconversion, HBeAg
loss and HBV DNA loss during LAM treatment (Fig. 2).
Long-term LAM therapy may induce YMDD mutations and
drug resistance, which would limit the effects of LAM treatment.
LAM therapy is effective in suppressing HBV replication to
undetectable levels by PCR, in relieving liver disease and in
achieving HBsAg seroconversion (25). The rate of YMDD mutations in patients
after one year of LAM therapy was 14–32% (7,8,10,16,20).
However, the rate of YMDD mutation after one year LAM therapy of
the present study was 12.12%, which is slightly lower than a
previous study (15). Allen
(8) classified YMDD mutations into
two categories: rtM204V + rtL180M and rtM204I, and the study
suggested that rtM204V and rtL180M do not occur alone. rtM204I +
rtL180M was identified in a study by Lok et al (26) Kobayashi et al (27) detected YMDD mutations in HBV
asymptomatic carriers who did not receive LAM therapy. Furthermore,
the YMDD mutation was detected in patients with chronic hepatitis B
that underwent pre-therapy with LAM (28,29),
which differs from the previous conclusion that LAM therapy induced
YMDD mutations (30). In the present
study, no YMDD mutations were detected at baseline and after 6
months of LAM therapy, while two common types of mutations: rtM204V
+ rtL180M (2 cases) and rtL180M alone (2 cases) were identified
after 12 months LAM therapy, and no rtM204I was found. However,
other types of mutations including rtV207 L, rtC188S, rtP177R,
rtS176R, rtV173 L, rtG172R, rtH160Q, rtF151Y and rtL145 M were
detected, whose characteristics are require further study.
As observed in previous studies, the mutations are
commonly found among patients with genotype C, which is prevalent
in East Asia and associated with more advanced liver disease
(5,31). However, the overall incidence rate of
rtM204I mutation in genotype B (45.61%, 26/57) was more frequent
compared with genotype C (20.59%, 7/34; P<0.05), even if the
incidence rate of other mutation patterns did not differ
significantly between genotypes B and C (19). In addition, a review analysis of 29
studies reported that 827 patients with a known hepatitis B
genotype who underwent LAM treatment and developed resistance
mutations (32). The present data
revealed that genotype A is associated with the rtM204V, unlike the
other major genotypes (P<0.001), and corresponds to a
significant difference in the mutation pattern of genotypes endemic
in Asian populations. Furthermore, the rtL180M mutation is
significantly associated with the rtM204V in genotypes A, B and C
(18). The above studies show that
HBV genotypes differ in their mutation pattern. In the present
study, two rtM204V mutations were detected in genotype C, and a
total of four rtL180M mutations appeared, three in genotype C, and
one in genotype D. The present findings also suggest that these
mutations are commonly found among patients with genotype C.
It has previously been demonstrated in clinical
studies of LAM therapy that drug resistance may appear following
YMDD mutation (33). The YMDD motif
is the binding site of LAM (8,9); thus,
the occurrence of YMDD mutations may affect LAM binding with the
YMDD motif. In the present study, the possible mechanism underlying
this process was investigated by analyzing the secondary structure
of YMDD mutations. The secondary protein structure was analyzed
prior to and following the occurrence of rtM204V + rtL180M. When
the mutation appeared, although the β turn corresponding to rtL180M
did not change, the β turn corresponding to rtM204V changed to β
sheet. Thus, the change in secondary protein structure may affect
the three-dimensional structure of the protein, then affect the
combination of LAM with the protein and lead to drug resistance.
However, in the present study, drug resistance only appeared in one
of the two patients with rtM204V + rtL180M. Recently, one report
has demonstrated that therapeutic effects continue to be associated
with LAM therapy, even after emergence of YMDD mutations (28). It remains unclear why no drug
resistance developed in the other patient with the same mutations.
LAM remains a viable antiviral therapy for HBeAg negative patients
with chronic hepatitis B. The rates of maintained virological and
biochemical responses to LAM decrease in time due to the selection
of drug-resistant mutants, leading to viral survival and
duplication (25). The potential
mechanism underlying this process requires further study.
In conclusion, the study present study performed in
patients from Shenyang (China) indicated that there were no
significant differences among the genotypes in terms of improvement
of ALT normalization, HBeAg seroconversion, HBeAg loss and HBV DNA
loss following one year of LAM treatment. YMDD mutations induced by
LAM therapy appeared after 6 months of treatment, and the rate of
YMDD mutations in patients after one year of LAM therapy was
12.12%. Two common types of mutation were detected: rtM204V +
rtL180M and rtL180M alone. The mutations were also commonly found
among patients with genotype C. The β turn changed to β sheet
following the occurrence of the rtM204V mutation. This may be the
mechanism underlying LAM resistance. Further large-scale population
studies are required to elucidate the mechanism underlying LAM
resistance with YMDD mutations induced by LAM therapy.
Acknowledgements
This study was supported by the National Nature
Science Foundation of China (grant no. 81200834), Natural Science
Foundation of Liaoning Province (grant no. 2013021070) and Shenyang
Science and Technology Program (grant no. F10-205-1-10). In
addition, this study was supported by China National Fund of
Ministry of Science and Technology (grant no. 30972612), Liaoning
Provincial Fund of Provincial Department of Science and Technology
(grant no. 2009225010-7), Liaoning Provincial Science and
Technology plan Projects (grant no. 2011225020), Shenyang Science
and Technology Plan Program (grant no. F10-149-9-56) and the
Natural Science Foundation of Liaoning Province (grant no.
201202283).
References
|
1
|
Ghaziani T, Sendi H, Shahraz S, Zamor P
and Bonkovsky HL: Hepatitis B and liver transplantation: Molecular
and clinical features that influence recurrence and outcome. World
J Gastroenterol. 20:14142–14155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai CL, Ratziu V, Yuen MF and Poynard T:
Viral hepatitis B. Lancet. 362:2089–2094. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Niederau C, Heintges T, Lange S, Goldmann
G, Niederau CM, Mohr L and Häussinger D: Long-term follow-up of
HBeAg-positive patients treated with interferon alfa for chronic
hepatitis B. N Engl J Med. 334:1422–1427. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Enomoto M, Tamori A, Kohmoto MT, Morikawa
H, Habu D, Sakaguchi H, Takeda T, Seki S, Kawada N, Shiomi S and
Nishiguchi S: Mutational patterns of hepatitis B virus genome and
clinical outcomes after emergence of drug-resistant variants during
lamivudine therapy: Analyses of the polymerase gene and full-length
sequences. J Med Virol. 79:1664–1670. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
França PH, Coelho HS, Brandão CE, Segadas
JA, Quintaes RF, Carrilho FJ, Ono-Nita S, Mattos AA, Tovo C, Gouvea
VS, et al: The emergence of YMDD mutants precedes biochemical flare
by 19 weeks in lamivudine-treated chronic hepatitis B patients: An
opportunity for therapy reevaluation. J Med Biol Res. 40:1605–1614.
2007. View Article : Google Scholar
|
|
7
|
Yen YH, Lu SN, Chen CH, Wang JH, Wu CM,
Hung CH, Tseng PL, Hu TH, Changchien CS and Lee CM: Changes in
serum hepatitis B e antigen (HBeAg) levels associated with the
emergence of YMDD mutants in HBeAg non-seroconverted patients
during lamivudine therapy. Liver Int. 27:1349–1355. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allen MI, Deslauriers M, Andrews CW,
Tipples GA, Walters KA, Tyrrell DL, Brown N and Condreay LD:
Identification and characterization of mutations in hepatitis B
virus resistant to lamivudine. Lamivudine clinical investigation
group. Hepatology. 27:1670–1677. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Seta T, Yokosuka O, Imazeki F, Tagawa M
and Saisho H: Emergence of YMDD motif mutants of hepatitis B virus
during lamivudine treatment of immunocompetent type B hepatitis
patients. J Med Virol. 60:8–16. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lai CL, Chien RN, Leung NW, Chang TT, Guan
R, Tai DI, Ng KY, Wu PC, Dent JC, Barber J, et al: A one-year trial
of lamivudine for chronic hepatitis B. Asia hepatitis lamivudine
study group. N Engl J Med. 339:61–68. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song BC, Suh DJ, Lee HC, Chung YH and Lee
YS: Hepatitis B e antigen seroconversion after lamivudine therapy
is not durable in patients with chronic hepatitis B in Korea.
Hepatology. 32:803–806. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dienstag JL, Cianciara J, Karayalcin S,
Kowdley KV, Willems B, Plisek S, Woessner M, Gardner S and Schiff
E: Durability of serologic response after lamivudine treatment of
chronic hepatitis B. Hepatology. 37:748–755. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ito K, Tanaka Y, Orito E, Hirashima N, Ide
T, Hino T, Kumashiro R, Kato A, Nukaya H, Sakakibara K, et al:
Predicting relapse after cessation of Lamivudine monotherapy for
chronic hepatitis B virus infection. Clin Infect Dis. 38:490–495.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Menéndez-Arias L, Álvarez M and Pacheco B:
Nucleoside/nucleotide analog inhibitors of hepatitis B virus
polymerase: Mechanism of action and resistance. Curr Opin Virol.
8:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang ZM, Huang QW, Qin YQ, He YZ, Qin HJ,
Zhou YN, Xu X and Huang MJ: YMDD mutations in patients with chronic
hepatitis B untreated with antiviral medicines. World J
Gastroenterol. 11:867–870. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma Y, Ding Y, Juan F and Dou XG:
Genotyping the hepatitis B virus with a fragment of the HBV DNA
polymerase gene in Shenyang, China. Virol J. 8:3152011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Palumbo E: Hepatitis B genotypes and
response to antiviral therapy: A review. Am J Ther. 14:306–309.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Damerow H, Yuen L, Wiegand J, Walker C,
Bock CT, Locarnini S and Tillmann HL: Mutation pattern of
lamivudine resistance in relation to hepatitis B genotypes:
Hepatitis B genotypes differ in their lamivudine resistance
associated mutation pattern. J Med Virol. 82:1850–1858. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li SY, Qin L, Zhang L, Song XB, Zhou Y,
Zhou J, Lu XJ, Cao J, Wang LL, Wang J and Ying BW: Molecular
epidemical characteristics of Lamivudine resistance mutations of
HBV in southern China. Med Sci Monit. 17:PH75–PH80. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schalm SW, Heathcote J, Cianciara J,
Farrell G, Sherman M, Willems B, Dhillon A, Moorat A, Barber J and
Gray DF: Lamivudine and alpha interferon combination treatment of
patients with chronic hepatitis B infection: A randomised trial.
Gut. 46:562–568. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dienstag JL, Schiff ER, Wright TL,
Perrillo RP, Hann HW, Goodman Z, Crowther L, Condreay LD, Woessner
M, Rubin M and Brown NA: Lamivudine as initial treatment for
chronic hepatitis B in the United States. N Engl J Med.
341:1256–1263. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chien RN, Liaw YF and Atkins M: Lamivudine
as initial treatment for chronic hepatitis B in the United States.
Hepatology. 30:770–774. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim IH, Lee S, Kim SH, Kim SW, Lee SO, Lee
ST, Kim DG, Choi CS and Kim HC: Treatment outcomes of clevudine
versus lamivudine at week 48 in naïve patients with HBeAg positive
chronic hepatitis B. J Korean Med Sci. 25:738–745. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bae SH, Baek YH, Lee SW and Han SY:
Treatment efficacy of clevudine, entecavir and lamivudine in
treatment-naive patients with HBeAg-positive chronic hepatitis B.
Korean J Gastroenterol. 56:365–372. 2010.(In Korean). View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ismail S, Hafez HA, Darweesh SK, Kamal KH
and Esmat G: Virologic response and breakthrough in chronic
hepatitis B Egyptian patients receiving lamivudine therapy. Ann
Gastroenterol. 27:380–386. 2014.PubMed/NCBI
|
|
26
|
Lok AS, Hussain M, Cursano C, Margotti M,
Gramenzi A, Grazi GL, Jovine E, Benardi M and Andreone P: Evolution
of hepatitis B virus polymerase gene mutations in hepatitis B e
antigen-negative patients receiving lamivudine therapy. Hepatology.
32:1145–1153. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi S, Ide T and Sata M: Detection
of YMDD motif mutations in some lamivudine-untreated asymptomatic
hepatitis B virus carriers. J Hepatol. 34:584–586. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee SH, Kim HS, Byun IS, Jeong SW, Kim SG,
Jang JY, Kim YS and Kim BS: Pre-existing YMDD mutants in
treatment-naïve patients with chronic hepatitis B are not selected
during lamivudine therapy. J Med Virol. 84:217–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan Y, Ding K, Su J, Trinh X, Peng Z, Gong
Y, Chen L, Cui Q, Lei N, Chen X and Yu R: The naturally occurring
YMDD mutation among patients chronically infected HBV and untreated
with lamivudine: A systematic review and meta-analysis. PLoS One.
7:e327892012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rahimi R, Hosseini SY, Fattahi MR,
Sepehrimanesh M, Safarpour A, Malekhosseini SA, Nejabat M, Khodadad
M and Ardebili M: YMDD motif mutation profile among patients
receiving liver transplant due to hepatitis B virus infection with
long term lamivudine/immunoglobulin therapy. Hepat Mon.
15:e271202015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Orito E, Mizokami M, Sakugawa H, Michitaka
K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H and Iino S: A
case-control study for clinical and molecular biological
differences between hepatitis B viruses of genotypes B and C. Japan
HBV Genotype Research Group. Hepatology. 33:218–223. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Damerow H, Yuen L, Wiegand J, Walker C,
Bock CT, Locarnini S and Tillmann HL: Mutation pattern of
lamivudine resistance in relation to hepatitis B genotypes:
Hepatitis B genotypes differ in their lamivudine resistance
associated mutation pattern. J Med Virol. 82:1850–1858. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xu G, You Q, Pickerill S, Zhong H, Wang H,
Shi J, Luo Y, You P, Kong H, Lu F and Hu L: Application of
PCR-LDR-nucleic acid detection strip in detection of YMDD mutation
in hepatitis B patients treated with lamivudine. J Med Virol.
82:1143–1149. 2010. View Article : Google Scholar : PubMed/NCBI
|